Abstract
Reef fishes disperse primarily as oceanic “pelagic” larvae, and debate continues over the extent of this dispersal, with recent evidence for geographically restricted (closed) populations in some species. In contrast, moray eels have the longest pelagic larval stages among reef fishes, possibly providing opportunities to disperse over great distances. We test this prediction by measuring mitochondrial DNA (mtDNA) and nuclear DNA variation in 2 species of moray eels, Gymnothorax undulatus (N = 165) and G. flavimarginatus (N = 124), sampled at 14–15 locations across the Indo-Pacific. The mtDNA data comprise 632 bp of cytochrome b and 596 bp of cytochrome oxidase I. Nuclear markers include 2 recombination-activating loci (421 bp of RAG-1 and 754 bp of RAG-2). Analyses of molecular variance and Mantel tests indicate little or no genetic differentiation, and no isolation by distance, across 22 000 km of the Indo-Pacific. We estimate that mitochondrial genetic variation coalesces within the past about 2.3 million years (My) for G. flavimarginatus and within the past about 5.9 My for G. undulatus. Permutation tests of geographic distance on the mitochondrial haplotype networks indicate recent range expansions for some younger haplotypes (estimated within ∼600 000 years) and episodic fragmentation of populations at times of low sea level. Our results support the predictions that the extended larval durations of moray eels enable ocean-wide genetic continuity of populations. This is the first phylogeographic survey of the moray eels, and morays are the first reef fishes known to be genetically homogeneous across the entire Indo-Pacific.
Keywords: connectivity, gene flow, Gymnothorax, leptocephalus, panmixia, reef fish
Coral reefs occur sporadically in a matrix of open ocean that spans 70% of the planet. Reef inhabitants that disperse only as adults can show population genetic structuring on scales of tens of kilometers (Bernardi 2000). However, a pelagic (oceanic) larval stage permits most reef fishes to disperse across open ocean. Larvae either swim actively or drift passively in surface currents, sometimes modifying their buoyancy to use both surface and deep currents (Leis 1991; Montgomery et al. 2001; Atema et al. 2002; Kingsford et al. 2002). This ability to disperse across unsuitable habitat prior to settling on a suitable reef enables dispersal among regions separated by thousands of kilometers.
The geographic distance over which larvae disperse varies across taxa and geographic regions and is probably influenced by the amount of time spent in the pelagic stage (the pelagic larval duration [PLD]; Ekman 1953; Scheltema 1968; Crisp 1978). The simplest expectation is that long PLDs yield proportionately greater gene flow among populations (Jones et al. 2005; Taylor and Hellberg 2005). For many reef fishes, this prediction is upheld by population genetic patterns (Craig et al. 2007). However, PLD is not necessarily predictive (Weersing and Toonen 2009), and many reef species maintain greater population genetic structure than expected from their PLD (Planes and Fauvelot 2002; Swearer et al. 2002; Bernardi et al. 2003; Taylor and Hellberg 2003; Thacker et al. 2007). These findings have prompted research into the types of geographic, oceanographic, and biological factors that mediate larval dispersal, including variation in effective population size, habitat preference, and demographic changes in response to major climatic and environmental shifts (Hellberg 2007; Leis 2007; Leis et al. 2007; Rocha et al. 2007; Floeter et al. 2008).
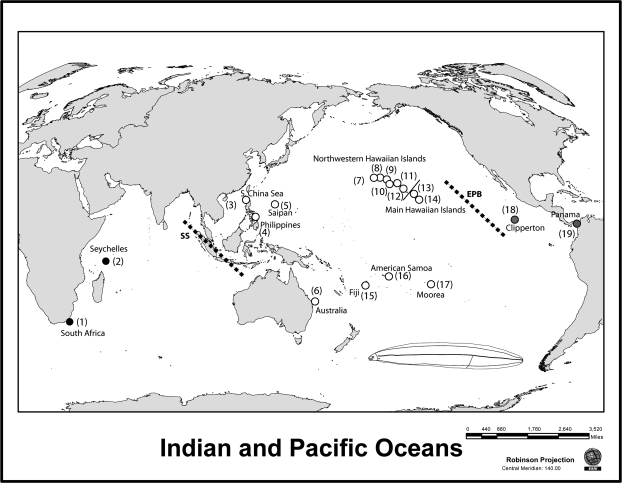
Genetic fragmentation among populations of reef fish species often coincides with major oceanic and geographic barriers (Briggs 1961; Springer 1982; Rocha et al. 2007). The 2 major phylogeographic breaks of the Indo-Pacific occur across the Eastern Pacific Barrier (EPB; 5000–7000 km of open ocean that separate eastern and central Pacific reefs) and the Sunda Shelf (SS; an area of shallow reefs, located just west of Indonesia, that are exposed during low sea levels and separate the Indian and Pacific oceans; Figure 1).
Figure 1.
Map of sampling localities and sketch of generalized moray leptocephalus larvae (bottom right). Each sampling locality is indicated by a number in parentheses that corresponds to localities in Figure 2 and Tables 1–3. Localities are categorized as Indian Ocean (black shading), western/central Pacific Ocean (open circles), or eastern Pacific Ocean (gray shading) to denote the 3 major subdivisions of the Indo-Pacific. See Table 1 for sample size at each locality. Dotted lines represent the biogeographic barriers of the Sunda Shelf and the Eastern Pacific Barrier.
Some reef fishes show high genetic connectivity across thousands of kilometers (Muss et al. 2001; Rocha et al. 2002; Bay et al. 2004), indicating gene flow across the SS (Horne et al. 2008) or the EPB (Lessios and Robertson 2006). However, no reef fish with a nonpelagic adult stage has been shown to maintain connectivity across both the SS and the EPB. Fish species showing geographically widespread populations typically have PLDs exceeding 45 days, whereas most reef fishes have PLDs between 15 and 30 days (Lester and Ruttenberg 2005). Surgeonfishes of the genus Naso are among the most extreme examples of reef fishes maintaining geographically widespread gene flow. Naso vlamingii (Klanten et al. 2007) and 2 congeners (Horne et al. 2008) demonstrate similarly high levels of mitochondrial haplotype diversity and lack of phylogeographic structure across their Indo-Pacific ranges west of the EPB. These 3 species share extended PLDs and similar life histories.
Moray eels (Muraenidae) are solitary predators in reef ecosystems. They comprise about 200 species globally and approximately 150 species within the Indo-Pacific. Many moray species span the entire Indo-Pacific, occupying coral reefs and rocky ledges from the intertidal zone to 200+ m deep. Moray eels are poor swimmers as juveniles and adults and maintain high site fidelity to a few square meters of reef (Böhlke et al. 1989). In a pattern typical of reef fishes, dispersal occurs by pelagic larvae. However, the morays and elopomorph fishes (which includes all true eels, tarpons, tenpounders, and ladyfish) are unique in having a slender elongate larval form called a “leptocephalus” (Figure 1). These larvae are among the simplest, long-lived, and self-sustaining vertebrate forms. They are transparent except for eye pigmentation, and the body wall may be only a few cells thick. They have no identifiable digestive activity or nutrient stores; yet, leptocephalus larvae can persist up to 2 years in pelagic environments sustained by consuming dissolved organic carbon and the fecal pellets and waste products of zooplankton and other larvae (Mochioka and Iwamizu 1996; Bishop and Torres 1999; Ishikawa et al. 2001).
Moray eels are difficult to sample because of their solitary, aggressive, reclusive habits, and this study represents the first phylogeographic survey of the group. Previous authors have suggested that they have extensive oceanic dispersal, based on the extended PLD of the leptocephalus larvae and “the broad distributions of many species within the vast Indo-Pacific region” (Randall 2007). Here, we test this hypothesis for the first time with phylogeographic surveys and inferred coalescence times for haplotype variation within Gymnothorax flavimarginatus and G. undulatus. Both species are ecological generalists on reefs from 0 to more than 100 m deep, with distributions that traverse the SS and the EPB. We sample G. undulatus and G. flavimarginatus from reefs across the Indo-Pacific, from South Africa to Panama, using portions of the mitochondrial genes encoding cytochrome b (CYB) and cytochrome oxidase subunit I (COI), plus portions of 2 nuclear recombination-activating loci, RAG-1 and RAG-2. Our results reveal the first cases of high gene flow through larval dispersal across both the SS and the EPB.
METHODS
Specimens were captured in fish traps and lobster traps, and by pole spears while snorkeling or scuba diving. Here, we report the collection of 289 specimens with only 2 injuries, a minor bite wound to the hand of author B.W.B. from G. undulatus and a bite to the foot of documentary filmmaker Ziggy Livnat. This is a notable accomplishment as moray eels are widely (and rightfully) feared for sudden and egregious acts of aggression (Riordan et al. 2004). In particular, G. undulatus is “especially vicious” (Hiatt and Strasburg 1960), and several bite victims are known to the authors (see also Randall 2007).
Fin clips or muscle samples were taken from live specimens and other tissue samples from tissue storage banks. Sampling includes 124 G. flavimarginatus specimens from 15 sites and 165 specimens of G. undulatus from 14 sites spanning the Indo-Pacific (Table 1, Figure 1). All samples were collected between 2001 and 2009. DNA was isolated using Viogene genomic DNA extraction column kits. Polymerase chain reactions (PCRs) were performed in 25-μl reactions of 5 μl of Promega (www.promega.com) 5× buffer, 2.5 μl of 25 mM MgCl2, 2.5 μl of 0.2 μM dNTPs, 2.5 μl of 0.2 μM of each primer, 0.125 μl (1 unit) of Promega GoTaq DNA polymerase, and 2 μl of template DNA at approximately 5 ng/μl. A 632-bp fragment of the gene encoding CYB was amplified using the primers L14725 (5′-GTG ACT TGA AAA ACC ACC GTT G-3′; Song et al. 1998) and H15573 (5′-AAT AGG AAG TAT CAT TCG GGT TTG ATG-3′; Taberlet et al. 1992) with an annealing temperature of 50°C. A 596-bp fragment of the gene encoding COI was amplified using primers FishF2 (5′-TCG ACT AAT CAT AAA GAT ATC GGC AC-3′) and FishR2 (5′-ACT TCA GGG TGA CCG AAG AAT CAG AA-3′; Ward et al. 2005) with an annealing temperature of 50°C. A 421-bp fragment of the nuclear recombination-activating gene RAG-1 was amplified using primers designed by author J.S.R. based on published marine fish RAG-1 sequences: RAG1-F3 (5′-GCC TCA GAA AAC ATG GTG CT-3′) and RAG1-R3 (5′-CCA CAC AGG TTT CAT CTG GA-3′) with an annealing temperature of 50°C. A 754-bp fragment of the nuclear recombination-activating gene RAG-2 was sequenced using primers RAG2-F3 (5′-AGG TGA CCC TTC GTT GTC AG-3′) and RAG2-R3 (5′-ATG AGG CTC CCT TCC AAA GT-3′), also designed by author J.S.R., with an annealing temperature of 52°C. The thermal profiles for PCR were as follows: 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, annealing temperature for 40 s, and 72°C for 45 s, with a final elongation at 72°C for 7 min.
Table 1.
Sample sizes at each location (with zeros indicated by a “—”), subdivided into 3 regions corresponding to the biogeographic barriers of the SS and the EPB
| Locality number | Gymnothorax flavimarginatus | Gymnothorax undulatus | Latitude | Longitude | |
| Indian Ocean | |||||
| 1 | South Africa | 7 | 6 | 33°44'S | 26°37'E |
| 2 | Seychelles | 5 | 7 | 8°21'S | 50°31'E |
| Western/central Pacific | |||||
| 3 | South China Sea | 1 | 3 | 18°40'N | 118°45'W |
| 4 | Philippines | 6 | — | 11°35'N | 123°49'W |
| 5 | Saipan | 5 | — | 15°7'N | 145°43'W |
| 6 | Australia | 7 | 7 | 24°41'S | 152°42'E |
| 7 | Midway (NWHI) | — | 3 | 28°12'N | 177°19'W |
| 8 | Pearl and Hermes (NWHI) | 3 | 3 | 27°50'N | 175°50'W |
| 9 | Laysan (NWHI) | 5 | — | 25°47'N | 171°45'W |
| 10 | Maro (NWHI) | 10 | 29 | 25°37'N | 170°33'W |
| 11 | Gardner Pinnacles (NWHI) | — | 1 | 25°0'N | 167°55'W |
| 12 | Necker (NWHI) | 3 | 34 | 23°36'N | 164°39'W |
| 13 | Oahu (MHI) | 7 | 19 | 21°30'N | 157°45'W |
| 14 | Hawaii (MHI) | 53 | 27 | 19°5'N | 155°29'W |
| 15 | Fiji | 1 | 5 | 17°39'S | 177°7'E |
| 16 | American Samoa | 5 | — | 14°12'S | 170°55'W |
| 17 | Moorea | — | 2 | 17°36'S | 149°38'W |
| Eastern Pacific | |||||
| 18 | Clipperton | 6 | — | 10°22'N | 109°31'W |
| 19 | Panama | — | 19 | 8°47'N | 79°0'W |
| Total | 124 | 165 | |||
NWHI = Northwestern Hawaiian Islands; MHI= Main Hawaiian Islands.
PCR products were visualized through agarose-gel electrophoresis and purified using Exo-Sap or Viogene gel purification kits (www.viogene.com). Sequences were generated on ABI 3130 and ABI 3330 automated DNA sequencers at the Washington University Genome Sequencing Center and the Smithsonian Museum Support Center. All loci were sequenced in both the forward and the reverse directions, and all data were generated and edited by the same researcher (J.S.R.). DNA sequences were manually edited using SEQUENCHER 4.8 (Ann Arbor, MI) and aligned by hand. For nuclear markers, heterozygous positions were identified as cases wherein a secondary peak in the electropherograms reached at least 25% of the intensity of the primary peak. Gametic phases of nuclear sequences with more than a single heterozygous site were estimated using a Bayesian approach implemented in the software program PHASE 2.1 (Stephens et al. 2001; Stephens and Donnelly 2003). All PHASE analyses were run through 5 iterations, each initiated using a different random-number seed and run for 1000 iterations with a single thinning interval and 100 burn-in iterations. Consistency of results was determined by examining allele frequencies and coalescent goodness-of-fit measures estimated for each of the 4 runs. In the case that the phase of a polymorphism could not be confidently estimated with 90% posterior probability, that site was coded as missing data. Less than 3% of all nucleotide characters were coded as missing data under this criterion. None of the markers for either species appear to be under selection according to Tajima's D (Tajima 1989) tests for selection (all P > 0.1) in ARLEQUIN 3.1 (Excoffier and Schneider 2005). Last, tests of linkage disequilibrium in ARLEQUIN between the nuclear loci RAG-1 and RAG-2 were nonsignificant (P > 0.1) for both species.
We constructed haplotype networks for the mitochondrial fragments from CYB and COI concatenated as a single genetic marker and separately for each of the nuclear markers RAG-1 and RAG-2. In all cases, haplotype networks were constructed with TCS 2.1 (Clement et al. 2000) under 95% statistical parsimony criteria. This approach permits a qualitative assessment of the geographic distributions of alleles. The standard molecular diversity measures of haplotype diversity (h) and nucleotide diversity (π) were calculated in ARLEQUIN accordingly to Nei (1987).
We tested for population structure and phylogeographic patterns with 3 independent analyses: 1) an analysis of molecular variance (AMOVA; Excoffier et al. 1992), 2) pairwise Φst and a Mantel test of geographic versus genetic distances (Φst), and 3) permutation tests of geographic distances on haplotype networks. Sample sizes less than N = 5 were excluded from any pairwise comparisons. After preliminary trials, the 2 mitochondrial DNA (mtDNA) fragments were analyzed as a single composite sequence. We subjected each marker (mtDNA, RAG-1, RAG-2) to AMOVA using ARLEQUIN based on haplotype frequencies and molecular distances among haplotypes. Individuals were grouped by localities and then by 3 larger subgroups comprising localities from the Indian Ocean, the western and central Pacific Ocean, and the eastern Pacific Ocean. These partitions correspond to the 3 oceanic regions demarcated by the SS and the EPB (Figure 1). Statistical significance of the corresponding ARLEQUIN statistic, Φsc, was determined by 10 000 permutations.
We conducted Mantel tests in the program PASSAGE 2.0 (Rosenberg 2001) to measure correlations between geographic and genetic distances among populations. Genetic distances for all loci were computed in ARLEQUIN as pairwise Φst values based on the number of nucleotide differences between alleles and allele frequency differences between populations. We report only uncorrected differences because corrected pairwise differences under a variety of mutation models yielded congruent results. Significance of pairwise Φst values was computed using the B-H (Benjamini and Hochberg 1995; Misawa et al. 2008) and the more conservative Bonferroni (1936) corrections for multiple comparisons. The pairwise matrix of geographic distances between populations was obtained with ARCGIS 9.3 as the shortest straight-line distance that did not cross dry land. We could not incorporate oceanic currents in this analysis because most available data are for surface currents, whereas moray eel larvae use both surface currents and counterflowing deeper currents (Böhlke et al. 1989). Geographic and genetic distance (Φst) matrices were tested for correlation separately for each molecular maker (mtDNA, RAG-1, and RAG-2).
Associations of geographic and genetic distances were resolved using a permuted chi-square test of genetic distances among hierarchically nested groups of haplotypes and geographic areas covered by distributions of grouped haplotypes (Templeton 1998). This approach is the initial step in nested clade phylogeographic analysis (NCPA; Templeton 1998) and mirrors the Mantel test, but instead of using single populations as the sole level of sampling, it tests for associations between geographic distance and hierarchically nested groups from the haplotype network. Geographic distances were identical to those of the Mantel tests. Tests were performed separately on each locus (mtDNA, RAG-1, and RAG-2) in the program GEODIS 2.6 (which includes the Dunn–Sidak correction for multiple tests; Posada et al. 2000) using 10 000 permutations. Inferences were made using the 2008 inference key available at http://darwin.uvigo.es/software/geodis.html. Loops denoting ambiguous groupings in haplotype networks were treated as follows: all possible nested combinations of resolutions to the loops were computed, and analyses were repeated for each possible network configuration. Alternative resolutions of loops did not alter inferences in this study.
There is little information available on the time since divergence from a most recent common ancestor of moray eels and their closest relatives, the genera Conger and Anguilla (Inoue et al. 2003). Without a strong fossil record or obvious cases of vicariance, one cannot derive a species-specific mutation rate for estimating coalescence times. For the mitochondrial gene CYB, we used a divergence rate of 2% per million year (My), which has been widely applied to phylogeographic studies of reef fishes (Bowen et al. 2001, 2006b) and is supported by measurements for goby (Gnatholepis) species separated by the Isthmus of Panama (1.95–2.17% divergence per My; Rocha et al. 2005). We used a mutation rate for the COI data of 1.2% sequence divergence per My as calibrated by Bermingham et al. (1997) for marine fishes. We estimated the time to coalescence for standing genetic variation in each of the 2 study species and for haplotype groups with significant inferences in NCPA, using the Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in BEAST 1.5.2 (Drummond and Rambaut 2007). We used a congeneric outgroup and determined the target species (G. undulatus or G. flavimarginatus) to be monophyletic for haplotypic variation. Models of evolution were computed in MrModelTest (Posada and Buckley 2004), and the combined mtDNA data were partitioned by gene region by codon position, for a total of 6 partitions. A fixed mutation rate was estimated at 1.6% divergence per My (the weighted average of 2% and 1.2% for CYB and COI, respectively) and input as 0.008 substitutions per site per lineage per My. A coalescent tree prior of expansion growth was used, as recommended for estimating intraspecific coalescence times when population growth is inferred (see results of mismatch distributions; Drummond and Rambaut 2007). Simulations ran for 30 million generations, with sampling every 3000 generations. Two independent runs were computed for each species, and runs were then combined with a 10% burn-in using LogCombiner 1.5.2 (Drummond and Rambaut 2007). Adequate parameter estimation and convergence of chains in the MCMC run were computed in the program Tracer 1.4 (Rambaut and Drummond 2007). The effective Sample Size values for each parameter were more than 200, and 95% highest posterior density (HPD) intervals are reported as encompassing the uncertainty around coalescence time estimates within each species. This estimate of confidence intervals is directly proportional to and does not account for variation in the mutation rates used.
Frequency distributions of numbers of mutational differences between haplotypes sampled within a species (=mismatch distribution) were computed in ARLEQUIN using the least-square method (Schneider et al. 2000). To compute the confidence intervals around these values and the ability to reject a model of demographic expansion through time, we performed 100 000 parametric bootstraps and calculated Harpending's raggedness index (HRI; Harpending 1994). We also computed Fu's (1997) FS statistics to identify cases of demographic expansion, which are indicated by significant negative values.
RESULTS
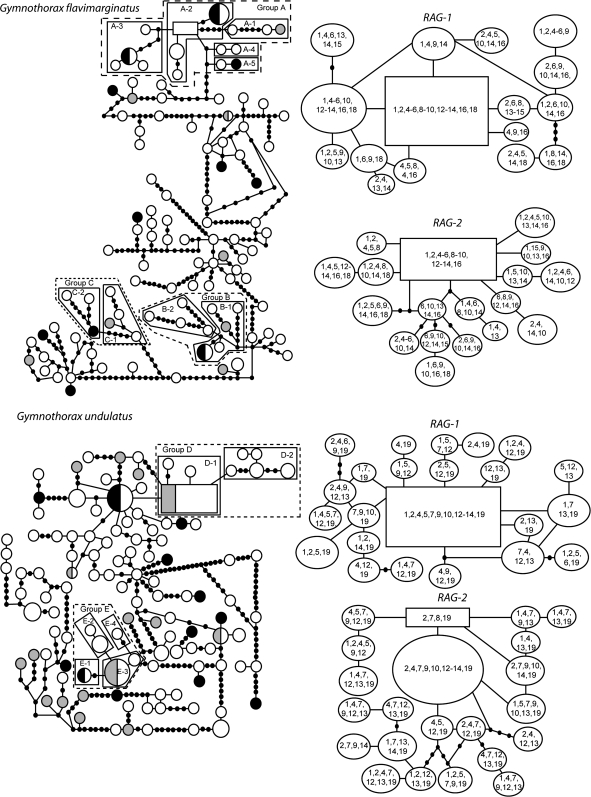
Nuclear and mitochondrial haplotype networks for G. flavimarginatus and G. undulatus are presented in Figure 2. Notably, haplotypes from the major geographic subdivisions of the Indo-Pacific are dispersed throughout the networks. Combining CYB and COI, we identify 104 mtDNA haplotypes for G. flavimarginatus (N = 124; GenBank accession number GU175445-GU175541) and 112 haplotypes for G. undulatus (N = 165; GU175576-GU175657). No mitochondrial haplotype occurs in samples of all 3 major oceanic regions (Indian Ocean, western/central Pacific Ocean, and eastern Pacific Ocean), but 4 haplotypes in G. flavimarginatus and 6 in G. undulatus occur in adjacent regions; all other haplotypes are observed in a single region. The nuclear markers RAG-1 and RAG-2 reveal less variation than the combined mitochondrial markers, as expected from their slower mutation rate and larger effective population size. RAG-1 sequences resolved 16 haplotypes for G. flavimarginatus (GU175542-GU175557) and 23 haplotypes for G. undulatus (GU175658-GU175680). RAG-2 sequences included 18 haplotypes for G. flavimarginatus (GU175558-GU175575) and 22 haplotypes for G. undulatus (GU175681-GU175702). The most common nuclear haplotypes occur in most localities, with haplotype sharing among the 3 major oceanic regions (Figure 2). Groupings of haplotypes endemic to 1 of the 3 major oceanic regions occur only in the mitochondrial haplotype networks. Molecular diversity indices for G. flavimarginatus at mtDNA were h = 0.997 and π = 0.012; at RAG-1, h = 0.706 and π = 0.0050; and at RAG-2, h = 0.912 and π = 0.0043. Molecular diversity indices for G. undulatus at mtDNA were h = 0.972 and π = 0.012; at RAG-1, h = 0.808 and π = 0.0053; and at RAG-2, h = 0.907 and π = 0.0067.
Figure 2.
Haplotype networks for mtDNA (left) and for RAG-1 and RAG-2 for both Gymnothorax flavimarginatus and G. undulatus. Squares represent inferred ancestral haplotypes; circles, additional haplotypes; and small black-filled circles, missing haplotypes needed to connect observed haplotypes. The size of the circle or square is proportional to the number of individuals sampled that share that haplotype. For mitochondrial haplotypes, shading denotes oceanic region as in Figure 1: Indian Ocean (black fill), western/central Pacific Ocean (open circles), and eastern Pacific Ocean (gray fill). Localities are listed for each nuclear haplotype as numbered in Figure 1 and Tables 1–3, increasing from west to east. For mitochondrial haplotype networks, groups A–E show regions for which hypotheses of panmixia could be rejected by NCPA in favor of either restricted gene flow or contiguous range expansion (Table 4).
Results of a locus-by-locus AMOVA revealed no population structure among the 3 geographic groupings demarcated by the EPB and the SS. For G. flavimarginatus, the mtDNA yields a nonsignificant Φsc = 0.0088 (P = 0.29) and the nuclear markers a nonsignificant Φsc < 0.00001 (P = 0.89) for RAG-1 and Φsc < 0.00001 (P = 0.73) for RAG-2. The mtDNA results for G. undulatus reveal a low but significant Φsc = 0.039 (P = 0.007). The nuclear markers yield nonsignificant Φsc < 0.00001 (P = 0.30) for RAG-1 and Φsc < 0.00001 (P = 0.99) for RAG-2. Results for both species identify a vast majority of the variation (95–100%) occurring within populations.
The Mantel tests for correlations between geographic distances and Φst values are not significant for mtDNA, RAG-1, and RAG-2 (all R < 0.0001, P values >0.2). Tables 2 and 3 present mtDNA pairwise Φst values for G. flavimarginatus and G. undulatus, respectively. Structure in G. flavimarginatus was especially low, with 0 of 55 significant comparisons for mtDNA (Table 2) and no significant comparisons for RAG-1 or RAG-2. Similarly, G. undulatus showed low levels of structure, with 4 of 36 significant comparisons for mtDNA (Table 3) and none for RAG-1 or RAG-2.
Table 2.
Pairwise Φst values calculated from mtDNA haplotypes (below diagonal) and the mean value for nuclear loci RAG-1 and RAG-2 (above diagonal) for Gymnothorax flavimarginatus
| 1 | 2 | 4 | 5 | 6 | 9 | 10 | 13 | 14 | 16 | 18 | |
| 1 | — | −0.05 | −0.07 | −0.09 | −0.01 | −0.02 | −0.06 | −0.01 | −0.03 | −0.07 | −0.04 |
| 2 | −0.07 | — | −0.06 | −0.08 | −0.01 | −0.02 | −0.03 | −0.02 | 0.01 | −0.06 | 0.01 |
| 4 | −0.05 | −0.12 | — | −0.09 | 0.01 | −0.02 | −0.04 | −0.03 | −0.03 | −0.07 | −0.03 |
| 5 | −0.02 | −0.03 | −0.02 | — | 0.01 | −0.01 | −0.05 | −0.04 | −0.02 | −0.07 | −0.04 |
| 6 | −0.08 | −0.10 | −0.07 | −0.04 | — | −0.06 | −0.03 | 0.09 | 0.04 | 0.01 | 0.04 |
| 9 | −0.05 | −0.10 | −0.09 | −0.03 | −0.03 | — | −0.05 | 0.06 | 0.02 | −0.09 | 0.01 |
| 10 | −0.01 | −0.06 | 0.00 | 0.01 | 0.02 | −0.07 | — | 0.0 | −0.03 | −0.06 | 0.02 |
| 13 | −0.06 | −0.05 | −0.05 | −0.07 | −0.06 | −0.04 | 0.03 | — | −0.01 | 0.0 | 0.04 |
| 14 | −0.03 | −0.05 | 0.00 | 0.01 | −0.01 | −0.02 | 0.02 | −0.01 | — | −0.03 | −0.04 |
| 16 | −0.04 | −0.07 | −0.07 | −0.01 | −0.08 | −0.01 | 0.09 | −0.02 | 0.02 | — | −0.07 |
| 18 | −0.04 | −0.11 | −0.08 | −0.04 | −0.06 | −0.08 | −0.05 | −0.01 | 0.00 | −0.05 | — |
None of the pairwise comparisons were significantly greater than zero for mtDNA, RAG-1, or RAG-2 after B-H or Bonferroni correction for multiple comparisons. Populations are numbered as in Figure 1 and Table 1. Values less than zero have no biological meaning and should be considered indistinguishable from zero.
Table 3.
Pairwise Φst values calculated from mtDNA haplotypes (below diagonal) and the mean value for nuclear loci RAG-1 and RAG-2 (above diagonal) for Gymnothorax undulatus, with values significantly greater than zero in bold type
| 1 | 2 | 6 | 10 | 12 | 13 | 14 | 15 | 19 | |
| 1 | — | −0.06 | −0.05 | −0.04 | −0.03 | 0.04 | 0.02 | −0.04 | −0.04 |
| 2 | 0.01 | — | −0.05 | −0.02 | −0.03 | 0.05 | 0.02 | −0.06 | −0.03 |
| 6 | 0.08 | −0.05 | — | −0.05 | −0.05 | −0.01 | 0.0 | −0.05 | −0.04 |
| 10 | 0.06 | 0.00 | 0.00 | — | −0.03 | −0.01 | −0.01 | −0.02 | −0.03 |
| 12 | −0.03 | 0.07 | 0.08 | 0.04 | — | 0.01 | −0.01 | −0.03 | −0.01 |
| 13 | 0.04 | −0.02 | −0.03 | 0.00 | 0.05 | — | −0.03 | 0.07 | 0.02 |
| 14 | 0.03 | 0.11 | 0.14 | 0.06 | −0.01 | 0.10 | — | 0.04 | 0.01 |
| 15 | −0.05 | −0.03 | −0.02 | −0.02 | −0.02 | −0.02 | −0.01 | — | 0.03 |
| 19 | 0.03 | 0.00 | −0.05 | 0.06 | 0.09 | 0.05 | 0.13 | 0.01 | — |
Populations are numbered as in Figure 1 and Table 1. The mtDNA comparison between Maro Island (no. 10) and Hawaii (no. 14) was significant after the B-H correction but not after the more conservative Bonferroni correction. None of the pairwise comparisons for RAG-1 or RAG-2 were significantly different from zero after B-H or Bonferroni correction for multiple comparisons. Values less than zero have no biological meaning and should be considered indistinguishable from zero.
The NCPA of the combined mtDNA sequences in G. flavimarginatus revealed only 3 nested groups that refute the hypothesis of panmixia. None of the resolutions of loops altered the chain of inference or the significance of pairwise comparisons; Figure 2 shows resolved loops only for clades with significant inferences. Group A (Figure 2, Table 4) supports an inference of contiguous range expansion, estimated to have occurred within the past 0.6 My (95% HPD 0.3–0.9 My). Tip haplotype groups in group A show expansion from the internal and mostly Hawaiian group to tip groups with haplotypes in the Seychelles, South Africa, and the eastern Pacific. Groups B and C yield inferences of restricted gene flow with isolation by distance coalescing within the past 0.5 My (95% HPD 0.2–0.8 My) and 0.4 My (95% HPD 0.3–0.7 My), respectively. Within group B, tip subgroup B-2 is observed only in Hawaii, whereas the more interior subgroup B-1 from which it derives includes haplotypes distributed across the Indo-Pacific. Both subgroups of group C include haplotypes from Hawaii, but they differ in having haplotypes from the Indian Ocean (subgroup C-2) versus the eastern Pacific Ocean (subgroup C-1).
Table 4.
Tests for geographically nonrandom distributions of nested groupings of mitochondrial haplotypes (NCPA; Templeton 1998) using the December 2008 inference key. Groups are listed as marked on Figure 2. All statistically significant departures from random distributions indicate range expansions or geographic fragmentation among relatively young haplotype groups (within the past 0.3–0.6 My). No significant departures from randomness were detected for nuclear markers in either species or for older groupings of mitochondrial haplotypes. Also reported are coalescence times (and 95% HPD intervals) in millions of years (My) as estimated under a coalescent model of demographic expansion in the program BEAST
| Group | Chain of inference | Inference | Coalescence time (My) |
| Gymnothorax flavimarginatus | |||
| A | 1-2-11-12-No | Contiguous range expansion | 0.6 (0.3–0.9) |
| B | 1-2-3-4-No | Restricted gene flow with IBD | 0.5 (0.2–0.8) |
| C | 1-2-3-4-No | Restricted gene flow with IBD | 0.4 (0.3–0.7) |
| G. undulatus | |||
| D | 1-2-11-12-No | Contiguous range expansion | 0.6 (0.4–0.8) |
| E | 1-2-11-12-No | Contiguous range expansion | 0.3 (0.2–0.5) |
IBD, isolation by distance.
The NCPA for G. undulatus includes 2 nesting levels (groups D and E) that refute the null model of panmixia, both involving range expansions of haplotypes (Figure 2, Table 4) that coalesce within 0.6 My (95% HPD 0.4–0.8 My) and 0.3 My (95% HPD 0.2–0.5 My), respectively. Range expansion in group D includes tip haplotypes sampled from Australia (subgroup D-1) or Fiji (subgroup D-2) from an inferred ancestral range in Hawaii, indicating some historical fragmentation followed by range expansion within the western Pacific. Group E shows a tip haplotype observed in Hawaii and the Indian Ocean (subgroup E-1) derived from an ancestral distribution in the Pacific Ocean (subgroup E-3).
The coalescence times for standing genetic variation based on mtDNA were 2.3 My (95% HPD 1.7–3.0 My) for G. flavimarginatus and 5.9 My (95% HPD 3.5–8.6 My) for G. undulatus. Estimates of coalescence times for G. flavimarginatus and G. undulatus are younger than those for other reef fishes with similar population structure (i.e., genus Naso; Klanten et al. 2007; Horne et al. 2008), but the confidence intervals surrounding those estimates broadly overlap.
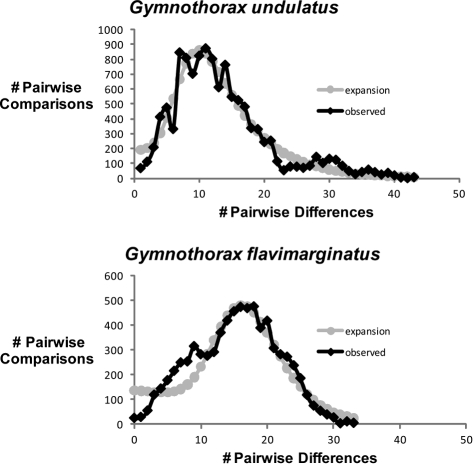
We evaluated evidence for population expansion using mismatch distributions and Fu's FS. Mismatch distributions test the null hypothesis of demographic expansion, whereas Fu's FS tests the null hypothesis of constant population size, with significantly negative values indicating demographic expansion. Therefore, a nonsignificant result for the mismatch distribution test and a significantly negative Fu's FS both indicate population expansion. The mismatch distributions for mtDNA are shown in Figure 3. Overall, mismatch analyses indicate a pattern of population growth in both species for all markers. In G. flavimarginatus, mismatch analyses fail to reject the null hypothesis of demographic expansion for mtDNA (P = 0.78, HRI = 0.10; P = 0.72), RAG-1 (P = 0.10, HRI = 0.04; P = 0.55), and RAG-2 (P = 0.22, HRI = 0.28; P = 0.52). Similar results were obtained in mismatch analyses of G. undulatus for mtDNA (P = 0.42, HRI = 0.017; P = 0.51), RAG-1 (P = 0.5, HRI = 0.03; P = 0.68), and RAG-2 (P = 0.99, HRI = 0.004; P = 0.99). The results of the Fu's FS analyses support these conclusions, with significant negative values in G. flavimarginatus for mtDNA (−24.0; P < 0.001) and RAG-1 (−14.8; P < 0.001) but a nonsignificantly negative value for RAG-2 (−3.2; P = 0.15). Fu's FS analyses in G. undulatus indicated demographic expansion with significant negative values for mtDNA (−24.0; P < 0.001) and RAG-1 (−7.2; P = 0.019) but with a nonsignificant value for RAG-2 (0.57; P = 0.65). In summary, all analyses support conclusions of demographic expansion except Fu's FS for RAG-2 in both species. Peak pairwise differences of approximately 10 substitutions (G. undulatus) versus 17 substitutions (G. flavimarginatus) between paired haplotypes for the 1228 bp of mtDNA are consistent with transitory fragmentation of populations between 300 000 and 600 000 years before present (the estimated coalescent times for clades with significant inferences in NCPA). These events yielded divergent haplotype lineages that have persisted within each species through subsequent climatic cycles of approximately 100 000 years (Paillard 2006).
Figure 3.
Mismatch distributions for mitochondrial haplotypes (1228 bp, black line) superimposed on the expected distribution calculated for the assumption of a demographically expanding population. Observed distributions do not differ significantly from expectations either in magnitude or in raggedness (see Results). Peak values of approximately 17 (Gymnothorax flavimarginatus) and 10 (G. undulatus) differences between haplotypes are consistent with an interpretation that some geographically isolated haplotype lineages formed during transitory geographic fragmentation of populations approximately 0.3–0.6 My ago and have persisted through subsequent merging of populations.
DISCUSSION
Several reef fishes are known to have highly dispersive pelagic stages, including soldierfishes (genus Myripristis; Bowen et al. 2006b; Craig et al. 2007), pygmy angelfishes (genus Centropyge; Bowen et al. 2006a; Schultz et al. 2007), and unicornfishes (genus Naso; Klanten et al. 2007; Horne et al. 2008); however, even these species show genetic partitions (or distribution limits) at major biogeographic barriers. In the first moray species subject to phylogeographic analyses, we reveal through 3 independent tests a prevailing pattern of very little to no genetic structure among populations separated by the major biogeographic barriers of the eastern Pacific and SS. Leptocephalus larvae in general are extremely long lived in the pelagic environment and can delay metamorphosis until appropriate conditions are available (Schmidt 1923; Castonguay 1987; Tseng 1990; Crabtree et al. 1992), and it is likely that these capabilities have enabled the high levels of connectivity that we observe throughout the Indo-Pacific.
Haplotypes from the Indian, western/central Pacific, and eastern Pacific oceans are interspersed on the networks for all 3 loci (mtDNA, RAG-1, and RAG-2), with geographic clustering of haplotypes occurring only near some tips of the mtDNA network (Figure 2). Nuclear haplotypes are shared among localities separated by more than 22 000 km and across 2 major biogeographic barriers. This high genetic continuity among populations with low adult vagility strongly implicates the leptocephalus larva (especially the long pelagic stage) as the dispersal engine.
The AMOVA results indicate a lack of persistent biogeographic barriers to gene flow. Only the mitochondrial locus of G. undulatus reveals a significant Φsc among regions delineated by the SS and EPB, with biogeographic barriers explaining only 3.9% of the overall variation. Mantel tests of genetic and geographic distances likewise fail to reject panmixia among geographic populations. Although the SS and EPB represent species boundaries in many taxa (Briggs 1961), and substantial obstacles to gene flow within species that cross them (Randall 1998), they appear porous to gene flow among populations of moray eels. Notably, the pairwise Φst comparisons show no consistent breaks across the SS, but G. undulatus has low/significant structure across the EPB in 2 comparisons with Hawaiian Islands. We conclude that the oceanic EPB can be a substantial barrier under contemporary conditions, whereas the SS seems to be important in a historical context but not a barrier under contemporary conditions. Leptocephalus dispersal capabilities quickly erase genetic separations developed across the SS during glacial maxima, whereas the signature patterns of partial isolation across the EPB are ongoing and detectable at low levels. Although sampling error should increase the variance on estimates of genetic divergence relative to actual divergence, our finding of widespread genetic homogeneity among populations is robust to the relatively small sample sizes within populations of the Indian and eastern Pacific oceans (Table 1).
Our NCPA results for the mitochondrial DNA indicate a pattern of transitory fragmentation among populations followed by genetic merging within 0.3–0.6 My. The NCPA is sensitive to historical events such as range expansion and previous fragmentation, and the mitochondrial locus is the only one variable enough to reveal such patterns. Three groupings of mitochondrial haplotypes from G. flavimarginatus, each group estimated to be less than 0.6 My old, show evidence of geographic fragmentation. Within group A (Figure 2, Table 4), mitochondrial haplotypes of G. flavimarginatus show evidence of a range expansion across the SS within the past approximately 0.6 My. Haplotypes in groups B and C also show some evidence of fragmentation on the timescale of 0.5–0.3 My, respectively. Within G. undulatus, mtDNA haplotype groups D and E show evidence of range expansion on this same timescale across the SS (group E) and between Australian and Hawaiian regions of the western/central Pacific Ocean. Results of the mismatch distributions and occurrence of 4 low but statistically nonzero pairwise Φst values for the mitochondrial markers of G. undulatus (Table 3) are consistent with this hypothesis of transitory geographic genetic structure within the past 0.6 My. Based on the estimated coalescence times within the past 2.3 My in G. flavimarginatus and 5.9 My in G. undulatus, the prevailing pattern seems to be genetic homogeneity of populations over the longer (My) timeframe, with transitory population fragmentation on the shorter timescale of 0.3–0.6 My.
Recent controversy regarding NCPA (Garrick et al. 2008; Knowles 2008; Petit 2008; Templeton 2008) concerns whether significant genetic associations of haplotype groupings with geographic distances justify inferences of isolation by distance or contiguous range expansion, as these are the 2 most common inferences from data sets generated under panmictic conditions by Panchal and Beaumont (2007). It should be noted that the limitations described by Petit (2008) and Panchal and Beaumont (2007) have been corrected (Templeton 2008, 2009). Our results from NCPA involve 3- and 4-step clades with significant nonrandom associations of genetic and geographic distances. Our interpretations from NCPA are consistent with the development of transient, geographically localized population genetic structure on the order of a few hundred thousand years, and these are supported in part by significant pairwise Φst comparisons and mismatch distributions. At broader scales and the entire cladogram level, NCPA, AMOVA, Mantel tests, and pairwise Φst all yield the same result: inability to reject panmixia. At best, NCPA has identified evidence for population genetic structure at a level that is lost to more reductionist approaches such as AMOVA. At worst, it has yielded a false inference, and all populations of G. undulatus and G. flavimarginatus surveyed here are panmictic. In either case, haplotypes likely to have arisen within the past few hundred thousands of years have not all acquired ocean-wide distributions, but genetic continuity of populations is nonetheless the prevailing pattern when results are averaged over the multi-My history covered by mtDNA markers.
Klanten et al. (2007) apply the term “temporal genetic partitioning” to Indo-Pacific reef fishes to describe a pattern of population genetic homogeneity on a My timescale, with transitory fragmentation and demographic expansion on shorter timescales. “Each expansion episode is likely to have been accompanied by re-colonization of distant reef habitats resulting in repeated and widespread secondary contacts amongst previously isolated, genetically divergent demes” (Klanten et al. 2007). Global climatic fluctuations provide the most obvious source of recurring cycles of fragmentation followed by expansion of populations for reef fishes having widespread larval dispersal. Randall (1998) hypothesizes 3–6 episodes within the past 700 000 years during which sea level would have been low enough to preclude dispersal of marine species between the Indian and Pacific oceans. A review of geological climatic data by Paillard (2006) indicates sea-level fluctuation occurring on a cycle of approximately 100 000 years throughout the past My. These results are highly congruent with our conclusion that the 2 Gymnothorax species are emerging from a transitory fragmentation event, with some remnants of fragmentation and recent range expansions over the past 0.3–0.6 My shown by some of the younger mitochondrial haplotypes.
This interpretation is consistent with our evidence for population growth in the form of mismatch distributions and significant negative Fu's FS values for the mitochondrial loci of each species and for RAG-1 of G. undulatus. As proposed for other reef fishes and invertebrates, this population expansion likely followed the last period of low sea levels that exposed portions of the SS (Barber et al. 2006; Klanten et al. 2007; Gaither et al. 2010).
The Hawaiian Archipelago has a special biogeographic significance for moray eels, and this is apparent in several facets of our data. First, morays are common and abundant on these islands, one of the most isolated tropical archipelagos in the world, whereas common reef predators such as groupers (Serranidae) and snappers (Lutjanidae) are nearly absent. Thus, the high dispersal evident in our survey has allowed the morays to colonize and to proliferate in Hawaii, to the exclusion of many west/south Pacific competitors (Randall 2007). Second, Hawaii may be the biogeographic crossroads between the tropical eastern Pacific and western Pacific. This is indicated by the distribution of shared haplotypes from Hawaii to the western Pacific and Indian oceans (subgroup C-2 and group B in G. flavimarginatus, subgroup E-1 in G. undulatus), and Hawaii to the eastern Pacific (subgroups A-1 and C-1 in G. flavimarginatus; Figure 2, Table 4). Based on the latter network configuration, NCPA reveals dispersal from Hawaii to the eastern Pacific in the past 0.3 My (directionality inferred from haplotype network rooting and historical biogeography; see Briggs 1999). Many tropical species show very shallow phylogeographic histories in the eastern Pacific, indicating that much of this fauna is extirpated during glacial conditions (Bowen and Karl 2007). Hence, Hawaii may be the essential source of moray diversity in the East Pacific. Third, the only significant mtDNA partitions on a local scale for either species are within the Hawaiian Islands (Table 3, populations number 10 and 14), and one of them is between the high islands of the southeastern Hawaiian Archipelago and the low islands to the northwest. The latter region is now protected as the Papahānaumokuākea Marine National Monument. Emerging data indicate that the jurisdictional and ecological differences between these regions are matched by population genetic differences in some other reef species (Rivera et al. 2004; Ramon et al. 2008; Eble et al. 2009).
Gymnothorax flavimarginatus and G. undulatus represent extreme cases among reef fishes in maintaining gene flow across two-thirds of the planet. Several other fishes found on or near coral reefs maintain high levels of connectivity across similar spatial scales (e.g., whale sharks, Castro et al. 2007; tunas, Theisen et al. 2008), but these all have highly vagile adult forms that are not restricted to coral reef habitat. Our favored hypothesis for the geographic–genetic homogeneity of these Gymnothorax species is that their extended pelagic larval phase permits widespread dispersal of individuals after the removal of transitory oceanic barriers to pelagic dispersal. The genetic connectivity of both Gymnothorax species across the EPB and SS indicates an ability to disperse across even the strongest obstacles to other reef fishes.
Funding
This work was supported by a United States National Science Foundation grant (OCE-0454873) and the Hawaii Institute of Marine Biology Northwestern Hawaiian Islands Coral Reef Research Partnership grant (NMSP MOA 2005-008/6682) to B.W.B.; National Geographic Young Explorers Award, the National Science Foundation Doctoral Dissertation Improvement Grant (DEB 0909756), the Society of Systematic Biologists Mini-Partnerships for Enhancing Expertise in Taxonomy Award, the DeepFin PEET Award, and the Professional Association of Diving Instructors Foundation grant to J.S.R; summer undergraduate research fellowships from the Howard Hughes Medical Institute to V.G. and K.J.
Acknowledgments
For logistic assistance, expertise, and sound advice we thank Randall Kosaki, Jeff Eble, Jane Ball, Jo-Ann Leong, Matt Iacchei, Yannis Papastamatiou, Carl Meyer, Luiz Rocha, Matthew Craig, David Smith, Michelle Gaither, Robert Moffitt, Joe O'Malley, Sheila Conant, Senifa Annandale, Toby Daly-Engel, Zoltan Szabo, Kelly Gleason, Robert Toonen, James Lakey, Malia Rivera, Elizabeth Keenan, Derek Skillings, Tim Clark, the officers and crew of the R/V Hi'ialakai, the Stock Assessment Program at the NOAA Pacific Islands Fisheries Science Center, and Matt Furtado of Koolau Pets. Thanks to Kevin Conway for the leptocephalus illustration in Figure 1. We thank the Papahānaumokuākea Marine National Monument, US Fish and Wildlife Services, and Hawaii Division of Aquatic Resources for coordinating research activities and permitting procedures for collections in the Northwestern Hawaiian Islands. This is HIMB contribution no. 1367 and the School of Earth Science and Technology contribution no. 7825. Collections made in the Northwestern Hawaiian Islands National Monument were made under permits PMNM 2007-32 to B.W.B. and PMNM 2008-31 to J.S.R. This study complied with current laws in the United States and was conducted in accordance with the regulations of the Washington University and the University of Hawaii Institutional Animal Care and Use Committee.
References
- Atema J, Kingsford MJ, Gerlack G. Larval reef fish could use odour for detection, retention and orientation to reefs. Mar Ecol Prog Ser. 2002;241:151–160. [Google Scholar]
- Barber PH, Erdmann MV, Palumbi SR. Comparative phylogeography of three codistributed stomatopods: origins and timing of regional lineage diversification in the coral triangle. Evolution. 2006;60:1825–1839. [PubMed] [Google Scholar]
- Bay LK, Choat HJ, Van Herwerden L, Robertson DR. High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar Biol. 2004;144:757–767. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Proc B. 1995;57:289–300. [Google Scholar]
- Bermingham E, McCafferty SS, Martin AP. Fish biogeography and molecular clocks: perspectives from the Panamanian isthmus. In: Kocher TD, Stepien CA, editors. Molecular systematics of fishes. New York: Academic Press; 1997. pp. 113–128. [Google Scholar]
- Bernardi G. Barriers to gene flow in Embiotoca jacksoni, a marine fish lacking a pelagic larval stage. Evolution. 2000;54:226–237. doi: 10.1554/0014-3820(2000)054[0226:BTGFIE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Holbrook SJ, Schmitt RJ, Crane NL. Genetic evidence for two distinct clades in the French Polynesian population of coral reef three-spot damselfish Dascyllus trimaculatus. Mar Biol. 2003;143:485–490. [Google Scholar]
- Bishop RE, Torres JJ. Leptocephalus energetics: metabolism and excretion. J Exp Biol. 1999;202:2485–2493. doi: 10.1242/jeb.202.18.2485. [DOI] [PubMed] [Google Scholar]
- Böhlke EB, McCosker JE, Böhlke JE. Family Muraenidae. In: Böhlke EB, editor. Fishes of the Western North Atlantic. New Haven (CT): Sears Foundation for Marine Research; 1989. pp. 104–206. [Google Scholar]
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probability a`. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- Bowen BW, Bass AL, Muss A, Carlin J, Robertson DR. Phylogeography of two Atlantic squirrelfishes (Family Holocentridae): exploring links between pelagic larval duration and population connectivity. Mar Biol. 2006a;149:899–913. [Google Scholar]
- Bowen BW, Bass AL, Rocha LA, Grant WS, Robertson DR. Phylogeography of the trumpetfishes (Aulostomus): ring species complex on a global scale. Evolution. 2001;55:1029–1039. doi: 10.1554/0014-3820(2001)055[1029:pottar]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Karl SA. Population genetics and phylogeography of sea turtles. Mol Ecol. 2007;16:4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Muss A, Rocha LA, Grant WS. Shallow mtDNA coalescence in Atlantic Pygmy Angelfishes (Genus Centropyge) indicates a recent invasion from the Indian Ocean. J Hered. 2006b;97:1–12. doi: 10.1093/jhered/esj006. [DOI] [PubMed] [Google Scholar]
- Briggs JC. The East Pacific Barrier and the distribution of marine shore fishes. Evolution. 1961;15:545–554. [Google Scholar]
- Briggs JC. Coincident biogeographic patterns: Indo-West Pacific Ocean. Evolution. 1999;53:326–335. doi: 10.1111/j.1558-5646.1999.tb03769.x. [DOI] [PubMed] [Google Scholar]
- Castonguay M. Growth of American and European eel leptocephali as revealed by otolith microstructure. Can J Zool. 1987;65:875–878. [Google Scholar]
- Castro ALF, Steward BS, Wilson SG, Hueter RE, Meekan MG, Motta PJ, Bowen BW, Karl SA. Population genetic structure of Earth's largest fish, the whale shark (Rhincodon typus) Mol Ecol. 2007;16:5183–5192. doi: 10.1111/j.1365-294X.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Crabtree RE, Cyr E, Bishop RE, Falkenstein L, Dean JM. Age and growth of tarpon, Megalops atlanticus, larvae in the eastern Gulf of Mexico, with notes on relative abundance and probable spawning areas. Environ Biol Fish. 1992;35:361–370. [Google Scholar]
- Craig MT, Eble JA, Bowen BW, Robertson DR. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae) Mar Ecol Prog Ser. 2007;334:245–254. [Google Scholar]
- Crisp DJ. Genetic consequences of different reproductive strategies in marine invertebrates. In: Battaglia B, Beadmore JA, editors. Marine organisms: genetics, ecology and evolution. New York: Plenum; 1978. pp. 257–273. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214–221. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble JA, Toonen RJ, Bowen BW. Endemism and dispersal: comparative phylogeography of three surgeonfish species across the Hawaiian Archipelago. Mar Biol. 2009;156:689–698. [Google Scholar]
- Ekman S. Zoogeography of the sea. London: Sidgwick & Jackson Ltd; 1953. [Google Scholar]
- Excoffier L, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–625. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floeter S, Rocha LA, Robertson DR, Joyeux JC, Smith-Vaniz W, Wirtz P, Edwards AJ, Barreiros G, Ferriera JP, Gasparini JL, et al. Atlantic reef fish biogeography and evolution. J Biogeogr. 2008;35:22–47. [Google Scholar]
- Fu Y- X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaither MR, Toonen RJ, Robertson DR, Planes S, Bowen BW. Genetic evaluation of marine biogeographic barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus spp.) J Biogeogr. 2010;37:133–147. [Google Scholar]
- Garrick RC, Dyer RJ, Beheregaray LB, Sunnucks P. Babies and bathwater: a comment on the premature obituary for nested clade phylogeographic analysis. Mol Ecol. 2008;17:1401–1403. doi: 10.1111/j.1365-294X.2008.03675.x. [DOI] [PubMed] [Google Scholar]
- Harpending RC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- Hellberg ME. Footprints on the water: the genetic wake of dispersal among reefs. Coral Reefs. 2007;26:463–473. [Google Scholar]
- Hiatt RW, Strasburg DW. Ecological relationships of the fish fauna on coral reefs of the Marshall Islands. Ecol Monogr. 1960;30:65–127. [Google Scholar]
- Horne JB, van Herwerde L, Choat HJ, Robertson DR. High population connectivity across the Indo-Pacific: congruent lack of phylogeographic structure in three reef fish congeners. Mol Phylogenet Evol. 2008;49:629–638. doi: 10.1016/j.ympev.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Tsukamoto K, Nishida M. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the "ancient fish". Mol Phylogenet Evol. 2003;26:110–120. doi: 10.1016/s1055-7903(02)00331-7. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Aoyama J, Tsukamoto K, Nishida M. Population structure of the Japanese eel Anguilla japonica examined by mitochondrial DNA sequencing. Fish Sci. 2001;67:246–253. [Google Scholar]
- Jones GP, Planes S, Thorrold SR. Coral reef fish larvae settle close to home. Curr Biol. 2005;15:1314–1318. doi: 10.1016/j.cub.2005.06.061. [DOI] [PubMed] [Google Scholar]
- Kingsford MJ, Leis JM, Shanks A, Lindeman KC, Morgan SG, Pineda J. Sensory environments, larval abilities and local self recruitment. Bull Mar Sci. 2002;70:309–340. [Google Scholar]
- Klanten OS, Choat HJ, Van Herwerden L. Extreme genetic diversity and temporal rather than spatial partitioning in a widely distributed coral reef fish. Mar Biol. 2007;150:659–670. [Google Scholar]
- Knowles LL. Why does a method that fails continue to be used? Evolution. 2008;62:2713–2717. doi: 10.1111/j.1558-5646.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Leis JM. The pelagic stage of reef fishes: the larval biology of coral reef fishes. In: Sale PF, editor. The ecology of fishes on coral reefs. New York: Academic Press; 1991. pp. 183–230. [Google Scholar]
- Leis JM. Behavior of fish larvae as an essential input for modeling larval dispersal: behavior, biogeography, hydrodynamics, ontogeny, physiology and phylogeny meet hydrography. Mar Ecol Prog Ser. 2007;347:185–193. [Google Scholar]
- Leis JM, Wright KJ, Johnson RN. Behavior that influences dispersal and connectivity in the small, young larvae of a reef fish. Mar Biol. 2007;153:103–117. [Google Scholar]
- Lessios HA, Robertson DR. Crossing the impassable: genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc R Soc B. 2006;273:2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester SE, Ruttenberg BI. The relationship between pelagic larval duration and range size in tropical reef fishes: a synthetic analysis. Proc R Soc B. 2005;272:585–591. doi: 10.1098/rspb.2004.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa K, Fujii s, Yamazaki T, Takahashi A, Takasaki J, Yanagisawa M, Ohnishi Y, Nakamura Y, Kamatani N. New correction algorithms for multiple comparisons in case-control multilocus association studies based on haplotypes and diplotype configurations. J Hum Genet. 2008;53:789–801. doi: 10.1007/s10038-008-0312-0. [DOI] [PubMed] [Google Scholar]
- Mochioka N, Iwamizu M. Diet of anguilloid larvae: leptocephali feed selectively on larvacean houses and fecal pellets. Mar Biol. 1996;125:447–452. [Google Scholar]
- Montgomery JC, Tolimieri N, Haine OS. Active habitat selection by pre-settlement reef fishes. Fish Fish. 2001;2:261–277. [Google Scholar]
- Muss A, Robertson DR, Stepien CA, Wirtz P, Bowen BW. Phylogeography of Ophioblennius: the role of ocean currents and geography in reef fish evolution. Evolution. 2001;55:561–572. doi: 10.1554/0014-3820(2001)055[0561:pootro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Oxford; New York: Columbia University Press; 1987. [Google Scholar]
- Paillard D. What drives the ice age cycle? Science. 2006;313:455–456. doi: 10.1126/science.1131297. [DOI] [PubMed] [Google Scholar]
- Panchal M, Beaumont MA. The automation and evaluation of nested clade phylogeographic analysis. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ. The coup de grâce for nested clade phylogeographic analysis? Mol Ecol. 2008;17:516–518. doi: 10.1111/j.1365-294X.2007.03589.x. [DOI] [PubMed] [Google Scholar]
- Planes S, Fauvelot C. Isolation by distance and vicariance drive genetic structure of a coral reef fish in the Pacific Ocean. Evolution. 2002;56:378–399. doi: 10.1111/j.0014-3820.2002.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over Likelihood Ratio Tests. Syst Biol. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Templeton AR. Geodis: a program for the cladistic nested analysis of geographical distribution of haplotypes. Mol Ecol. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. 2007 Tracer v.1.4 [Internet]. Available from: http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Ramon ML, Nelson PA, DeMartini E, Walsh WJ, Bernardi G. Phylogeography, historical demography, and the role of post-settlement ecology in two Hawaiian damselfish species. Mar Biol. 2008;153:1207–1217. [Google Scholar]
- Randall JE. Zoogeography of shore fishes of the Indo-Pacific region. Zool Stud. 1998;37:227–268. [Google Scholar]
- Randall JE. Reef and shore fishes of the Hawaiian Islands. Honolulu: University of Hawaii Sea Grant College Program; 2007. [Google Scholar]
- Riordan C, Hussain M, McCann J. Moray eel attack in the tropics: a case report and review of the literature. Wilderness Environ Med. 2004;15:194–197. doi: 10.1580/1080-6032(2004)15[194:meaitt]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Rivera MAJ, Kelley CD, Roderick GK. Subtle population genetic structure in the Hawaiian grouper, Epinephelus quernus (Serranidae) as revealed by mitochondrial DNA analyses. Biol J Linn Soc. 2004;81:449–468. [Google Scholar]
- Rocha LA, Bass AL, Robertson DR, Bowen BW. Adult habitat preferences, larval dispersal, and the comparative phylogeography of three Atlantic surgeonfishes (Teleostei: Acanthuridae) Mol Ecol. 2002;11:243–251. doi: 10.1046/j.0962-1083.2001.01431.x. [DOI] [PubMed] [Google Scholar]
- Rocha LA, Craig MT, Bowen BW. Phylogeography and the conservation of coral reef fishes. Coral Reefs. 2007;26:501–512. [Google Scholar]
- Rocha LA, Robertson DR, Rocha CR, Van Tassell JL, Craig MT, Bowen BW. Recent invasion of the tropical Atlantic by an Indo-Pacific coral reef fish. Mol Ecol. 2005;14:3921–3928. doi: 10.1111/j.1365-294X.2005.02698.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS. PASSAGE. Pattern analysis, spatial statistics, and geographic exegesis. Version 1.1. 2001 Tempe (AZ): Department of Biology, Arizona State University. [Google Scholar]
- Scheltema RS. Dispersal of larvae by equatorial currents and its importance to the zoogeography of shoal-water tropical species. Nature. 1968;217:1159–1162. [Google Scholar]
- Schmidt J. The breeding places of the eel. Philos Trans R Soc Lond Ser B. 1923;211:179–208. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Geneva (Switzerland): Genetics and Biometry Laboratory, University of Geneva; 2000. Arlequin ver. 2.000: a software for population genetics data analysis. [Google Scholar]
- Schultz JK, Pyle RL, DeMartini E, Bowen BW. Genetic connectivity among color morphs and Pacific archipelagos for the flame angelfish, Centropyge loriculus. Mar Biol. 2007;151:167–175. [Google Scholar]
- Song CB, Near TJ, Page LM. Phylogenetic relations among percid fishes as inferred from mitochondrial cytochrome b DNA sequence data. Mol Phylogenet Evol. 1998;10:343–353. doi: 10.1006/mpev.1998.0542. [DOI] [PubMed] [Google Scholar]
- Springer VG. Pacific plate biogeography, with special reference to shorefishes. Smithson Contrib Zool. 1982:1–182. [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swearer SE, Shima JS, Hellberg ME, Thorrold SR, Jones GP, Robertson DR, Morgan SG, Selkoe KQ, Ruiz GM, Warner RR. Evidence of self-recruitment in demersal marine populations. Bull Mar Sci. 2002;70:251–272. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Meyer A, Bouvet J. Unusually large mitochondrial variation in populations of the blue tit, Parus caeruleus. Mol Ecol. 1992;1:27–36. doi: 10.1111/j.1365-294x.1992.tb00152.x. [DOI] [PubMed] [Google Scholar]
- Taylor MS, Hellberg ME. Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Science. 2003;299:107–109. doi: 10.1126/science.1079365. [DOI] [PubMed] [Google Scholar]
- Taylor MS, Hellberg ME. Marine radiations at small geographic scales: speciation in neotropical reef gobies (Elacatinus) Evolution. 2005;59:374–385. [PubMed] [Google Scholar]
- Templeton AR. Nested clade analysis of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Reply: Nested clade analysis: an extensively validated method for strong phylogeographic inference. Mol Ecol. 2008;17:1877–1880. doi: 10.1111/j.1365-294X.2008.03731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Statistical hypothesis testing in intraspecific phylogeography: nested clade phylogeographical analysis vs. approximate Bayesian computation. Mol Ecol. 2009;18:319–331. doi: 10.1111/j.1365-294X.2008.04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker CE, Thompson AR, Roje DM, Shaw EY. New expansions in old clades: population genetics and phylogeny in a Gnatholepis species (Teleostei: Gobiodei) in the Pacific. Mar Biol. 2007;153:375–385. [Google Scholar]
- Theisen TC, Bowen BW, Lanier W, Baldwin JD. High connectivity on a global scale in the pelagic wahoo, Acanthocybium solandri (tuna family Scombridae) Mol Ecol. 2008;17:4233–4247. doi: 10.1111/j.1365-294x.2008.03913.x. [DOI] [PubMed] [Google Scholar]
- Tseng W. Relationship between growth rate and age at recruitment of Anguilla japonica elvers in a Taiwan estuary as inferred from otolith increments. Mar Biol. 1990;107:75–81. [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. DNA barcoding Australia's fish species. Philos Trans R Soc Lond Ser B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weersing KA, Toonen RJ. Population genetics, larval dispersal, and demographic connectivity in marine systems. Mar Ecol Prog Ser. 2009;393:1–12. [Google Scholar]