Abstract
The renal proximal tubule (RPT) is a central locale for Na+ reabsorption, and blood pressure regulation. Na+ reabsorption in the RPT depends upon the Na,K-ATPase, which is controlled by a complex regulatory network, including Salt-Inducible Protein Kinase (SIK). SIKs are recently discovered members of the AMP-activated Protein Kinase (AMPK) family, which regulate salt homeostasis and metabolism in a number of tissues. In the RPT, SIK interacts with the Na,K-ATPase in the basolateral membrane (BM), regulating both the activity and level of Na,K-ATPase in the BM. Thus, Na,K-ATPase activity can be rapidly adjusted in response to changes in Na+ balance. Long-term changes in Na+ intake affect the state of SIK phosphorylation, and as a consequence the phosphorylation of TORCS, Transducers of Regulated CREB (cAMP Regulatory Element Binding Protein). Once phosphorylated, TORCS enter the nucleus, and activate transcription of the ATP1B1 gene encoding for the Na,K-ATPase β subunit.
Keywords: proximal tubule; salt-inducible kinase; Na,K-ATPase; primary culture
1. Introduction
Na+ reabsorption by the kidney must be tightly regulated to maintain Na+ homeostasis, and blood pressure [1]. The RPT is particularly important, reabsorbing 70% of the glomerular filtrate. Filtered Na+ enters RPT cells by apical transporters including the Na+/H+ antiport system (NHE3), the Na+/glucose cotransport system (SGLT), the Na+/Phosphate (Pi) cotransport system (NPt2), and Na+/amino acids cotransport systems. The uptake of solutes by these transporters is driven by the Na+ gradient established by the Na,K-ATPase localized in the BM [2].
The activity of the Na,K-ATPase is subject to control by a diverse set of effectors [2]. The kidney responds to small increases in Na+ by regulating the Na,K-ATPase, causing an increase in Na+ excretion (natriuresis). When Na+ intake increases, the synthesis of natriuretic factors, including renal dopamine increases. Parathyroid Hormone (PTH) is also natriuretic. In contrast, when Na+ intake declines, the level of anti-natriuretic factors, such as Angiotensin II (Ang II), norepinephrine, and Prostaglandin E2 (PGE2) increase. These natriuretic and anti-natriuretic factors ultimately target Na,K-ATPase in the RPT.
When the regulatory network that controls Na,K-ATPase in the RPT is impaired, the ultimate result is hypertension, as illustrated in a number of animal models [1]. Included amongst these animal models is the Spontaneously Hypertensive Rat (SHR), with altered Na,K-ATPase regulation by dopamine [1], and the Milan hypertensive rat with a mutant α-adducin gene (ADD1) and an over-active SIK1 [3].
2.Salt Inducible Kinase
SIK was originally found in the adrenal glands of rats maintained on a high Na+ or a high K+ diet for a week [4]. Three SIK isoforms exist, whose genes are encoded on human chromosome 21 (sik1 gene), and human chromosome 11 (the sik2 and sik3 genes). The effects of SIKs are quite tissue-specific. For example, in the adrenal, SIK1 controls the expression of steroidogenic enzymes, whereas in the brain, both SIK1 and SIK2 regulate energy metabolism [5]. SIK2 also regulates gluconeogenesis in the liver, and lipogenesis in adipose tissue.
SIK homologs are present in species ranging from mice, chicken, flies, worms to plants [5; 6; 7]. Examples include the SIK-2 homolog KIN-29, which regulates chemosensory signals in worms [8], and Salt Overly Sensitive 2 (SOS2), which mediates the response of plants to salt stress [7]. All of these SIKs are members of the 5' AMP-activated Protein Kinase (AMPK) family [9]. However, unlike AMPK itself, which is an αβγ tetramer, an SIK consists of a single subunit.
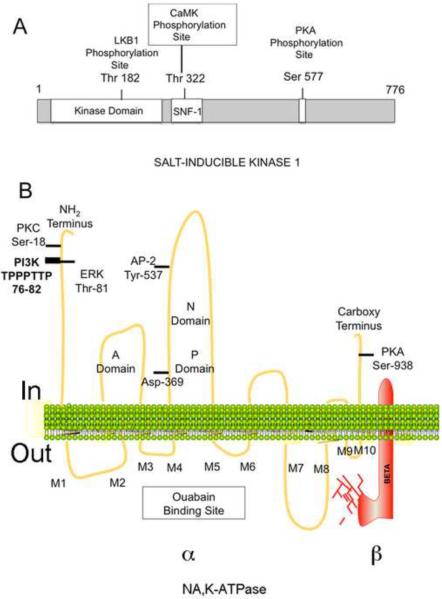
SIK1 is an 86 kDa protein with 3 major domains (Fig. 1A) [4]. Included amongst the domains is the amino terminal serine-threonine kinase domain, the adjacent yeast sucrose non-fermenting-1 (SNF-1) homology (SNH) domain, and a carboxy terminal domain with a Protein Kinase A (PKA) phosphorylation site. SIK, like other AMPK family members, has an Activation loop (A-loop) near the substrate-binding pocket in the kinase domain. When the A loop is phosphorylated by the LKB1 tumor suppressor kinase, the conformation of the catalytic site changes, increasing kinase activity [4]. SIK1 is also activated by phosphorylation of the SNH domain by Calmodulin-dependent protein kinase (CaMK) [7]. Phosphorylation by PKA causes SIK1 to translocate from the nucleus to the cytoplasm.
Figure 1.
Structure of SIK1 and Na,K-ATPase
SIK-1 has recently been found to play an important role in regulating Na,K-ATPase in the RPT [10]. Studies with the established RPT cell line Opposum Kidney (OK) have shown that SIK1 is associated with the Na,K-ATPase. When the level of intracellular Na+ is elevated, SIK1 is activated, resulting in increased Na,K-ATPase activity. A brief review of Na,K-ATPase is needed in order to understand how SIK1 regulates Na,K-ATPase in the RPT.
3. Mechanism of Action of the Na,K-ATPase
The Na, K-ATPase transports 3 intracellular Na+ ions out of the cell, and 2 extracellular K+ ions into the cells, while utilizing ATP [11]. During the transport reaction, Na,K-ATPase interconverts between 2 conformational states, E1 and E2. While in E1, 1) intracellular Na+ and ATP bind to Na,K-ATPase 2) Na,K-ATPase is phosphorylated on a conserved aspartate by the γ-Pi of ATP, and 3) Na+ is occluded. Na,K-ATPase then assumes the E2 conformation, during which 1) Na+ is released extracellularly, 2) extracellular K+ binds to Na,K-ATPase, and is occluded. Finally, 3) the asparate residue is dephosphorylated, and K+ is released intracellularly.
The Na,K-ATPase is a heterodimer consisting of a catalytic α subunit (110 Kd), and a highly glycosylated β subunit (55 Kd) (Fig. 1B) [11]. The β subunit is required for the integration of Na,K-ATPase into the plasma membrane (PM), and enzyme activity. Each subunit has 4 distinct isoforms, including α1 and β1, which are expressed in the kidney.
The α1 subunit has 10 transmembrane spanning segments [11] (Fig. 1B). There are 5 extracellular loops, including the ouabain-binding site (M3/M4 and M5/M6), and the M7-M8 loop (which interacts with β). Intracellular regions include the Actuator (A) domain (within the amino terminus and the M2M3 loop), the adjacent N domain (with the ATP binding site; in the M4M5 loop), and the P domain (with the ATP phosphorylation site, also in M4M5). Both the N and A domains approach the P domain as the pump functions.
The transport of Na+ and K+ by the Na,K-ATPase establishes an electrochemical gradient across the PM [11; 12]. This electrochemical gradient is necessary for cardiac and vascular contractility, neural transmission, and the transepithelial transport of solutes. In kidney tubule epithelial cells, the Na+, which enters the cells by apical transport systems, leaves the cells through the Na,K-ATPase, localized in the BM, resulting in Na+ reabsorption.
The Na,K-ATPase is subject to rapid (or acute), as well as chronic regulation [13]. Acute regulation may result from 1) changes affecting the kinetics of the enzyme, as well as 2) changes in the level of pre-existing Na,K-ATPases in the PM. Such acute changes are often caused by protein kinases, which bind to the Na,K-ATPase α subunit, and may eventually phosphorylate the enzyme (Fig. 1B) [14; 15].
The amino terminus of the α subunit contains a PI3K binding site, as well as PKC and ERK phosphorylation sites (Fig. 1B). The phosphorylation of Ser-18 by PKC reduces pump activity, and in addition may result in endocytosis. In contrast, phosphorylation of Thr-81 by ERK1/2 activates the pump. The α subunit also contains a recognition site for Adapter Protein-2 (AP-2) [16], and a PKA phosphorylation site (Fig. 1B).
4. Acute Regulation of Na,K-ATPase by SIK
4A. Activation by Intracellular Na+ (Na+in)
A number of hormones and growth factors stimulate Na,K-ATPase activity by increasing Na+in [13]. Increases in Na+in occur as a consequence of increased transport by NHE3 and other Na+ dependent transporters. Normally, the transport activity of Na,K-ATPase is sub-optimal because Na+in (9–14mM in RPTs) is below the Km value for Na+. However, when Na+in increases, the activity of Na,K-ATPase increases until the substrate binding sites are saturated.
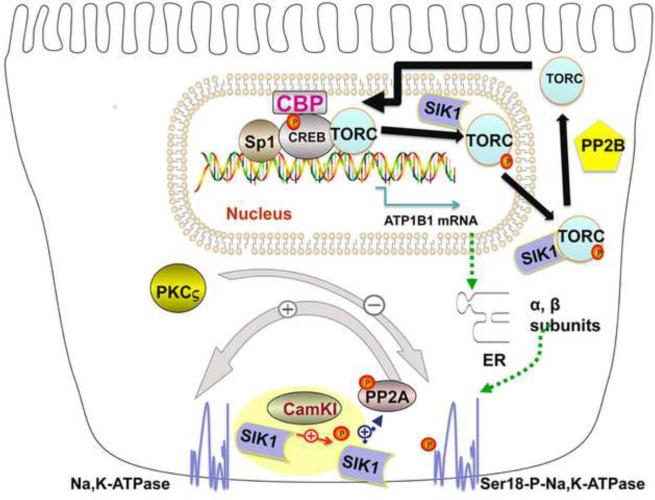
Recently, SIK was also found to be responsible for activating the Na,K-ATPase following increases in Na+in [10]. In Opossum Kidney (OK) cells, increases in Na+in caused by monensin (a Na+ ionophore), result in an increase in Ca2+ influx by a Na+/Ca2+ exchange system (NCE1). As a consequence, CaMK1 is activated. Activated CaMK1 phosphorylates, and activates SIK1 (which is associated with Na,K-ATPase). The activation of SIK1 results in an increase in Na,K-ATPase activity.
However, SIK1 does not directly increase Na,K-ATPase activity. Instead, SIK1 acts indirectly, via a Protein Phosphatase (PPase), Protein Phosphatase 2A (PP2A) (Fig. 2) [10]. PP2A is part of a multiprotein complex, which contains SIK1, the Na,K-ATPase, as well as another protein, PPase methylesterase-1 (PME-1). This methylesterase normally demethylates, and inactivates PP2A. However, when CaMK1 activates SIK1, SIK1 phosphorylates PME-1, causing PME-1 to dissociate from the SIK1,PP2A, Na,K-ATPase complex. As a consequence, PP2A is methylated by surrounding cellular methylases, and is activated. Activated PP2A dephosphorylates the Na,K-ATPase α subunit (at Ser18), causing an increase in the catalytic activity of the Na,K-ATPase.
Figure 2.
Acute and Chronic Regulation of Na,K-ATPase by SIK
4B. Response of the RPT to Natriuretic and Anti-Natriuretic Factors
4B1. The Initial Response
Following increases in dietary Na+, natriuretic factors are produced, including dopamine, which reduce basolateral Na,K-ATPase activity (by increasing endocytosis). In contrast, following decreases in dietary Na+, anti-natriuretic factors (norepinephrine, Ang II, PGs) are produced, which increase basolateral Na,K-ATPase activity (by recruiting pre-existing cytoplasmic Na,K-ATPases to the BM).
Initially, dopamine interacts with its receptors (primarily D1-like receptors, or D1Rs), causing additional D1Rs to enter the PM, and localize to caveolin-rich domains enriched with Na,K-ATPase [17]. Ang II type 1 receptors (AT1Rs) are localized in the same caveolin-rich regions. Following treatment with dopamine, the AT1Rs are rapidly internalized, into the cytoplasm, resulting in a complete loss of Ang II signaling. Conversely, extracellular Ang II causes additional AT1Rs to be recruited into the caveolin-rich domains, while D1Rs are rapidly internalized, such that D1R signaling is lost. The recycling and re-sensitization of D1Rs depends upon the dephosphorylation of D1Rs by PPases including PP2A, whose activation depends upon SIK1[18].
4B2. Role of Na+in
The actions of both natriuretic and antinatriuretic factors depend upon Na+in, and, thus, SIK1 [19]. In rat RPTs at normal Na+in, Ang II activates PKCβ, which results in the phosphorylation of the α subunit of cytoplasmic Na,K-ATPase (Ser 11 and Ser-18), and the recruitment of Na,K-ATPases to the PM. However, following an increase in Na+in, the inhibitory effect of dopamine on basolateral Na,K-ATPase becomes more substantial, and endocytosis of basolateral Na,K-ATPase occurs.
In order to exert its inhibitory effect, dopamine activates PKCς, which phosphorylates the Na,K-ATPase α1 subunit (at Ser-18) [16]. The 14-3-3 protein binds to this phospho-Ser motif, which facilitates the binding of PI3K to an adjacent Proline-Rich Domain in α1, resulting in PI3K activation. Activated PI3K promotes the binding of Adaptor Protein-2 (AP-2) to the α1 subunit, an event required for endocytosis.
4B3. Role of AP-2 in Endocytosis
In the absence of dopamine, an increase in Na+in causes the Na,K-ATPase α1 subunit to become dephosphorylated by SIK1. Dephosphorylated Na,K-ATPase has a low affinity for both PI3K and AP-2. However, in the presence of dopamine, AP-2 is phosphorylated by PKCς, and acquires an increased affinity for the basolateral α1 subunit (AP-2 binds to Tyr-537 in the M4/M5 loop) (Figure 1B) [16]. After phosphorylated AP-2 associates with the Na,K-ATPase, AP-2 recruits clathrin to the complex. A consequence is the formation of clatherin-coated pits, and the endocytosis of the Na,K-ATPase.
4B4. Altered Trafficking in the Milan Hypertensive Rat
The Milan hypertensive rat has an mutation in α-adducin (ADD1), similar to α-adducin mutations in humans [3]. This mutation causes an increase in the level and activity of SIK1 in the RPT, which in turn results in CaMK1 activation (independent of increased Na+/Ca2+ exchange). As a consequence, 1) PP2A is activated, and dephosphorylates the Na,K-ATPase α subunit, and 2) PI3K and AP-2 not longer associate with the α subunit, preventing endocytosis [20]. The result is increased Na,K-ATPase activity.
5. Chronic Regulation by Na,K-ATPase
5A. Role of SIK in Response to High Salt
The role of SIK signaling in response to increased Na+ was first studied in the adrenal [4]. The activation of SIK1 in the adrenal results in a decline in the level of the mRNAs for steroidogenic acute regulatory protein (StAR) and side chain cleavage P450 (CYP11A1). SIK causes a reduction in the level of these mRNAs by repressing the activation of their transcription via CREB. However, further studies indicated that the ability of SIK to cause transcriptional repression of CREB-regulated genes is tissue-specific.
5B. Classic Role of CREB
CREB plays a central role in transcriptional regulation in all mammalian cells. CREB is phosphorylated, and activated by PKA, as well as a number of other protein kinases, including PKC, Mitogen Activated Protein (MAP) Kinase, and CAMK [21]. Prior to its phosphorylation, CREB recognizes and binds to cAMP Regulatory Elements (CREs) (Fig. 3) via its carboxy terminal DNA binding domain (bZIP). The amino terminus of CREB contains a Kinase Inducible Domain (KID) with a PKA phosphorylation site at Ser-133. The phosphorylation of CREB at Ser-133 by PKA results in the binding of the CREB Binding Protein (CBP) to CREB. As a consequence CREB is activated, and increases the rate of transcription of CREB-regulated genes [22].
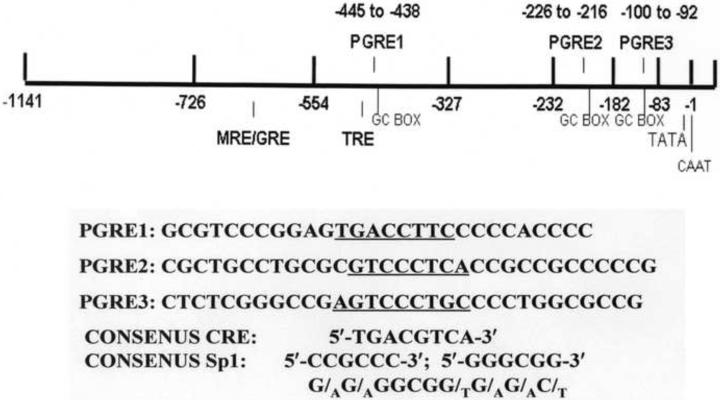
Figure 3.
Promoter of the Na,K-ATPase β Subunit Gene ATP1B1
5C. Activation of CREB by TORCs; Role of SIK
Recently, the classic view that CREB activation depends solely upon its interaction with CBP has changed. A genome-wide analysis of CREB target genes indicated that CBP is not recruited to all promoters with CREB binding sites even after CREB is phosphorylated and activated [23]. Subsequently, additional CREB binding partners were found. One such set of binding partners are TORCs, which interact with the bZIP DNA binding domain of CREB, and can activate CREB independently of Ser-133 phosphorylation. However, when Ser-133 phosphorylation does occur, TORCs can further increase the activation of CREB by CBP, in a tissue-specific manner [24].
SIK plays a central role in this process [4]. SIK may be either nuclear or cytoplasmic. While in the nucleus, SIK phosphorylates TORCs, which lose their affinity for CREB, and are exported from the nucleus [21]. All of these events depend upon the state of phosphorylation of SIK. In Y1 adrenal cells with elevated cAMP, SIK1 is phosphorylated at Ser577 by PKA, and inactivated. As a consequence, TORCs (localized in the cytoplasm) are dephosphorylated, and imported into the nucleus, where TORCS bind to CREB on TORC-sensitive genes (Fig. 2). In kidney cells, the Na,K-ATPase β1 subunit gene is included amongst these TORC-sensitive genes.
5D. Transcriptional Regulation of Na,K-ATPase by TORCs
5D1. Regulatory Elements
The Na,K-ATPase α and β subunit genes (ATP1A1 and ATP1B1, respectively) are subject to transcriptional control following chronic changes in Na+ balance and the hormonal milieu [13]. However, at the protein level, the quantity of β subunit is limiting to the formation of α/β heterodimers in many cell types. Thus, increases in transcription of ATP1B1 alone may have substantial affects on the cellular level of Na,K-ATPase, unlike the case with ATP1A. For this reason ATP1B1 gene regulation is of particular interest.
ATP1B1 gene regulation depends upon the regulatory elements in its promoter region. Included amongst these regulatory elements are a mineralocorticoid/glucocorticoid regulatory element (MRE/GRE)), a thyroid hormone regulatory element (TRE), as well as 3 Prostaglandin Regulatory Elements (PGREs) (Fig. 3) [22; 25]. Each of these regulatory elements is required in order to obtain a stimulatory effect in response to either thyroid hormone (T3), hydrocortisone, or PGE2, respectively.
The regulation of ATP1B1 transcription has been studied in kidney tubule epithelial cell cultures, including the established dog kidney cell line MDCK, a distal tubule model. Stimulatory effects of PGE1 and PGE2 on ATP1B1 transcription were observed, and were associated with increases in the level of β subunit mRNA, as well as in the overall level of the Na,K-ATPase, and in Na,K-ATPase activity [26]. The stimulatory effects of PGE1 and PGE2 on ATP1B1 transcription, are dependent upon the activation of PKA, PKC, and CaMK, as well as CREB [22; 27] [28]. Recently, PGE1, and PGE2 were also found to stimulate ATP1B1 transcription in differentiated primary rabbit RPT cell cultures [29] [30]. Norepinephrine and dopamine also regulated transcription [22; 30].
Two PGREs on the ATP1B1 promoter, PGRE1(−445 to −439; TGACCTTC) and PGRE3 (−100 to −92; AGTCCCTGC) (Fig. 3), are primarily involved in mediating the PGE effects [27]. In order to obtain a PGE-stimulation, must CREB bind to each of these 2 PGREs, and interact with Sp1 on an adjacent binding site (Fig. 3) [22; 31; 32]. Indeed, when the Sp1 binding site adjacent to PGRE3 is moved further upstream, the stimulatory effect of PGE1 is lost [32].
5D2. Role of TORCs
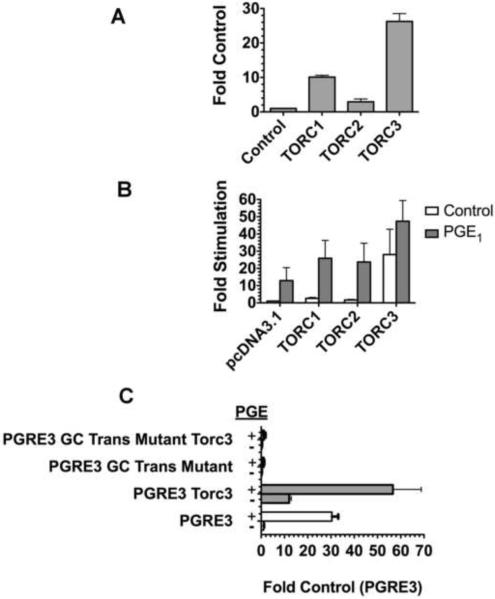
Recently, TORCs have also been found to control ATP1B1 transcription. In transient transfection studies conducted with pHβ1-1141Luc, a human ATP1B1 promoter/luciferase construct, 3 different TORC isoforms were observed to significantly stimulate transcription in primary RPT cells [30] [33] (Fig. 4). TORC3 had the largest effect. In MDCK cells (Fig. 4B), TORCs similarly increased basal transcription, and in addition increased the PGE1 stimulation(Fig. 4B). As with primary RPTs, TORC3 had the most substantial effect. Thus the effect of TORC3 on transcriptional regulation by PGRE3 in particular was studied, using the vector pLuc-MCS β72-167 (containing the PGRE3 region) (Fig. 4C) [22]. The results showed a stimulatory effect of TORC3 on transcription by pLuc-MCS β72-167. However, this stimulatory effect of TORC3 was lost when the Sp1 site adjacent to PGRE3 was moved further downstream (in pLuc-MCSβ72-167GCTrans) [22]. Thus, the stimulatory effect of TORC3 depends this Sp1 site as well as PGRE3. Overall the results are suggestive of a TORC/CREB/Sp1 interaction.
Figure 4.
Regulation of ATP1B1 Transcription by TORCs. A. Regulation in primary RPT cells. Primary RPT cells were transiently transfected as previously described ([30]) with 1 μg pH β1-1141Luc, 0.2 μg pSβGal (to monitor transfection efficiency), and 0.1 μg of either TORC1, TORC2, TORC3, or the empty vector pcDNA3.1/V5 (Control) [22]. After 24 hrs, luciferase activity was determined in quadruplicate. Values are averages (+/− SEM) in comparison to the control). B. Regulation in MDCK cells. MDCK cells were transfected (as above), and incubated 4 hrs either with 1.4 μM PGE1, or untreated, followed by luciferase determinations. Results are averaged from 3 experiments. C. Regulation via PGRE3. MDCK cells were transfected (as in A. above), and treated with PGE1, (or untreated), also as above, with the modification that the promoter/luciferase constructs were either pLuc-MCS β72–167 (PGRE3 vector) or pLuc-MCS β72–167 GC trans (PGE3 vector with GC trans mutation, as described previously [22]). Values are averages (+/− SEM) of quadruplicate determinations, as compared with the Control value (in cultures transfected with PGRE1 + pcDNA3.1/V5).
Thus, TORCS increase PGE1-stimulated transcription in kidney cells, which in turn is dependent upon the activation of PKA, PKC and CaMK. For this reason, TORCS appear to function in kidney cells in a manner similar to neurons [34], rather than adrenal cells, in which TORC1 antagonizes cAMP-induced transcription [6]. In unstimulated neurons, cytoplasmic SIK1 interacts with and sequesters TORC1 (Fig. 2). However, after stimulation with Ca2+ and cAMP, TORC1 is dephosphorylated by PPase 2B, is released from SIK1, and translocated to the nucleus [34]. Nuclear TORC1 binds to both CREB and CBP, and increases CREB activation over that obtained with CREB and CBP alone [34] (Fig. 2). The resulting increase in transcription caused by this TORC/CREB/CBP interaction depends upon the sequence and/or the orientation of the CREB binding site [34]. In the case of ATP1B1 transcription in the RPT, this TORC/CREB/CBP interaction depends upon PGREs as well as Sp1 (Figure 2).
6. Conclusion: Overall Role of SIK in Regulation of Na,K-ATPase in the RPT
Following increases in Na+in, SIK1 is activated in the RPT, causing the dephosphorylation and activation of the basolateral Na,K-ATPase. At the same time, cytoplasmic SIK1 sequesters TORCs in the cytoplasm, thereby reducing the ability of TORCs to enter the nucleus, and increase transcription of ATP1B1. In contrast, when SIK1 is inactive, cytoplasmic TORCs are dephosphorylated, and enter the nucleus, so as to increase the overall of level of expression of the Na,K-ATPase. In this manner, regulation at the level of SIK acts as a switch to change the level control of Na,K-ATPase from acute, to chronic regulation.
Acknowledgments
We thank Dr. Dong-Yan Jin (University of Hong Kong) for TORC vectors, Keikantse Matlhagela, Trivikram Rajkowa and Maryann Borsick for their research, as well as NHLBI 1R01HL6976-01 to M.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wang X, Armando I, Upadhyay K, Pascua A, Jose PA. The regulation of proximal tubular salt transport in hypertension: an update. Current Opinion in Nephrology & Hypertension. 2009;18:412–20. doi: 10.1097/MNH.0b013e32832f5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Al-Khalili L, Kotova O, Tsuchida H, Ehren I, Feraille E, Krook A, Chibalin AV. ERK1/2 mediates insulin stimulation of Na(+),K(+)-ATPase by phosphorylation of the alpha-subunit in human skeletal muscle cells. J Biol Chem. 2004;279:25211–8. doi: 10.1074/jbc.M402152200. [DOI] [PubMed] [Google Scholar]
- [3].Stenstrom K, Takemori H, Bianchi G, Katz AI, Bertorello AM. Blocking the salt-inducible kinase 1 network prevents the increases in cell sodium transport caused by a hypertension-linked mutation in human alpha-adducin. J Hypertens. 2009 doi: 10.1097/HJH.0b013e328330cf15. [DOI] [PubMed] [Google Scholar]
- [4].Okamoto M, Takemori H, Katoh Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends in Endocrinology & Metabolism. 2004;15:21–6. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- [5].Hietakangas V, Cohen SM. TORCing up metabolic control in the brain. Cell Metabolism. 2008;7:357–8. doi: 10.1016/j.cmet.2008.04.006. [DOI] [PubMed] [Google Scholar]
- [6].Katoh Y, Takemori H, Horike N, Doi J, Muraoka M, Min L, Okamoto M. Salt-inducible kinase (SIK) isoforms: their involvement in steroidogenesis and adipogenesis. Molecular & Cellular Endocrinology. 2004;217:109–12. doi: 10.1016/j.mce.2003.10.016. [DOI] [PubMed] [Google Scholar]
- [7].Bertorello AM, Zhu JK. SIK1/SOS2 networks: decoding sodium signals via calcium-responsive protein kinase pathways. Pflugers Archiv - European Journal of Physiology. 2009;458:613–9. doi: 10.1007/s00424-009-0646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van der Linden AM, Wiener S, You YJ, Kim K, Avery L, Sengupta P. The EGL-4 PKG acts with KIN-29 salt-inducible kinase and protein kinase A to regulate chemoreceptor gene expression and sensory behaviors in Caenorhabditis elegans. Genetics. 2008;180:1475–91. doi: 10.1534/genetics.108.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiological Reviews. 2009;89:1025–78. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- [10].Sjostrom M, Stenstrom K, Eneling K, Zwiller J, Katz AI, Takemori H, Bertorello AM. SIK1 is part of a cell sodium-sensing network that regulates active sodium transport through a calcium-dependent process. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16922–7. doi: 10.1073/pnas.0706838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–35. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- [12].Fâeraille E, Doucet A. Sodium-potassium-adenosinetriphosphatase-dependent sodium transport in the kidney: hormonal control. Physiol Rev. 2001;81:345–418. doi: 10.1152/physrev.2001.81.1.345. [DOI] [PubMed] [Google Scholar]
- [13].Therien AG, Blostein R. Mechanisms of sodium pump regulation. Am J Physiol Cell Physiol. 2000;279:C541–66. doi: 10.1152/ajpcell.2000.279.3.C541. [DOI] [PubMed] [Google Scholar]
- [14].Sweadner KJ, Feschenko MS. Predicted location and limited accessibility of protein kinase A phosphorylation site on Na-K-ATPase. American Journal of Physiology - Cell Physiology. 2001;280:C1017–26. doi: 10.1152/ajpcell.2001.280.4.C1017. [DOI] [PubMed] [Google Scholar]
- [15].Sweadner KJ, Donnet C. Structural similarities of Na,K-ATPase and SERCA, the Ca(2+)-ATPase of the sarcoplasmic reticulum. Biochemical Journal. 2001;356:685–704. doi: 10.1042/0264-6021:3560685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ogimoto G, Yudowski GA, Barker CJ, Kohler M, Katz AI, Feraille E, Pedemonte CH, Berggren PO, Bertorello AM. G protein-coupled receptors regulate Na+,K+-ATPase activity and endocytosis by modulating the recruitment of adaptor protein 2 and clathrin. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3242–7. doi: 10.1073/pnas.060025597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gildea JJ, Israel JA, Johnson AK, Zhang J, Jose PA, Felder RA. Caveolin-1 and dopamine-mediated internalization of NaKATPase in human renal proximal tubule cells. Hypertension. 2009;54:1070–6. doi: 10.1161/HYPERTENSIONAHA.109.134338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu P, Asico LD, Luo Y, Andrews P, Eisner GM, Hopfer U, Felder RA, Jose PA. D1 dopamine receptor hyperphosphorylation in renal proximal tubules in hypertension. Kidney International. 2006;70:1072–9. doi: 10.1038/sj.ki.5001708. [DOI] [PubMed] [Google Scholar]
- [19].Efendiev R, Budu CE, Cinelli AR, Bertorello AM, Pedemonte CH. Intracellular Na+ regulates dopamine and angiotensin II receptors availability at the plasma membrane and their cellular responses in renal epithelia. J Biol Chem. 2003;278:28719–26. doi: 10.1074/jbc.M303741200. [DOI] [PubMed] [Google Scholar]
- [20].Efendiev R, Krmar RT, Ogimoto G, Zwiller J, Tripodi G, Katz AI, Bianchi G, Pedemonte CH, Bertorello AM. Hypertension-linked mutation in the adducin alpha-subunit leads to higher AP2-mu2 phosphorylation and impaired Na+,K+-ATPase trafficking in response to GPCR signals and intracellular sodium. Circulation Research. 2004;95:1100–8. doi: 10.1161/01.RES.0000149570.20845.89. [DOI] [PubMed] [Google Scholar]
- [21].Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- [22].Matlhagela K, Borsick M, Rajkhowa T, Taub M. Identification of a Prostaglandin-responsive Element in the Na,K-ATPase {beta}1 Promoter That Is Regulated by cAMP and Ca2+: Evidence for an interactive role of cAMP regulatory element-bindin protein and Sp1. J Biol Chem. 2005;280:334–346. doi: 10.1074/jbc.M411415200. [DOI] [PubMed] [Google Scholar]
- [23].Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–64. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu W, Kasper LH, Lerach S, Jeevan T, Brindle PK. Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. The EMBO Journal. 2007;26:2890–2903. doi: 10.1038/sj.emboj.7601734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lingrel JB, Orlowski J, Shull MM, Price EM. Molecular genetics of Na,K-ATPase. Progress in Nucleic Acid Research & Molecular Biology. 1990;38:37–89. doi: 10.1016/s0079-6603(08)60708-4. [DOI] [PubMed] [Google Scholar]
- [26].Taub M, Borsick M, Geisel J, Matlhagela K, Rajkhowa T, Allen C. Regulation of the Na,K-ATPase in MDCK cells by prostaglandin E1: a role for calcium as well as cAMP. Exp Cell Res. 2004;299:1–14. doi: 10.1016/j.yexcr.2004.04.046. [DOI] [PubMed] [Google Scholar]
- [27].Matlhagela K, Taub M. Involvement of EP1 and EP2 receptors in the regulation of the Na,K-ATPase by prostaglandins in MDCK cells. Prostaglandins & other lipid mediators. 2006;79:101–13. doi: 10.1016/j.prostaglandins.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Matlhagela K, Taub M. Involvement of EP1 and EP2 receptors in the regulation of the Na,K-ATPase by prostaglandins in MDCK cells. Prostaglandins Other Lipid Mediat. 2006;79:101–13. doi: 10.1016/j.prostaglandins.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chung SD, Alavi N, Livingston D, Hiller S, Taub M. Characterization of primary rabbit kidney cultures that express proximal tubule functions in a hormonally defined medium. J Cell Biol. 1982;95:118–26. doi: 10.1083/jcb.95.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Herman M, Rajkhowa T, Cutuli F, Springate JE, Taub M. Regulation of Renal Proximal Tubule Na,K-ATPase by Prostaglandins. American Journal of Physiology - Renal Physiology. 2010 doi: 10.1152/ajprenal.00467.2009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Matlhagela K, Taub M. Regulation of the Na-K-ATPase beta(1)-subunit promoter by multiple prostaglandin-responsive elements. American Journal of Physiology - Renal Physiology. 2006;291:F635–46. doi: 10.1152/ajprenal.00452.2005. [DOI] [PubMed] [Google Scholar]
- [32].Matlhagela K, Taub M. Prostaglandins regulate transcription by means of prostaglandin response elements located in the promoters of mammalian Na,K-ATPase beta 1 subunit genes. Ann N Y Acad Sci. 2006;1091:233–43. doi: 10.1196/annals.1378.070. [DOI] [PubMed] [Google Scholar]
- [33].Siu YT, Chin KT, Siu KL, Yee Wai Choy E, Jeang KT, Jin DY. TORC1 and TORC2 coactivators are required for tax activation of the human T-cell leukemia virus type 1 long terminal repeats. Journal of Virology. 2006;80:7052–9. doi: 10.1128/JVI.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4700–5. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]