Abstract
Study Objectives
The regulation of sleep is poorly understood. While some molecules, including those involved in inflammatory/immune responses, have been implicated in the control of sleep, their role in this process remains unclear. The Drosophila model for sleep provides a powerful system to identify and test the role of sleep-relevant molecules.
Design
We conducted an unbiased screen for molecular candidates involved in sleep regulation by analyzing genome-wide changes in gene expression associated with sleep deprivation in Drosophila. To further examine a role of immune-related genes identified in the screen, we performed molecular assays, analysis of sleep behavior in relevant mutant and transgenic flies, and quantitative analysis of the immune response following sleep deprivation.
Results
A major class of genes that increased expression with sleep deprivation was that involved in the immune response. We found that immune genes were also upregulated during baseline conditions in the cyc01 sleep mutant. Since the expression of an NFκB, Relish, a central player in the inflammatory response, was increased with all manipulations that reduced sleep, we focused on this gene. Flies deficient in, but not lacking, Relish expression exhibited reduced levels of nighttime sleep, supporting a role for Relish in the control of sleep. This mutant phenotype was rescued by expression of a Relish transgene in fat bodies, which are the major site of inflammatory responses in Drosophila. Finally, sleep deprivation also affected the immune response, such that flies deprived of sleep for several hours were more resistant to bacterial infection than those flies not deprived of sleep.
Conclusion
These results demonstrate a conserved interaction between sleep and the immune system. Genetic manipulation of an immune component alters sleep, and likewise, acute sleep deprivation alters the immune response.
Keywords: Drosophila, Relish, immune response, NFκB, sleep deprivation
INTRODUCTION
Sleep is controlled by a homeostatic process, which determines how much sleep occurs depending on the length of time spent in prior wakefulness. Little is known about the molecular components of the sleep homeostatic system. Studies of differential mRNA expression associated with sleep deprivation, wakefulness and spontaneous sleep in mammals have identified candidate genes, many of which are involved in metabolism and synaptic plasticity.1, 2 In addition, functional studies of sleep homeostasis have implicated molecules involved in immune signaling, including cytokines such as interleukin-1 and tumor necrosis factor α (reviewed in Obal and Krueger3). However, precisely how each of these gene families functions in sleep homeostasis remains unclear. We sought to identify molecular candidates and their function in sleep by investigating sleep in a simpler organism, Drosophila melanogaster.
Rest behavior in Drosophila has features that qualify it as a sleep-like state. These features include reduced sensory responsiveness, consolidated periods of inactivity at a predictable time of day (indicating that it is under circadian control), a preferred location in both isolated and social settings, and a rebound, or recovery period, after a deprivation, indicating the presence of a homeostatic mechanism. A number of drugs, including caffeine and modafinil, that affect sleep in mammals also affect sleep in a similar manner in flies,4–6 indicating the existence of conserved neurochemical mechanisms.
Several mutants exhibit altered baseline levels of sleep and rebound responses to deprivation. For example, we have shown by genetic manipulation in flies that baseline sleep, or spontaneous undisturbed sleep, is inversely related to cAMP activity. Mutations that upregulate cAMP signaling, such as dunce, decrease baseline sleep. Conversely, mutations that decrease cAMP signaling, such as S162, which knocks out the dCREB2 gene, increase baseline sleep. Furthermore, the S162 mutants exhibit an exaggerated rebound response.7 Mice lacking a CREB gene also show increased baseline sleep,8 which further supports the notion that a conserved mechanism for sleep homeostasis exists between mammals and flies.
Studies of various clock mutants in Drosophila have revealed that cycle mutant flies (cyc01) show altered sleep behavior. The sleep phenotype in these flies consists of abnormal levels of baseline sleep as well as altered responses to sleep deprivation.9,10 cyc01 flies also express abnormal levels of some genes involved in the stress response as compared to a control strain.10 Although the relationship of cyc and other stress response genes is not understood, these studies are beginning to uncover the mechanisms of sleep homeostasis.
To identify molecular components of sleep homeostasis, we performed a genome-wide analysis of changes in gene expression that are associated with sleep deprivation in flies. We found that a large proportion of genes that change expression with sleep deprivation are those involved in the immune response and in energy metabolism. Further analyses were performed to determine the function of immune genes in the homeostatic system. The results indicated that altering a major player in the immune response, Relish, affects sleep and that acute sleep deprivation increases resistance to infection.
METHODS
Microarray Sample Preparation
Flies were grown on standard agar, corn meal, and molasses medium. At 1–3 days of age, flies were transferred to bottles containing 2% agar, 5% sucrose and entrained for a minimum of 3 days in a 12:12 light: dark schedule at 25° C. At the appropriate time, flies were removed from the incubators, and manually stimulated for 4 hours. A control group was also removed but not stimulated, to control for environmental exposure. At the end of the 4-hour period, half of the stimulated flies were collected immediately on dry ice, as was half of the control group. The remaining flies were allowed to recover for 3 hours and collected on dry ice. All populations of flies described in the main text consisted of both males and females.
Flies were collected on dry ice at times described in the main text and heads separated in a liquid N2-chilled sieve. mRNA was isolated from the heads using the Poly-A pure microRNA isolation kit (Ambion). cRNA was synthesized according to the Affymetrix protocol. Briefly, 2 μg of purified mRNA of each sample was used for double stranded cDNA synthesis. An HPLC purified oligo-(dT)24 primer containing a T7 RNA polymerase binding site sequence (Genset) was used for priming the first-strand reaction. Upon completion of the second strand reaction, samples were cleaned by a standard phenol/chloroform extraction followed by ethanol precipitation. In vitro transcription for cRNA synthesis was performed using the Enzo BioArray High Yield RNA transcript labeling Kit, and the products were subsequently cleaned up using RNeasy columns (Qiagen). Each sample was fragmented, and then hybridized to Affymetrix Drosophila microarrays. Microarray samples were processed either at the Massachusetts Institute of Technology or at the University of Pennsylvania core facilities. Data from y w flies represent 3 independent biological replicates. Data from rosy and cyc01 flies represent 2 biological replicates.
Microarray Data Analysis
Raw expression values from the MicroArray suite v 5.0 were background corrected and normalized using Robust Multiarray Analysis (RMA) software.11 For experiments in which comparisons were made with handled control (HC), expression values were normalized using the perfect match approach in dCHIP.12,13 Normalized expression values were filtered by discarding genes containing values scored as “absent” on >2 microarray chips across all arrays in the experiment. For comparing DEP to REB, this narrowed the analysis to 4,788 genes in y w, and to 4,791 genes in rosy. For comparing HC to DEP in rosy, 6051 genes were selected, and for comparing rosy and cyc01 baseline, 6455 genes fulfilled these criteria. Normalized expression values were tested for statistical significance using Significance Analysis of Microarray (SAM) software (http://www-stat.stanford.edu/~tibs/SAM/), where values are subjected to random permutations to determine false discovery rates and to nonparametric t-tests.14 Delta was adjusted to achieve a fairly high number of significant genes with a false discovery rate of ≤ 30%.
Genes were grouped into functional categories using updated gene ontology information from both the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://apps1.niaid.nih.gov/DAVID/) and Flybase.15 Although multiple functions can be attributed to single genes, we grouped them into single categories according to their primary function.
Validation of Microarray Data
Microarray data were validated by RNase protection assay. Samples were prepared by collecting tissue at appropriate time points as described above, and total RNA was extracted from heads using the Biotecx reagent following the manufacturer’s recommended procedure. RPAs were performed using the RPA III kit (Ambion).32 P-labeled antisense riboprobes were synthesized and hybridized at 42°C in a solution containing 10 μg total RNA template per reaction. RNase-digested samples were run on a 5% poly-acrylamide gel for subsequent quantitation using a phosphorimager (Molecular Dynamics). A tubulin probe16 was used as the loading control. Primers used for amplifying selected transcripts for making antisense probes were designed as follows: Relish, forward 5′ CCTGAAAAACCCGTGAGTCATC 3′, reverse 5′ AACGCCGAAACTAACGCCAG 3′; Cactus, forward 5′ AAGCACGAAAATGCCGAGCC 3′, reverse 5′ GCTGTGGAGGATTGAACCTTGC 3′; Drosocin, forward 5′ CGATTTGTCCACCACTCCAAG 3′, reverse 5′ GCTGTCTTTCGTGTGTTTATTGC 3′.
Primer sets were used to amplify cDNA synthesized from fly heads. Products were verified by sequencing and cloned into a TOPO vector (Invitrogen).
Quantitative PCR and Genotyping
To determine relative mRNA expression of each of the NFκB genes, total RNA was extracted from flies collected at ZT 8 as described above (ZT = Zeitgeber time). PCR reactions on cDNA samples were performed on the Mx4000 Multiplex system using incorporation of SYBR green dye (Stratagene). Primers used to amplify NFκB genes were designed as follows: Dorsal, forward 5′ GGATACGCCATATCGTCCTCAT 3′, reverse: 5′ CAGTGTACAGACGCCCTTCTT G 3′; and Dif forward: 5′ CGTTCGGTACTACCCGAATCC 3′, reverse: 5′ CTTTAGGCTTTCAACTGTTTTTTGG 3′. Actin was used as the control, forward 5′ GCGCGGTTACTCTTTCACCA 3′, reverse 5′ ATGTCACGGACGATTTCACG 3′.
To ensure that flies were carrying the E20 mutation, standard PCR was performed on genomic DNA extracted from candidate E20/E20 lines and a wild type control. Briefly 1–2 flies were mashed with a pipette tip containing 50 μL of buffer comprised of 10 mM Tris-Cl pH 8.2, 1 mM EDTA, 25 mM NaCl, and 200 μg/mL Proteinase K. Remaining buffer was then expelled, and the solution incubated at 37° C for 30 minutes. The sample was then heated to 95° C for 10 minutes, and left at room temperature overnight. Relish primers used for the PCR reaction were the same as those used for RPAs (see above), which targets a 500 bp product in the 5′ UTR. No gene product is detected in E20/E20 mutants, as this is the site of the deletion.17 To ensure the integrity of the genomic DNA in the E20/E20 mutants, we tested for the presence of a positive control by performing a second reaction using primers against NAD-dependent methylenetetrahydrofolate dehydrogenase- methenyltetrahydrofolate cyclohydrolase (Nmdmc), forward: 5′ GGACAAGGATGTGGATGGCTTC 3′, reverse: 5′ ATACCTGTGGCAGATGGTCACC 3′.
Behavior and Infection
Locomotor activity was measured in adult flies using the Trikinetics Drosophila Activity Monitoring System (Waltham, MA), model 2 (DAM2). At 1–3 days of age, flies were loaded individually into 5 × 65 mm glass tubes using CO2 anesthesia. Sleep is defined as a minimum of 5 consecutive minutes of inactivity.18 Sleep deprivation was performed by attaching monitors to a multi-tube vortexer (Corning 4010). The vortexer was attached to a computer-controlled power source (Trikinetics) that was programmed to stimulate for 2-s pulses at random intervals lasting 6–12 s. Usually, 100% of the flies survived sleep deprivation for up to 10 h with 80–100% loss of sleep. Analysis of activity and resting behavior was performed using Matlab (Mathworks, Inc.) based custom software (“Insomniac”, generous gift of Dr. Lesley Ashmore, University of Pennsylvania). Average day and nighttime sleep and other measures reported for each group represents averages within individual flies across 3 days in the behavioral assay (days 2–4). In cases where comparisons were made between three or more groups, one-way ANOVA (http://statpages.org/) was performed followed by Scheffe post hoc analysis using online software available to the public from the Department of Obstetrics and Gynecology, The Chinese University of Hong Kong (http://department.obg.cuhk.edu.hk/researchsupport/statmenu.asp).
For bacterial infection, ampicillin-resistant E. coli were grown to saturating concentrations (OD600 = 0.5) in LB-ampicillin medium. The broth was diluted 1:10 in PBS and food coloring. Groups of flies were subjected to sleep deprivation by manual stimulation at appropriate times of day as indicated in the main text. CS and rosy flies were sleep deprived from ZT 14–18. Sleep deprived (4h) and handled control flies were CO2 anesthetized and injected using small glass pipettes (tip diameter ~50 μm) connected to a large syringe for applying positive pressure. Food coloring was used as an indicator of the approximate volume per inoculation per fly. All injections were in the dorsal thoracic region. Food coloring was metabolized within several hours in individual flies. Injury, if any, related to the injection was limited to melanization in the area of the injection site. Flies which died or which showed signs of remaining food coloring after 24 h were discarded. Generally, most flies (>90%) survived the inoculation and were used for the following steps described below.
Twenty-four hours following the injection, groups of 10 flies were homogenized in 400 μL LB-ampicillin medium and spread onto LB/amp/agar plates in dilutions of 1:10, 1:100, and 1:1000. Plates were incubated at 37° C overnight. The number of colony forming units were determined for each plate, averaged for each condition, and reported as cfu/fly. A similar method has been described previously.19,20
RESULTS
Expression of Immune Response Genes Is Upregulated During Sleep Deprivation
Although the activity of a population of flies can synchronize to light:dark cycles, absolute homogeneity among individuals of long consolidated periods of spontaneous rest or activity is difficult to achieve. Our goal was to compare gene expression in a group of flies that had been active for >3 hours with one that had been sleeping for up to 3 hours. We decided to compare gene expression in flies that were forced awake, or sleep deprived, with those who were allowed to sleep for 3 hours following sleep deprivation, thus undergoing what is known as rebound sleep.
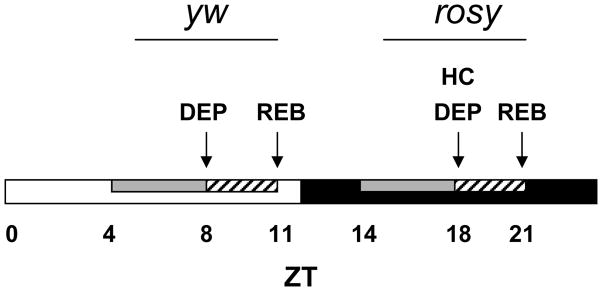
The stock of yellow white (y w) flies, a commonly used background strain, used for these experiments exhibited maximum consolidated rest during the daylight hours; sleep deprivation during the daytime was most effective at producing a rebound (Figure S1—which can be accessed on the web at www.journalsleep.org). We consistently observed that when flies were sleep deprived from ZT 4–8, most flies required a minimum of three hours to recover to normal activity levels. Samples for oligonucleotide microarray analysis were therefore collected immediately following a 4-h deprivation at ZT 8, and following a 3-h rebound period, at ZT 11 (Figure 1). In order to identify sleep specific genes common to different fly strains, we also analyzed gene expression in another commonly used laboratory fly strain carrying the rosy marker. These flies are mostly active during the day,9 and so sleep deprivation was performed during the nighttime, from ZT 14–18. Samples of rosy flies were collected at ZT 18 for the deprived group (DEP) and at ZT 21 for the rebound group (REB). An additional group was collected at ZT 18. These flies were exposed to the same environmental conditions as the DEP group during handling, but were not mechanically stimulated. This group is the handled control (HC, Figure 1).
Figure 1.
Timeline indicating experimental design of the microarrays. Flies were maintained in a light:dark cycle (ZT 0–12, open bar, = lights on; ZT 12–24, black bar, = lights off). y w or rosy flies were sleep deprived for 4 hr (gray bars) and allowed to recover for 3 hr (hatched bar). Collections were performed at the times designated by arrows. DEP = deprived, HC = handled control, REB = rebound.
Changes in gene expression were compared in both y w and rosy flies between DEP and REB groups. A summary of the results is reported in Table 1. A total of 272 and 217 genes changed expression between DEP and REB in y w and rosy, respectively. Of these, 247 genes in y w and 199 genes in rosy showed increased expression in DEP, and only 25 genes in y w and 18 in rosy increased expression in REB. A complete description of these genes along with statistical results is listed in Table S1 (which can be accessed on the web at www.journalsleep.org). The genes that increased during DEP fall into one of several general functional categories that include metabolism, redox, immune response, signaling molecules, DNA/RNA binding, cytoskeletal/structure, transporters, chaperones, and development. Precise overlap of genes was 20%; 43 genes were common between y w and rosy. Several factors may account for this observation. These 43 common genes may represent the core genes involved in sleep homeostasis. Alternatively, the difference in expression profile may be due to the genetic variability between the 2 fly strains or the different circadian times at which the experiments were performed. However, the distribution of genes into functional categories between the 2 genotypes is similar. Of the common genes, the most noticeable were the immune response genes, in particular Relish, a key player in the immune response (Table S1). As described below, flies deficient in Relish expression exhibited altered levels of sleep, which supports the notion that immune genes are components of the sleep homeostatic system.
Table 1.
Classification of Genes Affected by Sleep Deprivation
| Category | Increase with Deprivation |
Decrease with Deprivation |
Rosy vs cyc01 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vs HC (Ry) | vs REB (Ry) | Vs REB (y w) | vs HC (Ry) | vs REB (Ry) | vs REB (y w) | Increase in cyc01 | Increasein Ry | |||||||||
| % | N | % | n | % | n | % | n | % | n | % | n | % | n | % | n | |
| Metabolism | 37.36 | 34 | 23.62 | 30 | 26.01 | 45 | 50.00 | 24 | 45.45 | 5 | 35.29 | 6 | 27.38 | 89 | 39.13 | 18 |
| Immune | 28.57 | 26 | 10.24 | 13 | 2.89 | 5 | 4.17 | 2 | 18.18 | 2 | 35.29 | 6 | 6.46 | 21 | 0.00 | 0 |
| Redox | 7.69 | 7 | 7.09 | 9 | 19.08 | 33 | 14.58 | 7 | 27.27 | 3 | 11.76 | 2 | 7.69 | 25 | 8.70 | 4 |
| Signaling Molecules | 6.59 | 6 | 23.62 | 30 | 21.97 | 38 | 20.83 | 10 | 9.09 | 1 | 17.65 | 3 | 22.46 | 73 | 17.39 | 8 |
| DNA/RNA Binding | 4.40 | 4 | 12.60 | 16 | 5.78 | 10 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 10.77 | 35 | 17.39 | 8 |
| Cytoskeletal | 6.59 | 6 | 5.51 | 7 | 6.36 | 11 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 9.85 | 32 | 0.00 | 0 |
| Transporter | 2.20 | 2 | 11.02 | 14 | 11.56 | 20 | 8.33 | 4 | 0.00 | 0 | 0.00 | 0 | 7.38 | 24 | 6.52 | 3 |
| Development | 3.30 | 3 | 4.72 | 6 | 1.73 | 3 | 2.08 | 1 | 0.00 | 0 | 0.00 | 0 | 4.92 | 16 | 8.70 | 4 |
| Chaperone | 3.30 | 3 | 1.57 | 2 | 4.62 | 8 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 3.08 | 10 | 2.17 | 1 |
| Subtotal | 91 | 127 | 173 | 48 | 11 | 17 | 325 | 46 | ||||||||
| Unknown | 36.81 | 53 | 36.18 | 72 | 29.96 | 74 | 27.27 | 18 | 38.89 | 7 | 32.00 | 8 | 39.25 | 210 | 47.13 | 46 |
| Total | 144 | 199 | 247 | 66 | 18 | 25 | 535 | 87 | ||||||||
Genes that change with deprivation are grouped into categories as indicated in the table. Genes that “Increase with Deprivation” or “Decrease with Deprivation” are further grouped by comparison to handled control (vs HC) or rebound (vs Reb) in either rosy (Ry) or yellow white (y w) flies. The last 2 columns are genes that are affected by the presence of the cyc01 mutation in the rosy background. Genes are distributed into each category by percent (%) of genes of known function (subtotal). n columns denote the number of genes.
Since DEP and REB are 3 hours apart, circadian differences between the 2 time points could underlie any perceived sleep related changes. Furthermore, detection of some DEP-induced genes may be missed if they are not fully recovered in a 3-h rebound, despite the recovery in behavior. The circadian artifact was eliminated, and all possible deprivation-induced changes were identified, by comparing HC and DEP. In rosy 145 genes were elevated in DEP as compared to HC, and 66 genes were increased in HC. A complete description of statistical results is listed in Table S2 (which can be accessed on the web at www.journalsleep.org). Genes were grouped into the same functional categories as described above. A large proportion of genes, approximately 2/3, upregulated in DEP were devoted to metabolism or immunity. This supported the results of the comparison between DEP and REB which indicated that the expression of immune genes increases in response to sleep deprivation.
Expression of Immune Response genes is Upregulated during Baseline Conditions in the cyc01 Sleep Mutant
The circadian mutant, cyc01, shows abnormal sleep behavior, such that flies have significantly reduced levels of baseline daily sleep.9 Additionally, there is a sexually dimorphic phenotype in response to sleep deprivation.9,10 Males tend to have little or no rebound while females show an exaggerated rebound following a deprivation period. The mechanism of the effect of cyc01 on sleep is not well understood. Because baseline sleep is similarly affected in both male and female cyc01 mutants,9 we therefore performed an initial comparison of HC flies at ZT18 to determine whether there was a difference in baseline gene expression between cyc01 and its background strain, rosy.
622 genes showed a greater than 2-fold change in expression; 87 of these were elevated in rosy and 535 were elevated in cyc01 (Table S3—which can be accessed on the web at www.journalsleep.org). The distribution of genes that change expression in cyc01 is listed in Table 1. Of the genes that are increased in cyc01, we found 40 that overlapped with those that increase expression with sleep deprivation in rosy (Table S2). Immune related genes, which included Relish, accounted for 26% of these 40 genes, which is the largest proportion of genes of known function that overlap. These common genes are highlighted in Supplementary Tables S2 and S3. These data suggest that cyc01 flies exist in a chronically sleep deprived state. The notion that cyc01 flies are chronically sleep deprived is also supported by the observation that they require more than twice the stimulation required by rosy to disrupt sleep.9 We have consistently observed, during manual stimulation of wild type flies, that with increasing deprivation periods, more stimulation is required to prevent sleep.
Upregulation of Immune Genes in Response to Sleep Deprivation
Genes involved in the immune response comprised nearly one-third of the genes of known function whose expression was increased by sleep deprivation as compared to handled control. The Nuclear Factor kappa B (NFκB) transcription factor Relish and the I kappa B (IκB) repressor cactus were significantly affected in both strains tested. We also found that all of the NFκB genes in Drosophila, Dorsal, Dorsal-related immunity factor (Dif), and Relish, were significantly increased in the cyc01 mutants as compared to rosy. Other genes that changed expression included components of both the Toll and Immune deficiency (Imd) signaling pathways, such as necrotic, pelle, kenny, several peptidoglycan recognition proteins, and antimicrobial peptides. When compared to rebound, expression of more genes was upregulated in the deprived condition in rosy than in y w flies. However, in y w, several antimicrobial peptide genes were elevated during rebound as compared to DEP. One possibility is that the kinetics of the response differ between the two genotypes, where most genes are in a recovery phase during rebound in rosy, while in y w, the antimicrobial peptides are likely induced with deprivation and continue to rise during rebound (see below).
To confirm the upregulation of immune gene expression during sleep deprivation, we performed RNase protection assays (RPAs) on selected transcripts. In these experiments we also sought to determine the extent to which expression of these genes was due to loss of sleep rather than to stress or injury from the mechanical stimulation alone. Thus, we performed an additional control and collected RNA from y w flies that were mechanically stimulated at a time of day when they are most active. This time period is from the dark to light transition, ZT 22- ZT 2. During this time, flies gradually become more active as they anticipate the change to light. Our previous work established that stimulation of flies during times when they are most active does not produce a significant sleep rebound.4 Thus the same protocol that was used for the microarray study was used for RPA analysis of transcripts, except flies were deprived at 2 different times and collected after DEP and REB along with handled controls. The “active” groups consisted of deprived flies collected immediately following a 4- hour deprivation at ZT 2, and recovered flies collected at ZT 5. The “resting” groups consisted of those collected, following a deprivation at ZT 8, and following a rebound at ZT 11.
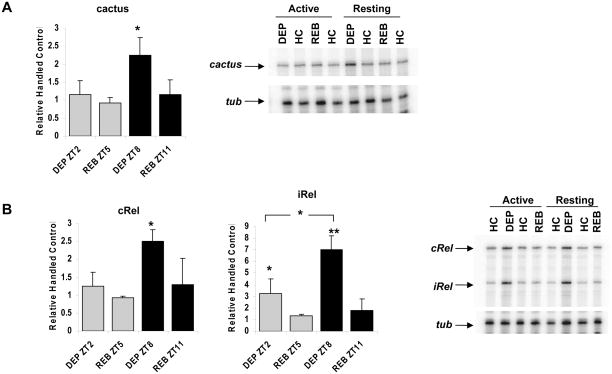
The results with 3 different transcripts are reported in Figure 2. All of these confirmed the results of the microarray analysis with variable degrees of specificity for dependence on sleep deprivation. Cactus, the Drosophila homolog of IκB, showed the strongest specificity for changes in expression associated with loss of sleep. No significant change in expression occurred with mechanical stimulation in active flies, but a 2-fold increase versus handled control was observed in flies stimulated during the resting period (P <0.05). This increase returned to baseline levels during the rebound period (Figure 2A).
Figure 2.
Up-regulation of immune response genes during sleep deprivation. RNase protection assays were performed to measure relative mRNA abundance of indicated genes relative to a control, tubulin (tub) under different experimental conditions. Flies were sleep deprived from ZT 22-2 (during their activity period, gray) or from ZT 4–8 (during their resting period, black) and collected at indicated time points. A. Levels of cactus are plotted relative to handled control (HC; left panel). Levels in DEP ZT 8 are higher than in all other conditions and time points (* = P< 0.05); representative RPA is shown in the right panel. B. Levels of cRel (left panel; * = P<0.01) and iRel (middle panel; * = P <0.05 and ** = P<0.001) are plotted relative to HC. mRNA levels of iRel in DEP ZT2 and DEP ZT8 are higher as compared to all other HC and REB groups as indicated by asterisks. The difference in iRel mRNA levels between DEP ZT2 and DEP ZT8 is also significant as shown by the bracket (* = P<0.05). A representative RPA is shown in the right panel. All experiments were performed in y w flies; each chart represents 3 independent experiments.
As mentioned above, Relish (Rel) is a Drosophila homolog of the mammalian transcription factor NFκB. NFκB was previously implicated in sleep, such that its protein activity increases with deprivation.21,22 In flies, Relish is an important component of an immune response and is required for expression of many antimicrobial peptides.23 Two isoforms of the Rel gene17 were detected by the RPA probe (Figure 2B). Inducible Relish (i-Rel) mRNA expression increased significantly in stimulated “active” and “resting” flies (ANOVA P<0.0001). While i-Rel expression was affected by the mechanical stimulation at ZT2, the magnitude of the effect was significantly larger in the resting group at ZT 8 (P <0.05). In contrast, constitutive Rel (c-Rel) showed no change with stimulation as compared to handled control in the “active” (ZT2) group, but showed a significant increase in the “resting” (ZT8) group (P <0.001). Although these experiments indicated a significant increase in baseline at ZT 2 as compared to the other handled control time points at ZT 5, 8, and 11 (P <0.05), more extensive circadian analysis of Relish expression did not reveal a significant oscillation in either of the Rel transcripts (data not shown). RPA analysis of Drosocin (Dro) mRNA, which encodes an antimicrobial peptide that requires activated Relish for its expression, 24 showed that mRNA expression is slightly higher in the HC condition at ZT 2 than in the same condition at ZT 8. Furthermore, Dro was induced during stimulation at both times, ZT 2 and ZT 8 with further increases in the following recovery period (n=2; data not shown).
Function of Immune Genes in Sleep Homeostasis
Because immune-related genes comprised a large proportion of those that were induced with sleep deprivation, we were interested in further defining a role of these genes in sleep homeostasis. The Drosophila innate immune response is mediated by 2 highly conserved pathways, Imd and Toll signaling. Relish is a central component of the Imd pathway, and another NFκB, Dif, and the IκB, cactus, are central components of the Toll pathway.25,26 We decided to focus our attention on Relish, because it was elevated with all manipulations that reduce sleep—with sleep deprivation in y w and rosy flies as well as in the cyc01 mutants. In addition, previous reports in mammals have suggested a role of NFκB in sleep.21,27,28
RelishE20 (E20) null mutants17 contain a second mutation, ebony, which is a commonly used third chromosome phenotypic marker that renders a dark cuticle. The ebony gene product is a beta-alanyl dopamine synthase, which is an enzyme involved in dopamine metabolism. Ebony mutants are known to be defective in circadian locomotor activity rhythms.29 Together with recent reports that dopamine has a role in Drosophila arousal mechanisms,30,31 these observations suggest that ebony may mask any sleep related behavioral effects of E20. Indeed, our initial observations in E20 flies showed no consistent change in overall sleep levels or in rebound responses to sleep deprivation periods of varying lengths (4 h, 6 h, and 10 h, data not shown). Ebony was therefore recombined away from the E20 mutation.
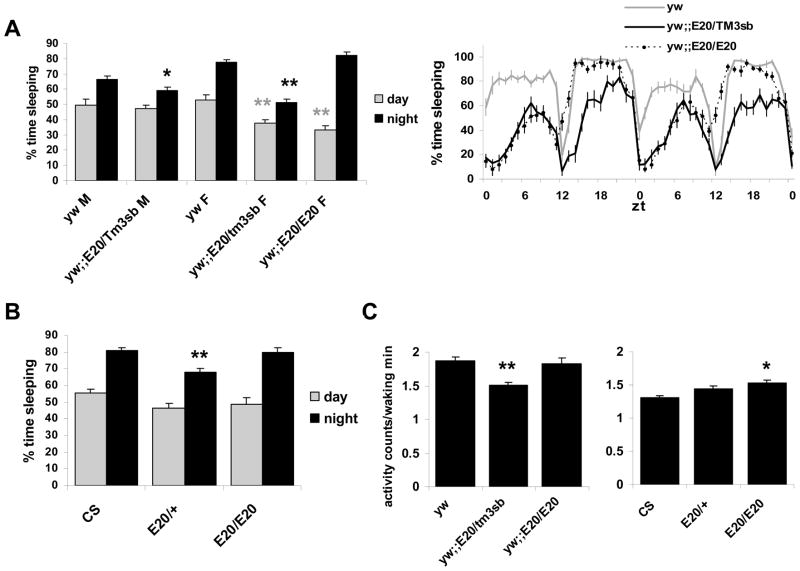
In order to have an appropriate background control for the E20 mutation, we crossed it into a y w strain, which we use routinely in the laboratory for sleep studies. This rendered the E20 mutation partially homozygous lethal in males. Surprisingly, we found that female y w;;E20/TM3sb flies slept significantly less than both y w and y w;;E20/E20 flies (Figure 3A). Heterozygous males also slept less than y w controls, but the effect was restricted to nighttime sleep, whereas in females, both day and nighttime sleep were reduced. Daytime sleep was also reduced in the female E20/E20 flies relative to the y w controls (P <0.01). Flies were also tested in a sleep deprivation assay and exhibited sleep rebound following a 10-h deprivation period that was comparable to wild type (data not shown). We therefore focused our analyses on the effect of Relish on baseline sleep.
Figure 3. E20 reduces sleep in flies.
A. Average percent time sleeping is reported for each genotype in males (M) and females (F). Heterozygous males (y w;;E20/TM3sb) sleep less than y w (* = P <0.05; n=51 y w and 56 y w;;E20/TM3sb, grouped from 3 independent experiments). Daytime sleep is significantly reduced in y w;; E20/TM3sb females (n=56) and in y w;;E20/E20 females (n=26) as compared to y w (n=51; ** = P <0.01). Nighttime sleep in E20 heterozygous females was also significantly lower than in y w and y w;; E20/E20 (P <0.01). % time sleeping per hour from a representative experiment in females is plotted in the right panel. B. Day and nighttime sleep in E20 females in a Canton-S (CS) background. Nighttime sleep is significantly reduced in E20/+ females (n=40) as compared to CS (n=42) and E20/E20 (n=26; p<0.01). C. Activity counts per waking minute are plotted for female genotypes indicated. y w;;E20/TM3sb heterozygotes have lower activity rates than their control strain, y w (P <0.01; left panel), whereas E20/E20 homozygotes have higher activity than their control strain, CS (P < 0.05; right panel). The stock of y w flies used here and in Figure 4 was diurnal like most other strains.
To ensure that the change in sleep was not attributable to a modifier in the genetic background, the E20 mutation was also crossed into a wild type Canton-S (CS) background without the use of a balancer chromosome, so as to allow recombination to occur. We confirmed the presence of the E20 mutation by PCR analysis and by measuring immune responses to bacterial infection (see methods). E20 is homozygous viable in both genders in the CS background. The CS E20 homozygotes were backcrossed into the CS stock, and behavior was examined in E20/E20 flies, F1 E20/+ heterozygotes and wild type. Analysis of day and nighttime sleep indicated that the E20 phenotype was dampened in the CS background (Figure 3B). In males, the phenotype seen in the y w background did not hold up in this new background, leading us to conclude that the E20 mutation does not produce a significant effect on sleep in males (Table 2). Females, on the other hand, showed no significant change in daytime sleep, but nighttime sleep was significantly reduced in E20/+ as compared to CS (Scheffe, P <0.01). Thus one copy of the E20 mutation reduces sleep in females with a consistent effect at night. Since females showed a consistent reduction in sleep, we focused our additional analyses in females.
Table 2.
Summary of effects of RelishE20 on sleep.
| Genotype | M/F | n | % time sleeping per 24 h |
|---|---|---|---|
| yellow white | F | 51 | 73.4 ± 1.7 |
| yw;;E20/TM3sb | F | 56 | 44.3 ± 2.1** |
| yw;;E20/E20 | F | 26 | 57.5 ± 2.4** |
| yellow white | M | 51 | 69.2 ± 1.7 |
| yw;;E20/TM3sb | M | 56 | 57.5 ± 1.8** |
| CS | F | 42 | 68.2 ± 1.7 |
| E20/+ | F | 40 | 56.9 ± 2.2** |
| E20/E20 | F | 30 | 64.2 ± 3.0 |
| CS | M | 42 | 63.2 ± 1.6 |
| E20/+ | M | 40 | 67.9 ± 1.3 |
| E20/E20 | M | 29 | 71.7 ± 1.5** |
| ElavC155-Gal4 | F | 37 | 57.6 ± 1.5 |
| ElavC155;;E20/+ | F | 47 | 49.4 ± 2.0* |
| ElavC155;UAS-Rel;E20/+ | F | 31 | 46.9 ± 1.9** |
| Yolk-Gal4 | F | 31 | 68.4 ± 1.8 |
| Yolk-Gal4/E20 | F | 37 | 50.7 ± 2.5‡ |
| UAS-Rel;Yolk/E20 | F | 42 | 62.7 ± 2.4 |
| UAS-iRel | F | 45 | 68.0 ± 1.4 |
| Elav;UAS-iRel | F | 41 | 55.8 ± 1.2‡‡ |
| UAS-iRel;yolk-Gal4 | F | 43 | 61.1 ± 1.3* |
= P <0.01;
= P <0.05 (Scheffe post hoc).
indicates significant difference from yolk-Gal4, P <0.01 and from UAS-Rel; Yolk/E20, P<0.05.
indicates significant difference only as compared to UAS-iRel (P <0.01), but not to parent Gal4 line, elavC155.
To determine whether E20 affected locomotor ability, we calculated the rate of activity per waking minute. Activity per waking minute was slightly but significantly reduced in y w;; E20/TM3sb (P <0.01). However, no significant change was detected between E20/+ and CS. Activity per waking minute was slightly increased in E20/E20 (Figure 3C, P <0.05). These data do not correlate with our observations on overall sleep in these flies. Therefore, the effects of E20 on sleep are likely not due to changes in locomotor ability.
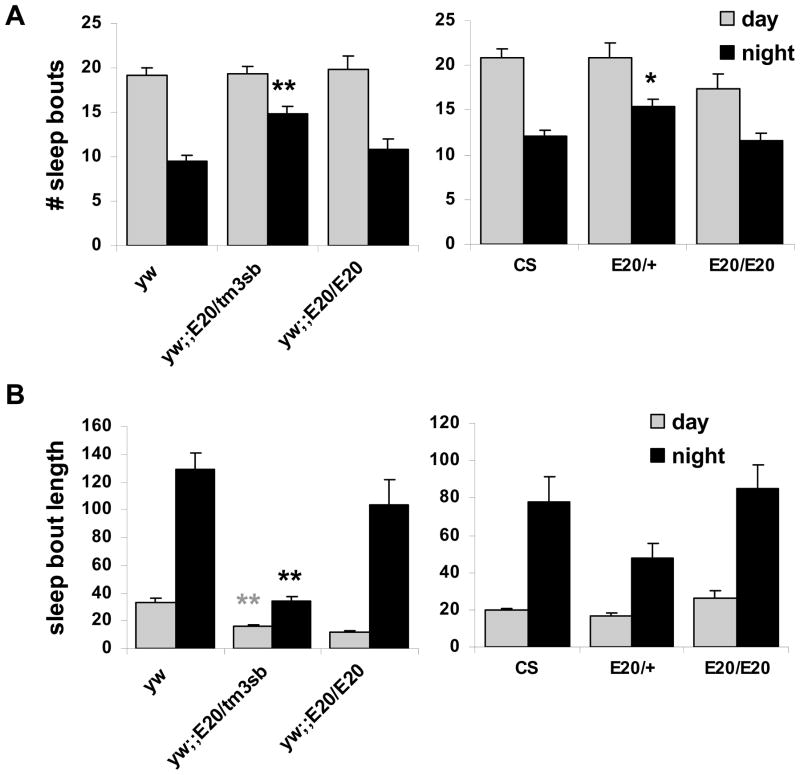
We also found that sleep was less consolidated in E20 heterozygotes. In y w;;E20/TM3sb, the number of nighttime sleep episodes (or bouts) was increased significantly (P <0.01) as compared to y w, and the mean bout duration was substantially reduced both during the day and at night (P <0.01, Figure 4). Similarly, the number of nighttime sleep bouts was increased in E20/+ in the CS background (P <0.05, Figure 4A). We found a significant effect of genotype on average nighttime bout duration in the CS background (P <0.05), but post hoc analyses revealed that this effect was not significant when comparing groups (Figure 4B). Taken together, despite the weakened effect in the CS background, these data indicate that nighttime sleep in the E20 heterozygotes is more fragmented.
Figure 4.
The E20 mutation causes sleep fragmentation. A. The number of sleep episodes (# bouts) significantly increases at night in E20 heterozygotes in both y w and CS backgrounds (P <0.05). B. Mean bout duration is decreased at night. This effect is significant in y w, but not in CS.
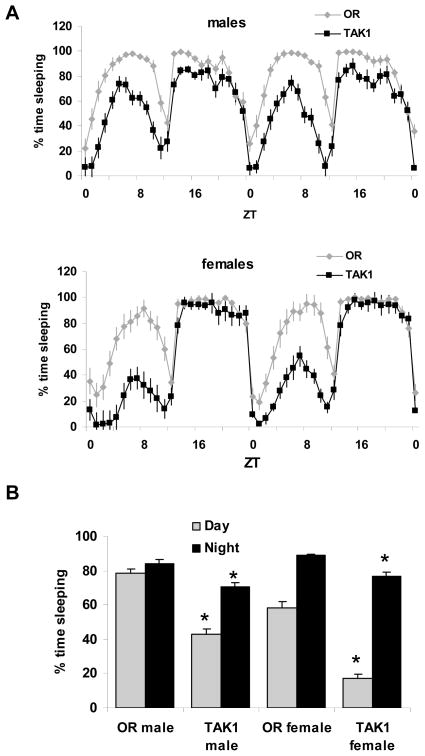
Why do E20 heterozygotes affect sleep and the E20/E20 mutants do not? Other heterozygous mutants have produced a stronger phenotype than their homozygous counterparts, such as I’m not dead yet (Indy),32 where heterozygous mutants produce a longevity phenotype but homozygotes do not. The most straightforward explanation is that a compensatory mechanism takes place when a gene such as Relish is absent, but not when its expression is reduced. To assess the possibility that other NFκB genes, Dorsal and Dif, compensate for the absence of Relish, we measured their mRNA expression levels using quantitative PCR and found no significant changes in either of the E20 homozyogous or heterozygous mutants (not shown). It is therefore unlikely that these other NFκBs account for the compensatory effects. A recent study demonstrated that Jun-N-terminal kinase (JNK) signaling acts cooperatively with the Imd pathway to affect antimicrobial peptide gene expression in the immune response.33 The JNK cascade is emerging as a major player in the immune response in both mammalian and Drosophila systems (reviewed in Stronach34). Because the observations of Delaney et al33 indicate an overlap between JNK and Relish dependent targets, we hypothesized that this pathway may account for the compensatory mechanism in E20/E20 mutants. We therefore tested flies deficient in dTAK1 (TGFβ activated kinase), which is necessary for JNK activation.33 Both male and female TAK1 mutants exhibit significantly reduced baseline sleep as compared to a control strain (Figure 5). Sleep in these flies was also less consolidated as indicated by an increase in the number of epochs and decrease in bout duration (not shown). Furthermore, activity rates were not affected. The sleep rebound response to a 10-h deprivation from ZT 14-0 was also unaffected (not shown), similar to the case in E20 mutants. Together, these data suggest a role of JNK in sleep and raise the possibility that this pathway compensates sleep in the absence of Relish.
Figure 5.
Sleep is reduced in TAK1 mutants. A. Percent time sleeping is plotted per hour from a representative experiment from male and female TAK1 mutants and a control strain, Oregon-R (OR). B. Both daytime and nighttime sleep are significantly reduced in TAK1 flies as compared to their corresponding control strains. n=32 TAK1 males, TAK1 females, n=25 OR males, 23 OR females. * = P<0.0005, student’s t-test.
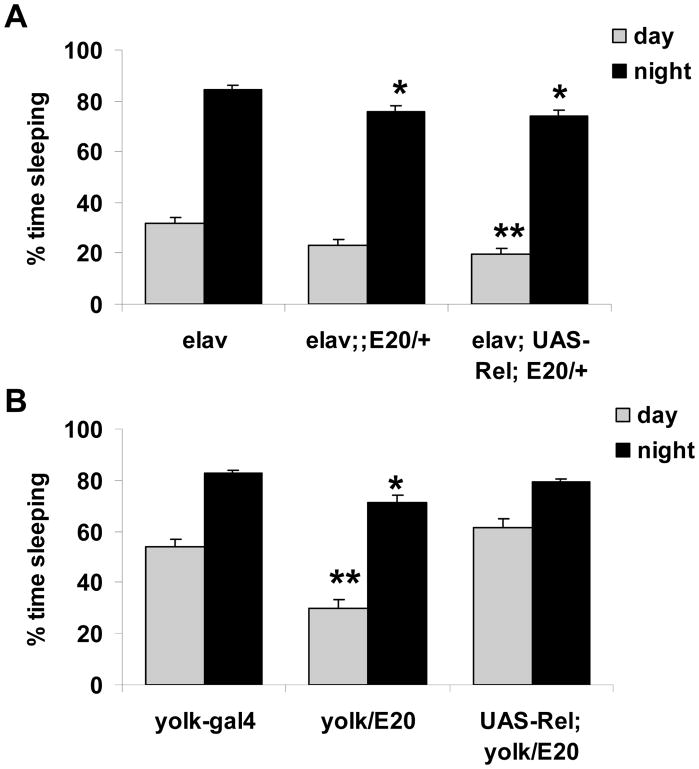
We next determined where Relish functions to affect sleep. UAS-iRel, which is an RNAi construct against Relish,35 was expressed in either neurons or fat body using elavC155-Gal4 or Yolk-Gal4 drivers, respectively. Both drivers produced weak but significant effects on sleep. The elavC155; UAS-iRel flies showed reduced sleep as compared to the UAS-iRel parent line, but not to the elavC155-Gal4 line. Thus the effect on sleep may be due to the effects of the background strain rather than reduced neuronal expression of Relish. On the other hand, when UAS-iRel was driven by Yolk-Gal4, which is known to drive strong expression specifically in adult female fat bodies,36 sleep was significantly reduced as compared to both parental lines (Table 2). Fat bodies are masses of adipose tissue located throughout the fly, including the head, and are a major site for the synthesis of Relish targets, the antimicrobial peptides.26 Since Drosophila contain fat bodies in the head as well as the abdomen, the fat bodies could have been a major source of Relish in the expression studies described above.
We also determined whether driving expression of UAS-Relish in E20/+ mutants could restore sleep behavior. Expression of UAS-Relish in neurons did not restore sleep in E20/+ mutants (Figure 6A and Table 2). However, UAS-Relish expression in fat body restored baseline sleep levels in E20/+ heterozygotes to those seen in control lines (Figure 6B and Table 2). Together, these data indicate that Relish is acting in the fat bodies to affect sleep.
Figure 6.
Expression of UAS-Relish in the fat body rescues the E20 mutant. A. Expression of UAS-Relish using elav-GAL4. Sleep is significantly reduced at night in both elav;;E20/+ (n=47) and elav; UAS-Rel; E20/+ flies (n=31) as compared to elav-GAL4 (n=37) and in daytime in elav; UAS-Rel; E20/+ as compared to elav-GAL4. B. Expression of UAS-Relish in fat body fully restores sleep in E20 heterozygotes (n=42). Sleep is significantly reduced in yolk-GAL4/E20 flies (n=37) as compared to yolk-GAL4 flies (n=31) and UAS-Rel; yolk/E20 (n=42). ** = P <0.01; * = P <0.05.
Effect of Sleep Deprivation on the Immune Response
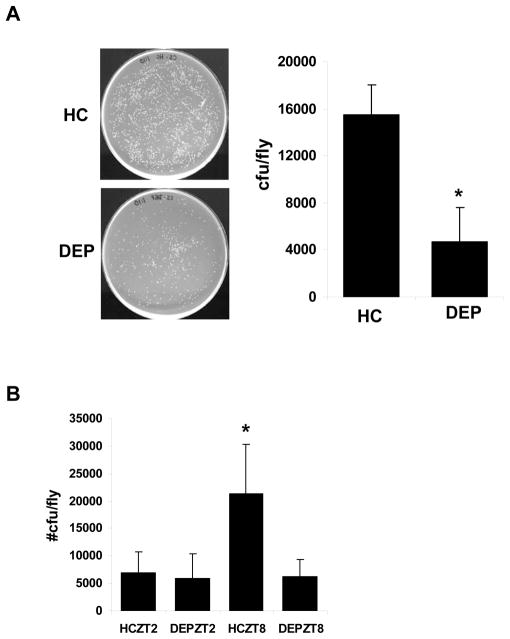
We have demonstrated an effect of sleep deprivation on immune- related genes and identified a sleep phenotype in flies mutant for one of these genes, Relish. We next assessed a possible function of immune-related gene expression in sleep deprivation. We quantified the immune response with and without sleep deprivation in several fly strains including CS (n = 3), y w (n=4), and rosy (n=2 independent experiments). Flies were subjected to 4- hour sleep deprivation by mechanical stimulation and inoculated with a solution containing ampicillin resistant E. coli. Handled control flies were also subjected to infection at approximately the same time as the deprived flies. Figure 7A shows the number of colony forming units (cfu) per fly in both the handled control and deprived conditions. Data from all nine independent experiments were averaged and plotted in Figure 7A. The number of cfu/fly decreased with deprivation by approximately 3 fold from 15500.89 ± 4241.78 to 4688.56 ± 1485.43 (P <0.02). These data indicate that short term sleep deprivation confers increased resistance to infection.
Figure 7.
Sleep deprivation increases resistance to infection. A. Left panel: Representative bacterial cultures obtained from Canton- S flies infected with E. coli immediately following a 4h deprivation (DEP; lower), or from handled controls (HC; upper). Plates are a 1:10 dilution of the homogenate. Right panel: the average number of colony forming units (cfu) per fly is plotted for flies from several strains including CS, y w, and rosy (see text; * = P <0.05; n=9) B. Effect of deprivation is limited to a time of day when flies are predicted to be resting. y w flies were sleep deprived and infected at the times indicated. Cfu/fly is significantly greater in HC as compared to DEP at ZT8 (* = P <0.05; n=4). No significant difference is observed between HC and DEP at ZT2, when flies are predicted to be active.
To determine whether the effect of sleep deprivation on the immune response was related to sleep loss as opposed to stress related to the mechanical stimulation, y w flies were stimulated at 2 different times of day as described above. When flies were stimulated at a time of day they were predicted to be active (ZT 22-2), no significant change in cfu/fly was observed from handled controls. In contrast, similar to the above findings, a significant decrease in cfu/fly was observed when flies were stimulated at a time of day they were predicted to be asleep (Figure 7B). ANOVA revealed a significant effect of handling on the immune response (P <0.001), and post hoc analysis (Fisher LSD) indicated a significant difference between the deprived group and handled control at ZT 8 (P <0.05). A difference between the handled control groups at ZT 2 and ZT 8 fell just short of statistical significance (P = 0.057), but it is formally possible that the immune response shows time-of-day variations. Taken together, these observations indicate that the increased resistance to infection is specific for sleep loss rather than due to stress related to the mechanical stimulation.
DISCUSSION
We have demonstrated an interaction between sleep and immunity in Drosophila. Genes involved in immune function constitute a major component of those affected by sleep deprivation. Many of these genes as well as others involved in immune function are also increased in the cyc01 mutant, which is known to decrease baseline sleep.9 We investigated a possible sleep-regulating role for one of the immune genes, Relish, and found that flies deficient in Relish expression have reduced sleep. This effect is sex dimorphic with more consistent effects in females, especially at night. Additionally, we found that heterozygous mutants display a stronger phenotype than homozygous E20 mutants. Although we do not have an explanation for this result, it is possible that redundancy in other immune related pathways, such as JNK, compensates behavior in homozygous mutants. This is supported by the observation that TAK1 mutants also exhibit reduced sleep. The Relish mutant phenotype is rescued by expression of a UAS-Relish transgene in fat body and is mimicked by RNAi knockdown of Relish in the fat body. Although we did not observe any changes in the ability of E20 mutants to recover from sleep deprivation, we demonstrated a function of immune-related genes in sleep deprivation. Flies subjected to short term sleep deprivation become more resistant to bacterial infection. Together, these data support a role of the NFκB, Relish, in sleep.
Our findings are supported by those in mammals. Several immune- related components are known to be associated with sleep loss in mammals.37 One example that is consistent with our data is the observation that the activity of the NFκB protein increases during sleep loss in mice21,22 and rats.22 Likewise, peptidoglycan recognition protein (PGRP) mRNA in the brain increases with sleep deprivation in rats.38 PGRP binds to peptidoglycan,39 a component of the bacterial cell wall, and is necessary for triggering specific immune responses.40,41 Four of the 12 known PGRPs in flies42 were elevated in DEP as compared to HC in our experiments. Genes involved in the immune response also reportedly increase expression with aging and with oxidative stress.43,44 Since both aging and oxidative stress have been linked to disturbances in sleep,45–47 it is possible that the effect on sleep underlies the changes in immune gene expression.
Upstream components of NFκB signaling, particularly cytokines, have effects on sleep. Exogenous application of IL1 or TNFα increases sleep in mammals, depending on the dose, time of day, and the site of injection (reviewed in Obal and Krueger3). In addition, loss of either the IL-1 Type I48 or the 55kDa TNF49 receptor in mice reduces baseline sleep during the light hours or during the dark hours, respectively. The mechanism underlying effects of these components on sleep remains unclear and is an important subject for future study. Although these data indicate a sleep promoting effect of NFκB signaling, NFκB-p50 knockout mice were reported to sleep more than control mice.50 Jhaveri et al50 surmised that this effect may result from a facilitation of expression of other NFκB genes in the absence of p50. Alternatively, since this study was carried out in males, it is possible that mammalian NFκB knockouts have a sex dimorphic effect. As mentioned above, Relish is a central component of the Imd signaling pathway, which is homologous to the TNFα pathway in mammals.25 That the TNF receptor knockout affects nighttime sleep49 is consistent with our observation that E20 also produces a stronger effect at night, and suggests conserved mechanisms between mammals and flies in sleep-immune interaction.
Other microarray studies on sleep-wake dependent gene expression also reported an increase in immune-related components during wakefulness in flies.51,52 Although none of the NFκB transcription factors were reported to increase with wakefulness in the study by Cirelli and Tononi,51 it is important to note that the duration of sleep deprivation was 8 h, whereas we analyzed changes in gene expression occurring after 4-h sleep deprivation. We speculate that Relish mRNA expression increases within the first few hours of sleep deprivation and then reverts to baseline or lower levels with prolonged periods of deprivation. Zimmerman et al52 also report increased Relish expression with sleep deprivation. However, they noted that Relish was induced with mechanical stimulation from ZT 10–14, which is a time of day that flies are predicted to be active. Our RPA data indicate that the inducible Relish transcript increased with mechanical stimulation in active flies from ZT 22-2, whereas the constitutive transcript did not. The RPA data suggest that a role of Relish in sleep may be specific to the constitutive transcript, and that results from microarray studies do not reflect this distinction.
It is particularly intriguing that expression of Relish in fat body rescues the E20 phenotype. We cannot necessarily rule out a role of neuronal Relish in controlling sleep, but one possible explanation for this result is the existence of an interaction between neurons and fat bodies. Neural interactions with non-neuronal cells have been well documented and have also been postulated to have an important role in sleep.53 A recent study also described a role of immune cells in learning.54 Our current finding has important implications for the contributions of non-neuronal cells in sleep, and therefore non-neuronal mechanisms of sleep should be considered in future studies.
The observation that flies show a decline in the infectivity produced by a bacterial challenge when they are sleep deprived is consistent with the gene expression profile, but may appear counterintuitive since one would expect immune resistance to be decreased by any stress, including sleep deprivation. We speculate that organisms occasionally have to be sleep deprived in order to survive and so may have evolved ways of coping with periods of acute sleep deprivation. Thus sleep deprivation is a type of stress that in small amounts may be tolerated, at least in part due to an increased resistance to infection. This is consistent with previous studies showing an augmentation of the immune response by acute stress or sleep deprivation.55,56
These data provide evidence for an interaction between sleep homeostasis and the immune system. It is widely accepted that a relationship between sleep and the immune system exists,37,57,58 but the nature of this interaction is not clear. Some studies have suggested that sleep deprivation weakens the immune response,59,60 but this is clearly a controversial issue.61,62 Bergman et al showed that tumor size was smaller in rats subjected to total sleep deprivation over several days as compared to controls.63 The authors concluded that tumor growth was impaired by sleep deprivation due to augmentation of the immune response. Additionally, Renegar et al demonstrated that short term sleep deprivation (6 h) in mice enhanced mucosal antiviral host defenses,56 a finding that is in agreement with our direct demonstration that 4-h sleep deprivation in flies enhances bacterial resistance.
CONCLUSION
Through a genome-wide screen for genes whose expression is associated with sleep and wake states in Drosophila, we have identified sleep-relevant genes. The expression of these genes is affected in the cyc01 sleep mutant and most likely underlies the behavioral phenotype of these flies. Of the genes that are upregulated during sleep deprivation, the inflammatory/immune response genes are of particular interest because they have previously been implicated in the regulation of sleep. To thoroughly investigate the nature of the interaction between the sleep and immune systems, we examined reciprocal effects of these systems. We find that a central immune component, Relish, affects sleep and that the homeostatic system has a profound effect on the immune response such that acute sleep deprivation increases resistance to infection.
Supplementary Material
Acknowledgments
The authors thank Dan Hultmark for providing RelE20 and UAS-Rel flies; Bruno Lemaitre for TAK1 mutants, and Herve Agaisse for UAS-iRel flies; Don Baldwin of the U. Penn. Microarray facility, the MIT microarray facility, Kevin Hellman and John Tobias for assistance and advice on statistical analyses of microarrays; Lesley Ashmore for computer software used for data analysis of sleep; Zhifeng Yue and Douglas Pike for technical assistance, JD Alvarez for sharing Actin primers, and members of the Sehgal Lab for helpful discussions. JCH was supported by NHLBI and NIA, AS is an Investigator in the HHMI, and JAW is supported in part by the UMDNJ Foundation.
Footnotes
Disclosure Statement
This was not an industry supported study. Dr. Sathyanarayanan is currently employed by Merck Inc. and is the Senior Research Biologist at the Department of Molecular Oncology in MRL Boston. Dr. Sehgal has participated in a speaking engagement for Merck Inc. Drs. Williams and Hendricks have indicated no financial conflicts of interest.
References
- 1.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 2.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–21. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 3.Obal F, Jr, Krueger JM. Biochemical regulation of non-rapid-eye-movement sleep. Front Biosci. 2003;8:d520–50. doi: 10.2741/1033. [DOI] [PubMed] [Google Scholar]
- 4.Hendricks JC, Finn SM, Panckeri KA, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 5.Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly drosophila melanogaster. Sleep. 2003;26:139–46. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- 6.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 7.Hendricks JC, Williams JA, Panckeri K, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 8.Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;90:1152–9. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 9.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 10.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flybase. The FlyBase database of the Drosophila genome projects and community literature. Nucl Acids Res. 2003;31:172–75. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–6. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 17.Hedengren M, Asling B, Dushay MS, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–37. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 18.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–39. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 19.Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Wu LP, Anderson KV. The antibacterial arm of the Drosophila innate immune response requires an I{kappa}B kinase. Genes Dev. 2001;15:104–10. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Gardi J, Kushikata T, Fang J, Krueger JM. Nuclear factor-kappaB- like activity increases in murine cerebral cortex after sleep deprivation. Am J Physiol. 1999;276:R1812–8. doi: 10.1152/ajpregu.1999.276.6.R1812. [DOI] [PubMed] [Google Scholar]
- 22.Basheer R, Rainnie DG, Porkka-Heiskanen T, Ramesh V, McCarley RW. Adenosine, prolonged wakefulness, and A1-activated NF-kappaB DNA binding in the basal forebrain of the rat. Neuroscience. 2001;104:731–9. doi: 10.1016/s0306-4522(01)00111-7. [DOI] [PubMed] [Google Scholar]
- 23.Khush RS, Leulier F, Lemaitre B. Drosophila immunity: two paths to NF-kappaB. Trends Immunol. 2001;22:260–4. doi: 10.1016/s1471-4906(01)01887-7. [DOI] [PubMed] [Google Scholar]
- 24.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–79. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–8. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 26.Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–83. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 27.Brandt JA, Churchill L, Rehman A, et al. Sleep deprivation increases the activation of nuclear factor kappa B in lateral hypothalamic cells. Brain Res. 2004;1004:91–7. doi: 10.1016/j.brainres.2003.11.079. [DOI] [PubMed] [Google Scholar]
- 28.Kubota T, Kushikata T, Fang J, Krueger JM. Nuclear factor-kappaB inhibitor peptide inhibits spontaneous and interleukin-1beta-induced sleep. Am J Physiol Regul Integr Comp Physiol. 2000;279:R404–13. doi: 10.1152/ajpregu.2000.279.2.R404. [DOI] [PubMed] [Google Scholar]
- 29.Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- 30.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–84. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–75. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Rogina B, Reenan RA, Nilsen SP, Helfand SL. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–40. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 33.Delaney JR, Stoven S, Uvell H, Anderson KV, Engstrom Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 2006 doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stronach B. Dissecting JNK signaling, one KKKinase at a time. Dev Dyn. 2005;232:575–84. doi: 10.1002/dvdy.20283. [DOI] [PubMed] [Google Scholar]
- 35.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5:441–50. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 36.Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-{kappa}B-dependent innate immune responses. Genes Dev. 2001;15:1900–12. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger JM, Majde JA, Obal F. Sleep in host defense. Brain Behav Immun. 2003;17 (Suppl 1):S41–7. doi: 10.1016/s0889-1591(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 38.Rehman A, Taishi P, Fang J, Majde JA, Krueger JM. The cloning of a rat peptidoglycan recognition protein (PGRP) and its induction in brain by sleep deprivation. Cytokine. 2001;13:8–17. doi: 10.1006/cyto.2000.0800. [DOI] [PubMed] [Google Scholar]
- 39.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. PNAS. 1998;95:10078–82. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–62. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 41.Michel T, Reichhart J-M, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–59. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 42.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. PNAS. 2000;97:13772–77. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landis GN, Abdueva D, Skvortsov D, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. PNAS. 2004;101:7663–68. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pletcher SD, Macdonald SJ, Marguerie R, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–23. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 45.Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles 10.1073/pnas.0605903103. PNAS. 2006;103:13843–47. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandi-Perumal SR, Seils LK, Kayumov L, et al. Senescence, sleep, and circadian rhythms. Ageing Res Rev. 2002;1:559–604. doi: 10.1016/s1568-1637(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 47.Ramanathan L, Gulyani S, Nienhuis R, Siegel JM. Sleep deprivation decreases superoxide dismutase activity in rat hippocampus and brainstem. Neuroreport. 2002;13:1387–90. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang J, Wang Y, Krueger JM. Effects of interleukin-1 beta on sleep are mediated by the type I receptor. Am J Physiol. 1998;274:R655–60. doi: 10.1152/ajpregu.1998.274.3.R655. [DOI] [PubMed] [Google Scholar]
- 49.Fang J, Wang Y, Krueger JM. Mice lacking the TNF 55 kDa receptor fail to sleep more after TNFalpha treatment. J Neurosci. 1997;17:5949–55. doi: 10.1523/JNEUROSCI.17-15-05949.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jhaveri KA, Ramkumar V, Trammell RA, Toth LA. Spontaneous, homeostatic, and inflammation-induced sleep in NF-{kappa}B p50 knockout mice. 10.1152/ajpregu.00262.2006. Am J Physiol Regul Integr Comp Physiol. 2006;291:R1516–26. doi: 10.1152/ajpregu.00262.2006. [DOI] [PubMed] [Google Scholar]
- 51.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–9. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman JE, Rizzo W, Shockley KR, et al. Multiple mechanisms limit the duration of wakefulness in drosophila brain. Physiol Genomics. 2006;27:337–50. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
- 53.Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45:347–60. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- 54.Ziv Y, Ron N, Butovsky O, et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9:268–75. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]
- 55.Dhabhar FS. Stress-induced augmentation of immune function-- The role of stress hormones, leukocyte trafficking, and cytokines. Brain Behav Immun. 2002;16:785–98. doi: 10.1016/s0889-1591(02)00036-3. [DOI] [PubMed] [Google Scholar]
- 56.Renegar KB, Crouse D, Floyd RA, Krueger J. Progression of influenza viral infection through the murine respiratory tract: the protective role of sleep deprivation. Sleep. 2000;23:859–63. [PubMed] [Google Scholar]
- 57.Krueger J, Walter J, Levin C. Factor S and related somnogens: an immune theory of for slow-wave sleep. In: McGinty DJ, Drucker-Colin R, Morrison A, Parmeggiani PL, editors. Brain mechanisms of sleep. New York: Raven Press; 1985. pp. 253–75. [Google Scholar]
- 58.Benca RM, Quintas J. Sleep and host defenses: a review. Sleep. 1997;20:1027–37. [PubMed] [Google Scholar]
- 59.Everson CA. Sustained sleep deprivation impairs host defense. Am J Physiol. 1993;265:R1148–54. doi: 10.1152/ajpregu.1993.265.5.R1148. [DOI] [PubMed] [Google Scholar]
- 60.Everson CA, Toth LA. Systemic bacterial invasion induced by sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2000;278:R905–16. doi: 10.1152/ajpregu.2000.278.4.R905. [DOI] [PubMed] [Google Scholar]
- 61.Benca RM, Kushida CA, Everson CA, Kalski R, Bergmann BM, Rechtschaffen A. Sleep deprivation in the rat: VII. Immune function. Sleep. 1989;12:47–52. doi: 10.1093/sleep/12.1.47. [DOI] [PubMed] [Google Scholar]
- 62.Rechtschaffen A, Bergmann BM. Sleep deprivation and host defense. Am J Physiol Regul Integr Comp Physiol. 2001;280:R602–3. doi: 10.1152/ajpregu.2001.280.2.R602. [DOI] [PubMed] [Google Scholar]
- 63.Bergmann BM, Rechtschaffen A, Gilliland MA, Quintans J. Effect of extended sleep deprivation on tumor growth in rats. Am J Physiol. 1996;271:R1460–4. doi: 10.1152/ajpregu.1996.271.5.R1460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.