Abstract
Characterization of glycosaminoglycans (GAGs), including chondroitin sulfate (CS), dermatan sulfate (DS) and heparan sulfate (HS), is important in developing an understanding of cellular function and in assuring quality of preparations destined for biomedical applications. While use of 1H and 13C NMR spectroscopy has become common in characterization of these materials, spectra are complex and difficult to interpret when a more heterogeneous GAG type or a mixture of several types is present. Herein a method based on 1H-15N two dimensional NMR experiments is described. The 15N- and 1H-chemical shifts of amide signals from 15N-containing acetylgalactosamines in CSs are shown to be quite sensitive to the sites of sulfation (4-, 6- or 4,6-), and easily distinguishable from those of DS. The amide signals from residual 15N-containing acetylglucosamines in HS are shown to be diagnostic of the presence of these GAG components as well. Most data were collected at natural abundance of 15N despite its low percentage. However enrichment of the 15N-content in GAGs using metabolic incorporation from 15N-glutamine added to cell culture media is also demonstrated, and used to distinguish metabolic states in different cell types.
Keywords: glycosaminoglycan, sulfation, metabolic labeling, heparin, heparan sulfate, chondroitin sulfate, dermatan sulfate
INTRODUCTION
Glycosaminoglycans (GAGs) are important components of the extracellular matrix (ECM).1–3 These glycans are commonly attached to a membrane protein core to form the cell surface proteoglycans found virtually in all mammalian cells.4 As a part of the ECM they influence numerous physiological processes, including organogenesis/growth control,5, 6 cell adhesion,7 angiogenesis,8 wound healing,9, 10 tumorigenesis,10, 11 morphogenesis,12, 13 inflammation,14–16 haemostasis,17, 18 and neural development/regeneration.19–22 GAGs also participate as receptors of various pathogens during the process of infection,23 and they have found applications in the treatment of diseases,24–26 for example, in the use of heparins for the treatment of acute coronary conditions.27–30 In many cases it is clear that biological function is dependent on specific structures (motifs) along the backbone of these complex polymeric carbohydrates.3, 31 Therefore, improvements in methods of characterization have become a high priority. The importance of this characterization was brought home in early 2008 when contamination of the medical supply of low-molecular-weight heparin, with what proved to be oversulfated chondroitin sulfate (OSCS),32–34 resulted in a worldwide withdrawal of the product. While characterization methods have improved, there is still a need for facile methods for rapid recognition of GAG types, particularly when mixtures of types are involved. These may occur in crude preparations or in isolates from cell surface proteoglycans where changes in sulfation, or distribution among GAG types with stages of cell development may be of interest. Here we describe methodology, based on the use of 1H-15N heteronuclear NMR methods, that we believe to be useful in the characterization of various GAG types. We also show that supplementation with a 15N-enriched metabolic precursor during cell growth can be used to monitor differences in GAG composition among various cell types and/or on changes in growth conditions.
GAGs are linear carbohydrate polymers built on alternating hexosamine and uronic acid units.3 For example, chondroitin sulfates (CSs), the most abundant GAGs in the human body,3 are composed of alternating β1-4-linked glucuronic acids (GlcA) and β1-3-linked N-acetylgalactosamines (GalNAc).31 Their major structural heterogeneity arises mostly from different sulfation patterns, which correlate with changes in biological actions.31, 35 The GalNAc residues can be mostly 4-sulfated (CS-A), almost entirely 6-sulfated (CS-C), and highly 4,6-di-sulfated (CS-E, or OSCS); sometimes sulfation at the 2-position of the GlcA units (CS-B or CS-D) can also occur.31
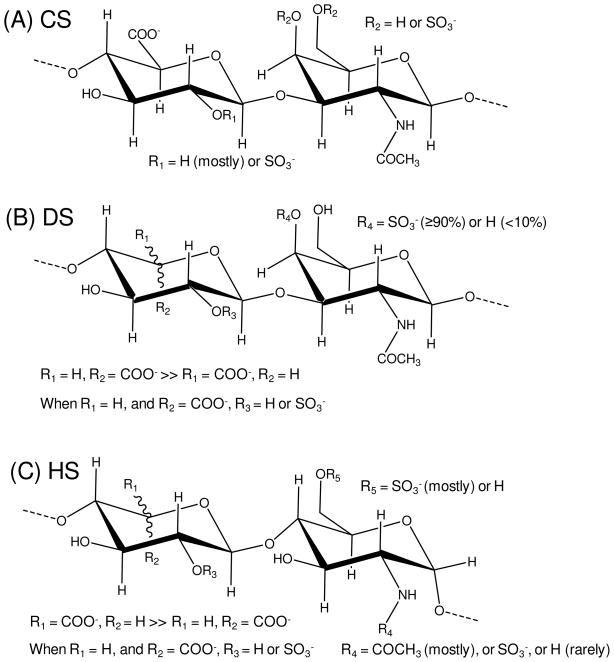
Restricting our attention to the most abundant GAGs of mammalian cell surface proteoglycans, two other species are of primary interest, dermatan sulfate (DS), and heparan sulfate (HS). Dermatan sulfate (DS), which also contains GalNAc as it’s amino sugar, is a close relative of CS, but the GlcA of CS is largely replaced by its 5′ epimer, iduronic acid (IdoA).31 Heparan sulfate (HS) is more complex, being characterized by mixtures of GlcA and IdoA (its uronic acid components) respectively β1-4 and α1-4 linked to an N-substituted glucosamine derivative (GlcNX). The GlcNX is α1-4 linked to the next uronic acid. Sulfation occurs at the nitrogen of the amino sugar in addition to various O-sulfonated sites (mostly at 6- and rarely at 3-positions) on the GlcNX residues. Sulfated regions tend to cluster in segments interspersed with stretches of non-sulfated N-acetylglucosamine residues.36 Representative structures of these GAGs are shown in Figure 1.
Figure 1.
Major repeating disaccharide units of the most abundant ECM GAGs found in mammalian cell surface proteoglycans. (A) The backbone of CS is homogeneously composed of [-4)-GlcA-β(1-3)-GalNAc-β(1-]n. (B) The backbone of DS (also known as CS-B) is mostly composed of [-4)-IdoA-α(1-3)-GalNAc-β(1-]n, but it also contains minor amounts of [-4)-GlcA-β(1-3)-GalNAc-β(1-]n. (C) The HS backbone is mostly composed of [-4)-GlcA-β(1-4)-GlcNAc-α(1-]n, but it also contains regions of [-4)-IdoA-α(1-4)-GlcNAc-α(1-]n.
Given that two of the three primary variations in ECM GAGs, are in the types of hexosamine (glucose or galactose based), and the different patterns of sulfation, mainly also on the hexosamines, it should not be surprising that a chemical group on these amino sugars would make a good probe of structural variations. 1H-15N Heteronuclear Single Quantum Coherence (HSQC) experiments are widely used as two dimensional (2D) NMR methods for structural characterization in proteins and carbohydrates. For example, in proteins, chemical shifts of both the nitrogen and the proton of the amide groups respond to the torsional angles of bonds connecting the amide group to the polypeptide chain, as well as substituents on the α-carbon bonded to the nitrogen.37, 38 Similar effects should exist for the amide 1H-15N pair in acetylated sugars, and influences from sites of glycosylation, anomeric configurations, neighboring residue types, and so forth, should also occur. In addition, charged groups, such as sulfates, that characterize HS, DS, and CS polymers, can have long-range effects on chemical shifts that might increase chemical shift sensitivity to sulfation patterns. A few recent reports on 15N-HSs and 15N-hyaluronic acids confirm sensitivity to some of these structural features,39–43 however, the latter studies have not explored sensitivity related to sulfation patterns.
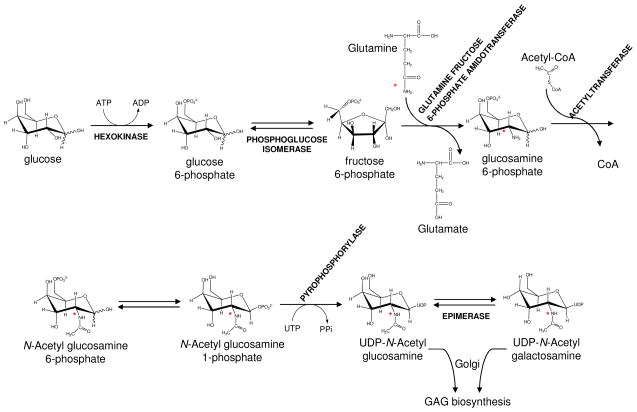
One of the primary limitations of GAG analysis by 15N-NMR spectroscopy is that of sensitivity. This can be improved through 15N isotopic enrichment of amide sites, something that has been accomplished for GAGs that can be synthesized beginning with bacterial products.40, 42 This is not an option for analysis of commercial products, but availability of adequate amounts of material is not usually a problem in these instances. For mammalian cell investigations, where large amounts of material are usually not available, there is however an alternative. Recently the incorporation of 15N from the side chain nitrogen of glutamine (Gln) has been demonstrated and used effectively in making Mass Spectroscopy (MS) analysis more quantitative.44 Basically the side chain nitrogen makes its way into hexosamines through the pathway illustrated in Figure 2. As will be further described, there are a few other destinations for these nitrogens, including bases of nucleic acids. However, widespread incorporation in protenacious materials seems relatively low. We will take advantage of metabolic labeling from Gln for 15N enrichment of cellular GAGs in the applications that follow.
Figure 2.
Schematic representation of the cytosolic biosynthesis of 15N-labeled N-acetyl hexosamines that will be further used inside the Golgi apparatus for building up the GAG backbones in proteoglycans. The names of the enzymes are fully written in upper cases while the substrates and products are mostly written in lower cases. The 15N-labeled sites are indicated with red asterisks. For more details about GAG biosynthesis see also Sugahara et al., 200331, or Esko and Lindahl, 200136.
Based on the above considerations we here undertake a systematic investigation of the effect of sulfation at different sites in one class of GAGs, namely CS polymers and oligomers, tracking positions of 1H-15N cross-peaks in HSQC spectra. Results show that the chemical shifts of 15N-resonances of amide protons of GalNAc residues in CS types are quite sensitive to different sulfation patterns, and that the positions of these resonances can be used to assess sulfate distributions in polymeric as well as oligomeric materials. The distinct positions of 1H-15N cross-peaks of other types of GAGs such as DS, and HS are also diagnostic, allowing identification of each type of these ECM GAGs in complex mixtures. Applications to the characterization of GAGs isolated from two types of cultured mammalian cells, mouse lung endothelium cells, and the Chinese hamster ovary (CHO) cells, are also presented. Sensitivity in these experiments was enhanced by taking advantage of the transfer of the side chain nitrogens of 15N-Gln to N-acetylhexosaminyl units (GalNAc and GlcNAc) of GAGs during their biosynthesis, resulting in 20–25 fold improvement in NMR detectability.
MATERIALS AND METHODS
Materials
The sodium salt of chondroitin sulfate A from bovine trachea (~65% of the residues have A type 4-O-sulfonation, and the balance have C type 6-O-sulfonation), the sodium salt of chondroitin sulfate C from shark cartilage (>95% of the residues have C type sulfation, and the balance have A type sulfation), the sodium salt of chondroitin sulfate B (dermatan sulfate) from porcine intestinal mucosa, hyaluronidase from sheep testes (type V), chondroitinase ABC lyase from Proteus vulgaris, N-acetyl-D-glucosamine (minimum 99%), N-acetyl-D-galactosamine (approx. 98%), and Sephadex G-15 resin (Fractionation Range of dextrans <1.5 kDa) were purchased from Sigma-Aldrich Co (St. Louis MO). A pre-packed Zorbax strong anion exchange (SAX) semi-preparative column (9.4 × 250 mm, 5 Micron) was purchased from Agilent Technologies (St. Louis MO). Bio-Gel P-10 Gel resin (Fractionation range of dextrans from ~1.5 to ~20 kDa) in fine polyacrylamide beads, and the polypropylene chromatographic columns (120 × 1.5 cm for size exclusion chromatography, and 1.0 × 50 cm for desalting) were purchased from Bio-Rad Life Science (Hercules, CA). 3 mm glass NMR tubes were purchased from VWR Scientific (Rochester, NY). Oversulfated chondroitin sulfate and mammalian heparan sulfate were provided by Dr. Roberto J.C. Fonseca, and Angélica M. Gomes, respectively, both from the Institute of Medical Biochemistry, Federal University of Rio de Janeiro, RJ, Brazil. L-Glutamine (AMIDE-15N, 98%+), and deuterium oxide “100%” (D 99,96%) were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). The endothelial cell line was derived from C57BL/6J mouse lung as reported previously.45 The CHO K1 cells were generously provided by Dr. Jeffrey D. Esko (University of California San Diego School of Medicine). The cell culture medium DMEM and L-glutamine-free DMEM, and related cell culture supplements including penicillin-streptomycin and non-essential amino acids were obtained from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA). ACS-grade 88% formic acid was purchased from J.T. Baker (Phillipsburg, NJ). HPLC-grade methanol was purchased from Fisher Scientific (Fair Lawn, NJ).
15N-Isotopic Labeling of Cellular GAGs
The mouse lung endothelial cells and CHO cells were cultured until confluent at established conditions as reported previously.45, 46 After initial washing with L-Gln-free DMEM (2 × 3 min), the confluent cells were further washed by incubating with L-Gln-free DMEM at 37 °C under 5% CO2 for 15 min. Following this the cells were cultured with L-Gln-free DMEM supplemented with 2 mM 15N-Gln at 37 °C under 5% CO2 for 24 hours. The culture media were collected and supernatants were pooled after centrifugation (2000 rpm for 5 min). The cell pellets and the monolayer cells in culture plates were lysed with 0.1 N NaOH. The cell lysates were adjusted to pH 7.0 with 1 N HAc and then pooled with the supernatant. The pooled solution was added with 0.166 volume protease solution [1mg/ml pronase in 0.24M NaAc (pH6.5) + 1.92M NaCl] and digested overnight at 40 °C. For GAG isolation, the digested solution was loaded to a DEAE-Sephadex column (3 ml bed volume, Pharmacia) that was pre-treated with equilibration buffer [20 mM NaAc (pH 6.0) + 0.25 M NaCl]. The column was initially washed with 30 ml of the equilibration buffer, the GAGs were then eluted from the column with 20 mM sodium acetate + 1M NaCl and collected. The GAGs (plus other high negatively charged non-GAG molecules) were desalted through a PD-10 column (Sephadex™ G-25 TM, GE Healthcare) following the manufacturer’s instructions.
Preparation of Unlabeled Samples and Cellular 15N-Labeled GAG Oligosaccharides
CS-A from bovine trachea (150 mg) was digested with 1 mg of chondroitinase ABC (0.33UI) in 5 mL digesting buffer (50 mM Tris-HCl, pH 8.0, 150 mM sodium acetate, 100 μg/mL of bovine serum albumin) at 37 °C for 150 min. The digested sample was heated (boiled) at 100 °C for 15 min to stop digestion. CS-C from shark cartilage (150 mg) was digested with 10 mg of hyaluronidase in 3 mL 50 mM sodium phosphate, 150 mM NaCl (pH 6.0) at 37 °C for 24 hours. The digested sample was heated (boiled) at 100 °C for 15 min. A 3 mL sample from each digestion (heterogeneous CS oligosaccharide mixtures) was subjected to size-exclusion chromatography on a Bio-Gel P-10 column (120 × 1.5 cm) using an elution solution of 10% ethanol, 1 M NaCl at a flow-rate of 1 mL/15 min. Peaks corresponding to fractionated oligosaccharides were pooled and desalted on a Sephadex G-15 column (1.0 × 50 cm) in distilled water. The respective masses of the oligosaccharides were determined by analysis on a ThermoFinnigan LCQ Advantage MS instrument. The lyase- or hydrolase-digested dimers and hexamers were selected for further separation and analysis. These samples were freeze-dried after desalting and ~5 mg of the powered material was dissolved in ~100 μL H2O with the pH previously adjusted to 5.0 with 0.1 M HCl. Then about ~ 50 μL of four different lyase- and hydrolase-treated samples (three CS hexamers from CS-A+ABC lyase, CS-A+hydrolase, or CS-C+hydrolase; and one CS dimers from CS-A+ABC lyase) were subjected to SAX-High Pressure Liquid Chromatography (HPLC) using a linear NaCl gradient from 0 to 2 M in H2O with pH previously adjusted to 5.0 with HCl, over a 50 min period at a flow rate of 3.0 mL/min. The separations were monitored by absorption at 214 nm and 232 nm for the hydrolase- and lyase-products respectively. The peaks were collected separately, desalted on a Sephadex G-15 column, lyophilized, and weighed. To provide samples having terminal sugars reduced to their corresponding galactitol forms (−ol), powdered samples from commercial sources were treated with an equivalent weight of NaBH4 in 1 mL water for 3 hours. The reactions were then stopped by adding a molar equivalent of acetic acid and incubating for 1 hour in an ice-bath, followed by desalting on a Sephadex G-15 column.
The respective structures of dimers and hexamers (ΔC4S, ΔC6S, ΔC444S-ol, C644S-ol, ΔC664S-ol, and C666S-ol) were determined by one and two dimensional NMR experiments (1D 1H, 1H/1H Double quantum filtered correlation spectroscopy (DQF-COSY), 1H/1H Total correlation spectroscopy (TOCSY), 1H/13C Heteronuclear Multiple Quantum Coherence (HMQC), and 1H/15N Heteronuclear Single Quantum Coherence (HSQC)). The structural nomenclature used above is exemplified with the ΔC4S, and C644S-ol. These respectively correspond to [ΔGlcA-β(1→3)GalNAc-4(SO3−)], and [GlcA-β(1→3)GalNAc-6(SO3−)-β(1→4)GlcA-β(1→3)GalNAc-4(SO3−)-β(1→4) GlcA-β(1→3)GalNAc-4(SO3−)-ol], where the symbol Δ indicates 4,5-unsaturation on the GlcA produced by ABC chondroitinase cleavage.
The GAG samples from endothelial and CHO cells were treated similarly. However, because of a suspicion that samples may carry contamination from released nucleic acids (another abundant negatively charged polymeric cellular component), half of the sample from the endothelial cellular preparation was treated with 200 μL of nucleases (1.32 mg/mL DNAse/RNAse) in Lysis buffer (75 mM tris, 300 mM NaCl, 200 mM EDTA, pH 7.7) plus 10 μL 1M MgSO4 and 10 μL 0.1 M phenylmethanesulphonylfluoride (PMSF) at room temperature for 19 hours. The other half was not treated. Both halves were dissolved in 200 μL of the ABC lyase buffer and treated with 0.33UI of the chondroitinase ABC at 37 °C for 19 hours. Both samples were then separately applied to a Sephadex G-15 column and the peaks eluted with 1% butanol in water were individually pooled, lyophilized and analyzed by 15N-gHSQC (digested material, Figure S2A) and TOCSY (for both undigested and digested materials, Figures S2B and S2C respectively). Curiously, the nuclease-untreated endothelial sample (pool of negatively charged molecules) was resistant to the chondroitinase ABC lyase. This lack of enzymatic activity is likely due to inhibition of the enzymes by nucleic acids which are competing with the natural substrate (Figure S2B vs. S2C). After some NMR analyses (Figure S3A-C), the nuclease/ABC-treated sample was applied to the SAX-HPLC, and the 4-sulfated dimers were separated from 6-sulfated dimers and from minor amounts (10%) of other oligosaccharides, mainly monomers and tetramers. The 4-sulfated chondroitin sulfate dimer was desalted on a Sephadex G-15 column and then analyzed by both NMR (Figure S3D) and MS (Figure 7).
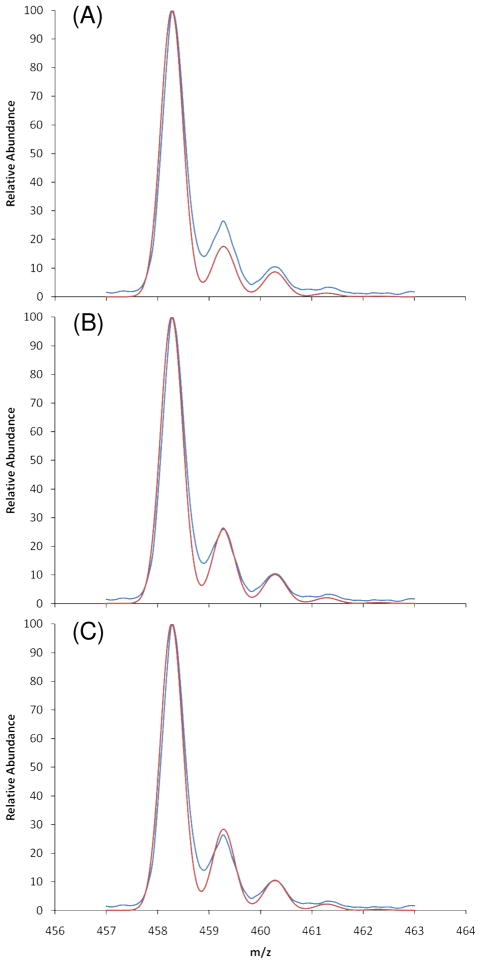
Figure 7.
MS spectra of the endothelial ΔC4S dimers. The blue peaks represent the measured spectra while the red peaks represent the simulated 15N-isotopic percentage in natural abundance of 0.37% (A), with (B) 8% of 15N incorporation (B), and with 10% 15N incorporation (C).
NMR Experiments
For 1D 1H, 1H/1H DQF-COSY, 1H/1H TOCSY, and 1H/13C gHSQC spectra, the samples were dissolved in 130 μL 100% D2O and transferred into 3 mm NMR tubes. For 1H/15N gHSQC spectra, the samples were dissolved in 130 μL 50 mM acetate buffer 12.5% D2O (pH 4.5) 0.1 % sodium azide and transferred into a 3 mm NMR tube.
All NMR experiments were recorded at 25 °C on a Varian Inova spectrometer (with a triple resonance cold probe) operating at 800 MHz for 1H, 200 MHz for 13C, and 80 MHz for 15N. The 1D 1H spectra were recorded with 128 scans with a spectral width of 7 kHz, carrier position at the HOD peak (4.773 ppm), acquisition time set to 2 sec, and water presaturation pulse (when used) set to the position of the carrier for a period equal to the recovery delay, 1.5 sec.
The DQF-COSY spectra were run with spectral widths of 6 kHz, and acquisition time of 0.175s using 32 scans per t1 increment (128 points) to achieve a time domain matrix of 2048 × 256 complex points (real and imaginary parts). The TOCSY spectra were run with spectral widths of 6 kHz, and acquisition time of 0.175s using 96 scans per t1 increment (64 points) to achieve a time domain matrix of 2110 × 128 complex points, using a spin-lock field of 9 kHz, and a mixing time of 60 ms. 1H-13C HMQC spectra were run with acquisition time of 0.128s using 144 scans per t1 increment (128 points) to achieve a time domain matrix of 1366 × 256 complex points. 1H-15N HSQC spectra were recorded with acquisition time of 0.095s and with192 scans per t1 increment (128 points) to achieve a time domain matrix of 1366 × 256 complex points. All processing was done with NMRPipe software.47 All spectra were apodized in both dimensions with the automatic function cosine-bells, together with automatic zero-filling to double the sizes followed by rounding to the nearest power of 2. Reported chemical shifts for 1H and 13C are relative to the trimethylsilylpropionic acid, and liquid ammonia for 15N.
MS analysis
Mass spectrometric analyses of GAG disaccharides derived from endothelial cells was performed by nanospray ionization in the negative ion mode, and analyzed with an LTQ-FT hybrid mass spectrometer (Thermo Scientific, Waltham, MA) operating in the linear ion trap only mode. One volume of GAG sample in 50 mM sodium acetate buffer was diluted with two volumes of methanol, to a final concentration estimated to be ~10 μM. The sample was infused through a hand-pulled silica capillary at a flow rate of ~0.6 μL/min, with an electrospray voltage of 2.6 kV. Spectra were accumulated for one minute and averaged to increase the signal-to-noise ratio. Isotopic distribution modeling was performed using IsoPro 3.0.
RESULTS
15 N-NMR analysis of galactosaminoglycans
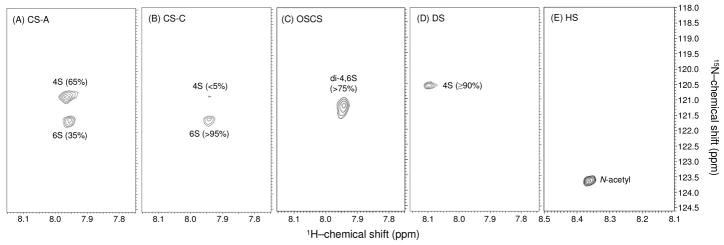
In order to establish the viability of 1H-15N HSQC spectra as a tool for recognition and characterization of GAGs, in particular their sulfation patterns or the linked uronic acid types, a series of GAG polymers were examined (Figure 3). These are available in substantial quantities and observation through HSQC spectra at natural abundance of 15N (0.3%) is possible. First, 15N-gHSQC spectra were run on the high-molecular weight CS standards at approximately 15 mg/ml, including CS-A (Figure 3A) which is primarily 4-sulfated (65%) and less 6-sulfation (35%), CS-C (Figure 3B) which is nearly completely 6-sulfated, and OSCS (Figure 3C) which is more than 75% sulfated at both 4- and 6-positions. Comparison of CS-A and CS-C spectra (Figure 3A vs 3B) shows that the 15N-chemical shifts of amide groups from 4-sulfated GalNAc units are significantly more upfield than 15N-resonances from 6-sulfated GalNAc residues; the 15N-resonance of the GalNAc residues of the OSCS, which have both types of sulfation shows a resonance at an 15N shift midway between that of the 4- and 6-sulfated species (Figure 3C). More specifically, these resonances occur at 120.9 ppm, 121.6 ppm and 121.2 ppm, respectively (Table 1). The 1H-resonances of the GalNAc units, in this series of GAGs, show a minimal dependence on sulfation position.
Figure 3.
Comparative NMR analysis of 1 H- and 15N-chemical shifts of amide protons from CS types (A-D), and mammalian HS (E) using 1H-15N-gHSQC spectra. (A) The CS-A from bovine trachea possesses ~ 65% of 4-sulfation, and ~ 35% of 6-sulfation GalNAc residues, whereas (B) the CS-C from shark cartilage has over 95% 6-sulfation GalNAc units. (C) The OSCS possesses over 75% of 4,6-di-sulfation GalNAc units. (D) The DS, also known as CS-B, from porcine intestinal mucosa is composed of ≥ 90% 4-sulfated GalNAc units. (E) The mammalian HS reveals only one peak that necessarily belongs to its 15N-acetylated glucosaminyl unit. Note the different 1H chemical shift scale in (E).
Table 1.
1H- and 15N-chemical shifts (ppm) of amide protons of 15N-acetylhexosamines from GAG types, unsaturated dimer derivatives, and commercial standards.
| GAG types or derivatives | major amino sugar | figure | chemical shifts a (ppm) |
|
|---|---|---|---|---|
| 1H | 15N | |||
| CS-A | β-GalNAc-4(SO3−) | 3A | 7.963 | 120.91 |
| CS-C | β-GalNAc-6(SO3−) | 3B | 7.942 | 121.61 |
| OSCS | β-GalNAc-4,6di(SO3−) | 3C | 7.941 | 121.21 |
| DS (CS-B) | β-GalNAc-4(SO3−) | 3D | 8.096 | 120.46 |
| HS | α-GlcNAc-6(SO3−) | 3E | 8.361 | 123.62 |
| ΔC4S dimer | α-GalNAc-4(SO3−) | 4A | 8.353 | 120.60 |
| β-GalNAc-4(SO3−) | 4A | 8.138 | 121.40 | |
| ΔC6S dimer | α-GalNAc-6(SO3−) | 4B | 8.241 | 121.80 |
| β-GalNAc-6(SO3−) | 4B | 8.092 | 122.75 | |
| GlcNAc (standard) | α-GlcNAc | S1A | 8.138 | 122.95 |
| β-GlcNAc | S1A | 8.039 | 123.72 | |
| GalNAc (standard) | α-GalNAc | S1B | 8.080 | 122.61 |
| β-GalNAc | S1B | 7.972 | 123.53 | |
Chemical shifts are relative to trimethylsilylpropionic acid at 0 ppm for 1H and 13C, and liquid ammonia for 15N, obtained in experiments at 25 °C and pH 4.5.
Dermatan sulfate (DS), which also belongs to the class of galactosaminoglycans like CS, still contains a GalNAc with β(1-4) linkage to an uronic acid. However, the uronic acid of DS is primarily iduronic acid (IdoA) with its linkage to GalNAc denoted α(1-3) rather than β(1-3) as in the GlcA of CSs. Note, as shown in Figure 1B vs.1A, that the change from β to α is the result of the dependence of nomenclature on the chirality of the C5 carbon rather than an actual change in linkage geometry. DS is predominantly 4-O-sulfonated (Figure 1B). So, in comparison to the 4-O-sulfonated CS, the main structural difference is actually epimerization at the C5 carbon (Figure 1A vs. 1B). Comparison of cross-peak positions should, therefore reflect simply the effect of moving the uronic acid carboxyl group from an equitorial to an axial position. Figure 3D shows a down field shift in the proton chemical shift of about 0.13 ppm compared to that of CS-A (Figure 3A), and a minimal change in the 15N chemical shift (Table 1).
15N-NMR analysis of HS polymers
Heparan sulfate (HS) is built on a different backbone than CS and DS polymers, having a β1-4 substituted (if glucuronic acid) and α1-4 linked GlcNAc instead of a β1-3 substituted and β1-4 linked GalNAc unit. It can also contain IdoA residues where its linkage nomenclature changes to α (Figure 1C). Many of the GlcNAc residues of HS are normally converted to N-sulfonated forms, for which an N-H signal in the HSQC spectrum is hard to observe. This cross-peak has been observed by Zhang and coworkers,48 but it is very weak in that report as well. It is possible that observation is difficult because of enhanced proton exchange among protonated sites resulting in peak broadening or transfer of magnetization from a saturated water resonance. This N-sulfonation is followed by epimerization of the linked GlcAs to IdoAs along with additional O-sulfonation. However, stretches of the original GlcNAc residues linked to unepimerized GlcAs usually persist, and N-H signals from these residues are easier to observe by NMR. Figure 3E shows the spectrum of a HS preparation with a resonance for the amide protons of the 15N-acetylated glucosamine residues that is more downfield than those of 15N-acetylated galactosamine residues. The 15N chemical shift is also about 2 ppm further downfield than GalNAc compounds (Table 1). Comparison with the cross-peak from the α-anomer in a 1H-15N gHSQC spectrum of the monomeric GlcNAc shows reasonable agreement (Figure S1A). Differences (~ 0.22 and ~ 0.66 ppm in H and N, respectively, Table 1) are assumed to arise from the linkage to an uronic acid and substitution at the 4-position. Hence, comparison of 15N-gHSQC spectra of different GAGs suggests that 1H-15N resonances can be exploited for diagnostic studies of GAG composition, clearly delineating GlcNAc and GalNAc composition (similarly to commercial standard acetylhexosamines as shown in Table 1), and clearly reporting on the extent of 4- and/or 6-sulfation of these same amino sugars (Figure 3A-D).
15N-NMR analysis of 4- and/or 6-sulfated CS oligomers
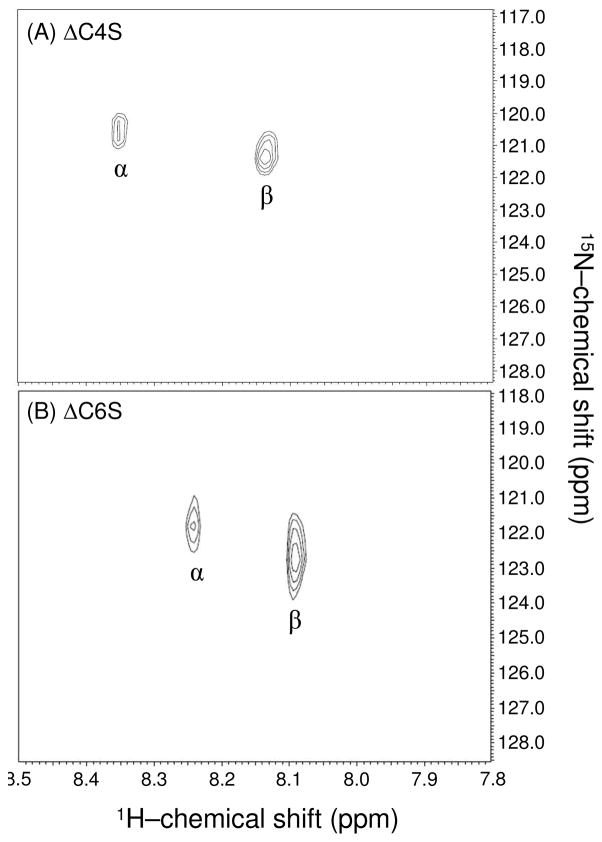
If short oligomers, rather than long polymers, are to be assayed, additional effects due to the absence of substitutions, or anomeric mutarotation at the reducing end enters. This can be seen in 15 N-gHSQC spectra of CS oligosaccharides with different sulfation patterns (disaccharides in Figure 4, and hexasaccharides in Figure 5). CS dimers were obtained by digestion of standard CS-A with chondroitinase ABC. Extensive digestion produces two disaccharides differing in sulfation position (4- or 6-) with an additional 4–5 double bond in the non-reducing end GlcA units. Both purified disaccharides revealed two 1H-15N cross-peaks which are related to the α- and β-anomeric equilibrium in aqueous solution (Figure 4). The ΔC4S disaccharide (Figure 4A) has its amide protons for α- and β-anomers at δH/δN = 8.353/120.60 ppm and δH/δN = 8.136/121.34 ppm respectively (Table 1). The ΔC6S disaccharide (Figure 4B) has its α- and β-amide protons at δH/δN = 8.241/121.80 ppm and δH/δN = 8.092/122.75 ppm respectively (Table 1). Both 4- and 6-sulfated CS dimers show higher populations of β-anomers in comparison to those of α-anomers in a ratio of 6.5/3.5. The β-anomer amide 1H-resonance in the disaccharide with GalNAc at the reducing end is 0.15 to 0.20 ppm downfield of the amide proton resonance in the polymeric CS (Table 1). The 15N-chemical shifts are downfield by about 1 ppm relative to the polymeric material (Table 1), however the difference between amide 15N chemical shifts for the β-anomers of 4- and 6-O-sulfonated disaccharides remains near 1 ppm (Table 1), suggesting that this difference remains diagnostically useful.
Figure 4.
Comparative NMR analysis of α- and β-1H/15N-resonances of unreduced unsaturated 4- (A), and 6-sulfated (B) CS dimers obtained from ABC lyase digestion, using 1H-15N gHSQC spectra. The β-/β-signals arises from mutarotation of anomerics in aqueous solution, which the equilibrium ratio is 3.5/6.5.
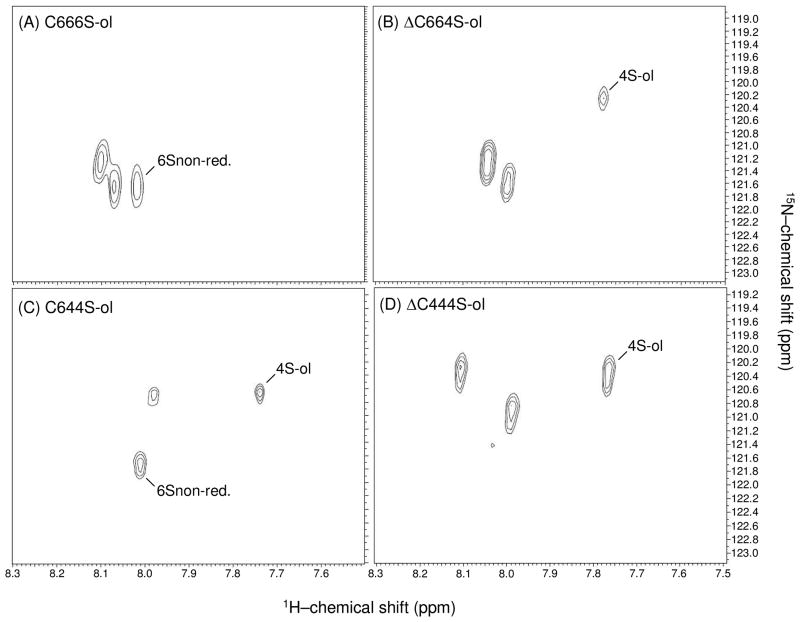
Figure 5.
Comparative NMR analysis of reduced CS hexamers from ABC lyase (A,C), and hyaluronidase (B,D) digestions using 1H-15N gHSQC spectra. (A) C666S-ol, (B) ΔC664S-ol, (C) ΔC644S-ol, and (D) C444-ol. The peaks assigned as 6Snon-red., and 4S-ol belong to the 6-sulfated and 4-sulfated GalNAc units from non-reducing and reduced terminals respectively.
The 15N-gHSQC spectra of the CS hexamers with different sulfation patterns obtained from ABC lyase or hyaluronidase digestion are very illustrative of the efficacy of the 15N-1H HSQC analysis for fast NMR assignment of sulfation types of CS derivatives (Figure 5). Before recording the 15N-gHSQC spectra, these hexamers were reduced to eliminate superposition of peaks from α- and β-anomers, and their structures were independently determined by a combination of 2D NMR experiments, including DQF-COSY, TOCSY and 13C-gHSQC (data not shown). The 15N-gHSQC spectra of these hexamers showed the same upfield and downfield relationship of 15N-resonances for the amide protons of 4- and 6-sulfated GalNAc units, supporting the fact that this pattern can be widely used for recognition and assignment of sulfate sites. All the 1H-15N cross-peaks of the 4-sulfated units have 15N-chemical shifts between 120.3–120.6 ppm, whereas the 15N-chemical shifts from 6-sulfated GalNAc units are between 121.2–121.6 ppm (Figure 5).
15N labeled GAGs from cell culture
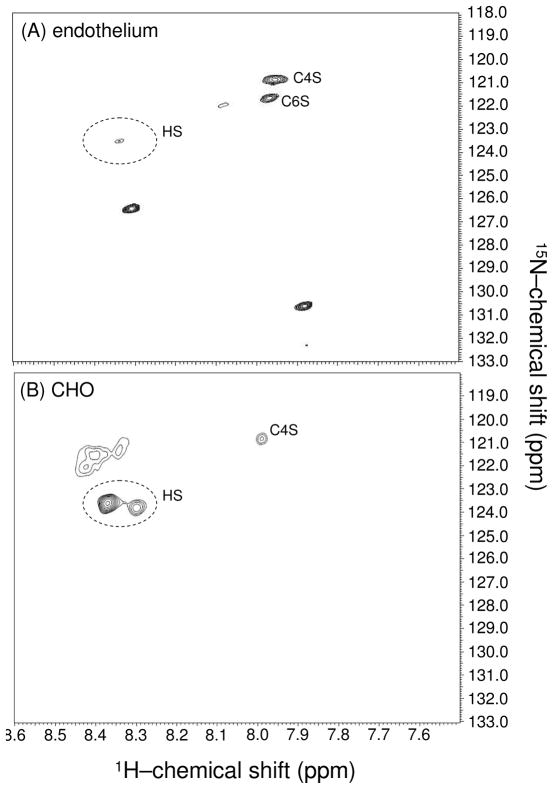
One of the potential applications of a 1H-15 N HSQC analysis of GAGs would be the determination of their composition as a function of cell type and growth conditions. However, the amount of material isolated from cell culture is apt to be small, and sensitivity is low at the 0.3% abundance of 15N. This limitation makes the development of an in vivo 15N-labeling technique worthwhile. After supplementation of media with 15N-Gln in mouse lung endothelium or CHO cell culture for 24h, GAG polymers were isolated as described in the methods section. Each preparation yielded approximately 600 μg of total 15N-labeled material allowing 15N-gHSQC spectra to be rapidly recorded (Figure 6). The endothelial labeled material (Figure 6A) clearly shows peaks characteristic of 4- and 6-sulfated CSs (compare to Figure 3A). However, other downfield 15N-resonances also are present. Treatment with DNase and RNAse, followed by separation from low-molecular-weight materials on a Sephadex-G15 column eliminated these additional peaks suggesting they arise from nucleic acid contaminants from the initial step of GAG isolation. Curiously, the cross-peak characteristic of HS in this cell type is very weak (Figure 6A) even though clear evidence for significant amounts of HS in endothelial cells has been previously demonstrated.49 The low intensity of the HS cross-peak expected near δH = 8.36 ppm, and δN = 123.6 ppm could be due either to a high level of N-sulfonation (since we detect through residual N-acetyls), to a low level of 15N-HS production under the conditions used, or to an unusual level of HSQC insensitivity to this particular type of GAG. However, a 1D 1H-NMR spectrum of the endothelial GAGs (Figure S4), taken in D2O without presaturation which should detect quite universally, also reveals very small α-anomeric1H signals in the region downfield of the water resonance. These downfield 1H-anomeric resonances must belong to HS-type GAGs since CS polymers have β-1H-anomerics with chemical shifts upfield of the water signal. This suggests that the small 1H-15N HSQC cross-peak in the HS region (Figure 6A) is at least partly the result of a low level of HS production by endothelial cells under our growth conditions.
Figure 6.
1H-15N gHSQC spectra of 15N-labeled negatively charged molecules extracted from the mouse lung endothelial cells (A), and (B) CHO cells.
Conversely, material isolated from CHO cells (Figure 6B) revealed significant resonances characteristic of N-acetyls in HS molecules together with measurable resonances characteristic of C4S (compare Figure 6B with 3A). There are some more dispersed peaks near the primary HS cross-peak. These multiple peaks (dashed ellipse at Figure 6B) may belong to GlcNAc residues from a more heterogeneous region in HS, characterized by a mixture of both 6-O-sulfonated and non-O-sulfonated GlcNAc units together with adjacent uronic acid residues modified or not by epimerization and/or 2-sulfation (see Figure 1C). Neither cell type showed evidence for expression of 15N-DS-proteoglycans.
The level of 15N-incorporation in the cellular GAG samples is clearly significant given the ease of observation. Using intensities of cross-peaks in comparison to the standard samples, labeling at a level of 5–10% was estimated. This represents enhancement over natural abundance by a factor of 15–30. To confirm this level of enrichment, the sample of endothelial CS was degraded to ΔC4S and ΔC6S disaccharides using nuclease treatment followed by chondroitinase ABC digestion, and subsequently analyzed by MS. These dimer products were purified by size exclusion and strong anion exchange chromatography, and then they were fully characterized by high resolution NMR spectroscopy (Figure S2 and S3 at supplementary material).
An MS spectrum of the pure ΔC4S dimers obtained from the last purification on the HPLC-SAX chromatography is presented in Figure 7. In Figure 7A the experimental spectrum is compared to a simulated isotopomer profile assuming natural abundances of 15N, 13C, and 2H. Note that the simulation of the M+1 peak in particular does not have sufficient intensity. Raising the 15N abundance to 8% produces a very good fit (Figure 7B), while assuming a 15N abundance of 10% produces a small overestimation. The 15N abundance of 8% calculated with MS agrees quite well with the previous NMR-based estimates.
DISCUSSION
Thus, the potential utility of GAG analysis using 15N-1H gHSQC NMR methods has been demonstrated. 15N-NMR methods do, however, have their own limitations. HS detection described here is currently through the residual N-acetyl groups, making characterization dependent on the extent of N-sulfonation, particularly in HS. However, detection of sulfated amides is possible under some conditions,48 and conditions might be adjusted to achieve this more routinely. NMR is also not a highly sensitive method and this is a potential problem here because of the low magnetogyric ratio and low natural abundance of the 15N isotope (0.37%). Although the natural abundance of the 15N-isotope remains a problem, observation by indirect detection through protons avoids problems with a low magnetogyric ratio and provides sufficient improvement in sensitivity to characterize materials even at natural abundance (~5 mg samples with about 120 min acquisition). The methods can therefore be of use in the characterization of commercial products where amounts of material are seldom a concern, including the low molecular weight heparins recently under scrutiny for OSCS contamination.51 For analyses of cell culture products, enrichment in 15N to levels approaching 10% has been demonstrated, greatly improving sensitivity. This opens the possibility of studying metabolic variations in the production of GAGs throughout the cell cycle and under the influence of various environmental factors.
It is not surprising that 15N and 1H chemical shifts would be sensitive to structural differences in hexosamine rings such as exist between GalNAc and GlcNAc. However, the sensitivity of amide 15N and 1H chemical shifts to changes in more remote groups is perhaps surprising. The 4-sulfate, 6-sulfate, and epimeric carboxylate of the substituted uronic acid are 5, 7, and 6 bonds removed respectively. However, in each case, we are dealing with a charged or partially charged group. Electric field effects on chemical shifts indeed can be quite long range.52, 53 Interpretation as the sum of field projections on bonds and field squared effects also makes them non-additive as observed in comparing 4-, 6-, and 4- plus 6-sulfations. Effects of titration of carboxylate groups in fatty acids on polarizable C-C double bonds can produce shifts of as much as a ppm even when olefinic carbons are 6 bonds removed from the carboxyl group.54, 55 Similar effects exist for N-H bonds of amides in peptides.53 The positions of sulfates and carboxyls depend on conformation, but in a low energy model of the fragments, they are approximately 5.7 and 4.3 Å away from the amide nitrogen of the 4-O-sulfonated GalNAc of CS. At this distance, assuming a charge of −1, an effective dielectric of 5, an optimum projection, and using the formulas given at the work of Hass et al.,53 a contribution to the 15N chemical shift of 1.4 ppm would be observed. Hence, it is possible to rationalize the sensitivity to sulfate placement. The effects are, of course, useful indicators of GAG composition regardless of their origin (Table 1 presents offsets due to addition of charged groups at various sites, or pairs of sites, that can be used as empirical guide). If the origin is electrostatic, an additional issue is raised, namely the importance of controlling factors which might modulate these effects, such as the types and concentration of counter-ions in assay solutions (50 mM Na+ and pH 4.5 in the data presented here).
The potential applications of the assay methods suggested herein are broad. The 15N-NMR spectra of GAGs might, for example, be used for auxiliary quality control procedures in checking for the presence of OSCS-contaminations in clinical samples of heparin,32, 51 since the 15N and 1H chemical shifts of glucosamine and galactosamine residues are quite distinct (Figure 3A-D vs. 3E, Table 1). In this case, the speed of the method could be considerably improved by replacing the 2D 1H-15N HSQC acquisitions with a 1D 15N-filtered 1H-observe acquisition that resolves primarily based on proton chemical shifts. Interpretation in terms of molecular characteristics of different species in mixed samples may also be more straightforward than methods currently suggested, such as diffusion ordered NMR spectroscopy,56 or the combination of 1D 1H-NMR spectra with polyacrylamide gel electrophoresis (PAGE).57
Applications to metabolic monitoring of GAG production in various cell types should also be possible. We provided a preliminary demonstration with applications to endothelial, and CHO cells. These applications were not pursued in a well controlled and systematic way that would allow biologically relevant conclusions, but the dramatic differences observed between cell types suggest the potential of these applications. The ECM GAGs are known to turn over in response to various environmental effects,58–60 and composition is known to vary with differentiation during development.61 In the work presented, the endothelium and CHO cells showed different patterns of 15N-labeled GAGs in their cell surface proteoglycans (Figure 6). The endothelial cells synthesized significant amounts of 15N-CS (Figure 6A) with the same 4-/6-sulfation ratio (6.2/3.8 based on cross-peak intensities) as the standard CS-A extracted from bovine trachea (Figure 3A), and very weak cross-peaks characteristic of HS. CHO cells, on the other hand, showed mostly cross-peaks of 15N-HSs. A peak characteristic of 4-sulfated CS is also observed, but a 6-sulfated peak is not. In addition, the labeled cellular GAG analyses showed clearly the absence (or a very low level) of 15N-DS proteoglycans in both cell types.
The low intensity of an HS cross-peak in isolates from endothelial cells (Figure 4A) is also intriguing since these cells have previously been reported to express significant levels of HS.49 As supported by weak, but diagnostic, α-1H anomeric resonances in 1D 1H-NMR spectrum of the isolated negatively charged molecules from the endothelial cells (Figure S4), HS is indeed present, although in much lower concentration than CSs. Based on 1D proton intensities, endothelial HSs comprise a bit less than 10% of the total GAGs, whereas CS is abundant (≥90%). Thus, CHO cells and endothelial cells under similar growth conditions are clearly metabolically different. Further studies of the time course of 15N-labeled GAG production in these cells, beginning with labeling at the initial stages of the cell growth, would certainly be of interest. The methods described here could easily be adapted to this kind of future analysis.
GAG composition analyses by conventional means usually begins with additional digestion and separation steps, after which identification of sulfation sites is normally done by additional HPLC-analyses and/or a combination of NMR and MS analyses.50 These procedures represent significantly more effort and can lead to uncertainties in quantitation because of incomplete digestion and molecule dependent MS and HPLC response. NMR, when based on simple 1D proton acquisitions with adequate relaxation recovery, can be very quantitative and quite universal in detection. Resolution is greatly improved by using 2D experiments, and when isotopically labeled metabolic precursors are used, even very crude isolates can be analyzed. This is well illustrated in the current work, utilizing the side chain 15N in glutamine to label newly synthesized GAGs in crude isolates from cell cultures. The 15N-based methods are, however, a class of NMR experiments seldom used for analysis of GAGs, and a general comparison of these 15N-methods to the more traditional 1H- and 13C-based NMR experiments is appropriate.
A comparison of 15N-1H HSQC experiments with more conventional NMR methods shows that each one has its particular advantages and disadvantages. 1D 1H-NMR spectroscopy, for example, is fast and sensitive. Well resolved diagnostic peaks (α-anomeric resonances of glucosamine derivatives in HS, the down field H4 resonance of 4-sulfated CS, and the down field shifted resonances of OSCS (Figures S5A, S5E and S5D, for example)) allow identification of certain components.62–64 However, subtle distinctions such as those between CS-C and DS (Figure S5C and S5B); and the sulfation at the 6-position in CSs, are very difficult to assess, particularly in GAG mixtures. 2D homonuclear experiments can improve the resolution and allow identification of more species, but cross-peak patterns remain very complex and acquisition times are not significantly different from those required for 15N-1H HSQC spectra on isotopically enriched samples. Examples of 1H-1H TOCSY spectra on enzymatically digested endothelial cells GAGs in Figure S2 illustrate the potential difficulties. Spectra of polymeric GAGs suffer additional broadening of resonances with associated sensitivity and resolution loses. 2D 13C-based NMR spectra (HSQC, HMQC, and Heteronuclear Multiple Bond Correlation (HMBC)) provide a more robust approach to identification of residue types, linkage patterns, and sulfation sites.65 However, at natural abundance, these spectra are less than a factor of three more sensitive than the 15N-1H HSQC spectra used in the present approach and the potential for peak overlap in complex mixtures is far greater when all protonated carbon sites contribute cross-peaks. The approach described herein has the advantage of being based on rather simple spectra with just few easily identifiable cross-peaks (Figure 3). Observation of this small number of peaks still allows the rapid identification of hexosamine types (GalNAc vs GlcNAc), of sulfation sites (4-O- and/or 6-O-), of anomeric ratios (α-/β-), and of uronic acid contents (DS and HS versus CS).
There is, however, a downside in application of these more complex 2D experiments, at least to polymeric materials. That is a loss of an ability to quantitatively relate cross-peak integrals to molar amounts of different constituents. HSQC spectra depend on the transfer and refocusing of magnetization over time intervals on the order of the reciprocal of the heteronuclear coupling constant. Magnetization is lost in these intervals in an inverse exponential relationship to proton line widths, something that is significant when line widths approach the value of scalar couplings (30 Hz, vs 90 Hz in our CS samples). Within a given GAG type, and GAG preparation, line widths for different amide protons, even if large, may be very similar and lead to small errors in quantitation (our measurements of 6- vs 4-sulfation in CS preparations is in close agreement with other analyses). However, across GAG types (CS vs DS vs HS) line widths can differ and lead to substantial errors in quantitation of relative amounts. While no attempt to improve quantitation was made in this work, there are likely paths to this improvement. Operation at higher temperatures and/or different pHs may minimize differential losses. In addition, a separate measurement of transverse relaxation profiles for amide protons may allow corrections to measured cross-peak intensities. These remain areas for future investigation.
Conclusions
Thus, in this work we have demonstrated that GAGs commonly found as components of cell surface proteoglycans of mammalian cells can easily be identified based on 1H-15N HSQC NMR experiments. CS, DS, and HS all give cross peaks in distinct regions of the NMR spectrum. In addition, the 4- and/or 6-sulfation patterns of CS polymers or oligomers can be identified. Analyses can also be carried out on crude isolates released from cell surfaces by lysis, pronase digestion, and retention on anion exchange chromatographic media. When combined with metabolic labeling with 15N via a pathway from the side chain nitrogen of glutamine to the amino nitrogen of the amino sugars in GAGs, direct analysis on these crude preparations becomes quite straightforward. We look forward to future applications in this area.
Supplementary Material
Acknowledgments
This work was supported by grant from the National Center for Research Resources of the National Institute of Health (NIH), RR005351. VHP was supported by a post-doctoral fellowship (PDE #201019/2008-6) from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. We are also grateful to Lidia Nieto for the NMR assignments of the standard unsaturated CS dimers, to John Glushka for his technical assistance on the NMR experiments, to Christian Heiss for the MS analysis of the CS dimers and hexamers, and to Laura Morris for the computational assistance.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures presenting NMR analysis of standards and of some samples from the endothelial cells are available free of charge via Internet at http://pubs.acs.org.
References
- 1.Heino J, Kapyla J. Curr Pharm Des. 2009;15:1309–1317. doi: 10.2174/138161209787846720. [DOI] [PubMed] [Google Scholar]
- 2.Yu F, Wolff JJ, Amster IJ, Prestegard JH. J Am Chem Soc. 2007;129:13288–13297. doi: 10.1021/ja075272h. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi NS, Mancera RL. Chem Biol Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 4.Iozzo RV. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 5.Beaulieu JF, Vachon PH, Chartrand S. Anat Embryol. 1991;183:363–369. doi: 10.1007/BF00196837. [DOI] [PubMed] [Google Scholar]
- 6.Cohn RH, Cassiman JJ, Bernfield MR. J Cell Biol. 1976;71:280–294. doi: 10.1083/jcb.71.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns RR, Gross J Exp Cell Res. 1980;128:1–7. doi: 10.1016/0014-4827(80)90379-1. [DOI] [PubMed] [Google Scholar]
- 8.Aviezer D, Hecht D, Safran M, Eisinger M, David G, Yayon A. Cell. 1994;79:1005–1013. doi: 10.1016/0092-8674(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 9.Gorio A, Lesma E, Vergani L, DiGiulio AM. Eur J Neurosci. 1997;9:1748–1753. doi: 10.1111/j.1460-9568.1997.tb01532.x. [DOI] [PubMed] [Google Scholar]
- 10.Cattaruzza S, Perris R. Matrix Biol. 2005;24:400–417. doi: 10.1016/j.matbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Muramatsu T, Muramatsu H. Proteomics. 2008;8:3350–3359. doi: 10.1002/pmic.200800042. [DOI] [PubMed] [Google Scholar]
- 12.Domowicz M, Mangoura D, Schwartz NB. Int J Dev Neurosci. 2000;18:629–641. doi: 10.1016/s0736-5748(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 13.Thesleff I, Jalkanen M, Vainio S, Bernfield M. Dev Biol. 1988;129:565–572. doi: 10.1016/0012-1606(88)90401-0. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan CD, O’Neill SK, Koreny T, Czipri M, Finnegan A. J Immunol. 2002;169:5851–5859. doi: 10.4049/jimmunol.169.10.5851. [DOI] [PubMed] [Google Scholar]
- 15.Koninger J, Giese NA, Bartel M, di Mola FF, Berberat PO, di Sebastiano P, Giese T, Buchler MW, Friess H. J Clin Pathol. 2006;59:21–27. doi: 10.1136/jcp.2004.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doodes PD, Cao YX, Hamel KM, Wang YM, Rodeghero RL, Kobezda T, Finnegan A. Arthritis Rheum. 2009;60:2945–2953. doi: 10.1002/art.24842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Mattos DA, Stelling MP, Tovar AMF, Mourao PAS. J Thromb Haemost. 2008;6:1987–1990. doi: 10.1111/j.1538-7836.2008.03145.x. [DOI] [PubMed] [Google Scholar]
- 18.He L, Giri TK, Vicente CP, Tollefsen DM. Blood. 2008;111:4118–4125. doi: 10.1182/blood-2007-12-127928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inatani M, Haruta M, Honjo M, Oohira A, Kido N, Takahashi M, Honda Y, Tanihara H. Brain Res. 2001;920:217–221. doi: 10.1016/s0006-8993(01)02856-6. [DOI] [PubMed] [Google Scholar]
- 20.Pettway Z, Domowicz M, Schwartz NB, BronnerFraser M. Exp Cell Res. 1996;225:195–206. doi: 10.1006/excr.1996.0170. [DOI] [PubMed] [Google Scholar]
- 21.Perris R, Perissinotto D, Pettway Z, BronnerFraser M, Morgelin M, Kimata K. Faseb J. 1996;10:293–301. doi: 10.1096/fasebj.10.2.8641562. [DOI] [PubMed] [Google Scholar]
- 22.Tully SE, Mabon R, Gama CI, Tsai SM, Liu XW, Hsieh-Wilson LC. J Am Chem Soc. 2004;126:7736–7737. doi: 10.1021/ja0484045. [DOI] [PubMed] [Google Scholar]
- 23.vanPutten JPM, Hayes SF, Duensing TD. Infect Immun. 1997;65:5028–5034. doi: 10.1128/iai.65.12.5028-5034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean DD, Muniz OE, Rodriquez I, Carreno MR, Morales S, Agundez A, Madan ME, Altman RD, Annefeld M, Howell DS. Arthritis Rheum. 1991;34:304–313. doi: 10.1002/art.1780340308. [DOI] [PubMed] [Google Scholar]
- 25.Todhunter RJ, Lust G. J Am Vet Med Assoc. 1994;204:1245–1251. [PubMed] [Google Scholar]
- 26.Fellstrom B, Backman U, Danielson B, Wikstrom B. World J Urol. 1994;12:52–54. doi: 10.1007/BF00182052. [DOI] [PubMed] [Google Scholar]
- 27.Siragusa S, Cosmi B, Piovella F, Hirsh J, Ginsberg JS. Am J Med. 1996;100:269–277. doi: 10.1016/S0002-9343(97)89484-3. [DOI] [PubMed] [Google Scholar]
- 28.Hull RD, Raskob GE, Pineo GF, Brant RF. Clin Appl Thromb Hemost. 1995;1:151–159. [Google Scholar]
- 29.Koopman MMW, Prandoni P, Piovella F, Ockelford PA, Brandjes DPM, vanderMeer J, Gallus AS, Simonneau G, Chesterman CH, Prins MH, Bossuyt PMM, deHaes H, vandenBelt AGM, Sagnard L, Dazemar P, Buller HR. N Engl J Med. 1996;334:682–687. doi: 10.1056/NEJM199603143341102. [DOI] [PubMed] [Google Scholar]
- 30.Fareed J, Hoppensteadt DA, Bick RL. Clin Appl Thromb Hemost. 2003;9:101–108. doi: 10.1177/107602960300900202. [DOI] [PubMed] [Google Scholar]
- 31.Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Curr Opin Struct Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang ZQ, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Nat Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishimoto TK, Viswanathan K, Ganguly T, Elankumaran S, Smith S, Pelzer K, Lansing JC, Sriranganathan N, Zhao GL, Galcheva-Gargova Z, Al-Hakim A, Bailey GS, Fraser B, Roy S, Rogers-Cotrone T, Buhse L, Whary M, Fox J, Nasr M, Dal Pan GJ, Shriver Z, Langer RS, Venkataraman G, Austen KF, Woodcock J, Sasisekharan R. N Engl J Med. 2008;358:2457–2467. doi: 10.1056/NEJMoa0803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blossom DB, Kallen AJ, Patel PR, Elward A, Robinson L, Gao GP, Langer R, Perkins KM, Jaeger JL, Kurkjian KM, Jones M, Schillie SF, Shehab N, Ketterer D, Venkataraman G, Kishimoto TK, Shriver Z, McMahon AW, Austen KF, Kozlowski S, Srinivasan A, Turabelidze G, Gould CV, Arduino MJ, Sasisekharan R. N Engl J Med. 2008;359:2674–2684. doi: 10.1056/NEJMoa0806450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A, Hsieh-Wilson LC. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 36.Esko JD, Lindahl U. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YJ, Jardetzky O. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 39.Blundell CD, DeAngelis PL, Day AJ, Almond A. Glycobiology. 2004;14:999–1009. doi: 10.1093/glycob/cwh117. [DOI] [PubMed] [Google Scholar]
- 40.Mobli M, Nilsson M, Almond A. Glycoconj J. 2008;25:401–414. doi: 10.1007/s10719-007-9081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almond A, DeAngelis PL, Blundell CD. J Am Chem Soc. 2005;127:1086–1087. doi: 10.1021/ja043526i. [DOI] [PubMed] [Google Scholar]
- 42.Blundell CD, Almond A. Magn Reson Chem. 2007;45:430–433. doi: 10.1002/mrc.1969. [DOI] [PubMed] [Google Scholar]
- 43.Sattelle BM, Shakeri J, Roberts IS, Almond A. Carbohydr Res. 2010;345:291–302. doi: 10.1016/j.carres.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orlando R, Lim JM, Atwood JA, Angel PM, Fang M, Aoki K, Alvarez-Manilla G, Moremen KW, York WS, Tiemeyer M, Pierce M, Dalton S, Wells L. J Proteome Res. 2009;8:3816–3823. doi: 10.1021/pr8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuster MM, Wang LC, Castagnola J, Sikora L, Reddi K, Lee PHA, Radek KA, Schuksz M, Bishop JR, Gallo RL, Sriramarao P, Esko JD. J Cell Biol. 2007;177:539–549. doi: 10.1083/jcb.200610086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei G, Bai XM, Gabb MMG, Bame KJ, Koshy TI, Spear PG, Esko JD. J Biol Chem. 2000;275:27733–27740. doi: 10.1074/jbc.M002990200. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Zhang ZQ, McCallum SA, Xie J, Nieto L, Corzana F, Jimenez-Barbero J, Chen M, Liu J, Linhardtt RJ. J Am Chem Soc. 2008;130:12998–13007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang LC, Fuster M, Sriramarao P, Esko JD. Nat Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 50.Sasisekharan R, Raman R, Prabhakar V. Annu Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 51.Zhang ZQ, Weiwer M, Li BYZ, Kemp MM, Daman TH, Linhardt RJ. J Med Chem. 2008;51:5498–5501. doi: 10.1021/jm800785t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bader R. J Phys Chem B. 2009;113:347–358. doi: 10.1021/jp807362v. [DOI] [PubMed] [Google Scholar]
- 53.Hass MAS, Jensen MR, Led JJ. Proteins. 2008;72:333–343. doi: 10.1002/prot.21929. [DOI] [PubMed] [Google Scholar]
- 54.Batchelo Jg, Cushley RJ, Prestega Jh. J Org Chem. 1974;39:1698–1705. doi: 10.1021/jo00925a023. [DOI] [PubMed] [Google Scholar]
- 55.Batchelo Jg, Prestega Jh, Cushley RJ, Lipsky SR. J Am Chem Soc. 1973;95:6358–6364. doi: 10.1021/ja00800a032. [DOI] [PubMed] [Google Scholar]
- 56.Sitkowski J, Bednarek E, Bocian W, Kozerski L. J Med Chem. 2008;51:7663–7665. doi: 10.1021/jm801198b. [DOI] [PubMed] [Google Scholar]
- 57.Zhang ZQ, Li BYZ, Suwan J, Zhang FM, Wang ZY, Liu HY, Mulloy B, Linhardt RJ. J Pharm Sci. 2009;98:4017–4026. doi: 10.1002/jps.21729. [DOI] [PubMed] [Google Scholar]
- 58.Gressner AM. Biol Chem Hoppe-Seyler. 1989;370:902–903. [Google Scholar]
- 59.Gressner AM, Schafer S. J Clin Chem Clin Biochem. 1989;27:141–149. doi: 10.1515/cclm.1989.27.3.141. [DOI] [PubMed] [Google Scholar]
- 60.Gressner AM, Pazen H, Greiling H. Hoppe-Seyler’s Z Physiol Chem. 1977;358:825–833. doi: 10.1515/bchm2.1977.358.2.825. [DOI] [PubMed] [Google Scholar]
- 61.Oohira A, Matsui F, Matsuda M, Shoji R. J Neurochem. 1986;47:588–593. doi: 10.1111/j.1471-4159.1986.tb04540.x. [DOI] [PubMed] [Google Scholar]
- 62.Guerrini M, Naggi A, Guglieri S, Santarsiero R, Torri G. Anal Biochem. 2005;337:35–47. doi: 10.1016/j.ab.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 63.Toida T, Toyoda H, Imanari T. Anal Sci. 1993;9:53–58. [Google Scholar]
- 64.Maruyama T, Toida T, Imanari T, Yu G, Linhardt RJ. Carbohydr Res. 1998;306:35–43. doi: 10.1016/s0008-6215(97)10060-x. [DOI] [PubMed] [Google Scholar]
- 65.Duus DØ, Gotfredsen CH, Bock K. Chem Rev. 2000;100:4589–4614. doi: 10.1021/cr990302n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.