Abstract
Summary: In bacteria, motility is important for a wide variety of biological functions such as virulence, fruiting body formation, and biofilm formation. While most bacteria move by using specialized appendages, usually external or periplasmic flagella, some bacteria use other mechanisms for their movements that are less well characterized. These mechanisms do not always exhibit obvious motility structures. Myxococcus xanthus is a motile bacterium that does not produce flagella but glides slowly over solid surfaces. How M. xanthus moves has remained a puzzle that has challenged microbiologists for over 50 years. Fortunately, recent advances in the analysis of motility mutants, bioinformatics, and protein localization have revealed likely mechanisms for the two M. xanthus motility systems. These results are summarized in this review.
INTRODUCTION
Myxococcus xanthus is a Gram-negative soil bacterium with a life cycle that includes vegetative growth, predation, and development (Fig. 1). M. xanthus cells are unable to swim in liquid culture; however, on solid surfaces they move at about 2 to 4 μm/min, almost 1,000 times slower than flagellated bacteria. The ability of M. xanthus cells to move on solid surfaces is very important for vegetative swarming and development. Indeed, during their hunting and food-gathering activities, M. xanthus cells use gliding motility to scavenge for insoluble nutrients in decomposing soils and detritus or for predation of prey microorganisms (54, 56, 57, 102, 113). M. xanthus cells move in a coordinated manner by forming organized groups called “swarms” (Fig. 1 and 2). The swarms consist of aligned cells that spread as a monolayer or as multilayered cells stacked in tiers. When the swarms encounter prey microorganisms, they kill and lyse the cells using antibiotics and lytic enzymes (32, 105). Digested prey cells provide a food source for growth. M. xanthus cells that encounter cell debris, peptidoglycan, or many other macromolecules display intriguing rhythmic movements referred to as “rippling,” since the waves look similar to the ripples that appear in water when it is disturbed by a pebble (Fig. 1 and 3). During rippling, the aligned cells form “accordion waves” that travel in convergent or divergent directions. When two converging waves meet, cell reversals are induced and transmitted to the entire wave so that the two waves then reflect off each other (106, 116) (Fig. 3). Berleman et al. (9) proposed that the rhythmic reversals during rippling enable the bacteria to more efficiently lyse and absorb the nutrients from prey cells. Cell-cell coordination of cell movements during rippling is thought to occur through direct side-to-side cell contacts (see below) (78).
FIG. 1.
Life cycle of M. xanthus. (Vegetative growth) On a solid surface with soluble nutrients, groups of M. xanthus cells (swarms) grow, divide, and move outward. On a solid surface in the presence of lysing cells or prey, M. xanthus cells form “accordion waves” known as ripples. (Low-nutrient development) On a solid surface upon nutrient step-down or starvation, 105 to 106 cells aggregate to form mounds and then fruiting bodies. The rod-shaped cells in the fruiting bodies undergo morphogenesis and form spherical spores that are metabolically inactive but more resistant to desiccation and heat. Peripheral rods, a subpopulation of stressed cells, remain outside fruiting bodies in search of food. When nutrients become available, the spores germinate and complete the life cycle.
FIG. 2.
Morphologies of vegetative and developmental cells. Wild-type M. xanthus strain DZ2 with both A and S motilities (A+S+), cells lacking S motility (A+S−), cells lacking A motility (A−S+), and cells lacking both motility systems (A−S−) are shown. S-motility assays were performed on plates containing an agar concentration of 0.3%. Under these conditions, A−S+ cells show a swarming rate comparable to that of A+S+ cells, whereas A+S− cells are virtually nonmotile. A-motility assays were done on hard agar (1.5%). On this medium, A+S+ cells move as groups but also as individuals, A+S− cells move primarily as individuals, A−S+ cells move only as groups, and A−S− cells are nonmotile. Fruiting body formation assays were done on CF fruiting medium containing 1.5% agar, with incubation for 72 h at 32°C. On this medium, A+S+ cells form fruiting bodies in 48 to 72 h, A+S− cells and A−S+ cells show very delayed fruiting (not apparent before 72 h), and A−S− cells are nonfruiting.
FIG. 3.
Coordinated movements of M. xanthus cells. (A) When M. xanthus cells (left) encounter and then penetrate an E. coli colony (right) they align, forming accordion waves (ripples). In contrast, cells that do not encounter E. coli cells starve and undergo fruiting body formation (dark structures on the left). (The picture of M. xanthus invading an E. coli colony was adapted from reference 8 with permission of Blackwell Publishing Ltd.) (B) Phase-contrast microscopy (left) and fluorescence microscopy (right) of wild-type M. xanthus cells mixed at a 50:1 ratio with GFP-labeled cells, showing that during rippling behavior, the rippling pattern is stably maintained, although individual cells change position. (The picture of the M. xanthus ripple structures was adapted from reference 9.) (C) Ripples in a monolayer culture. The pictures show that during a collision between two waves, cells in one wave penetrate the opposing wave by one cell length, followed by cell reversals. (The picture of the M. xanthus ripples was adapted from reference 116.) (D) M. xanthus cells expressing FrzCD-GFP. FrzCD forms clusters that align relative to each other when cells make side-to-side contacts (see inset). We propose that during rippling behavior, side-to-side contacts between cells stimulate the FrzCD receptor to trigger reversals.
When M. xanthus swarms are subjected to a step-down in nutrients (or reduced prey), they enter a developmental pathway that results in two populations of cells: most of the cells aggregate into fruiting bodies, while the remaining cells form a monolayer of cells called peripheral rods (94, 95). Significant cell lysis may occur during this process, although the extent of lysis can vary with different strain backgrounds and culturing conditions (8, 95). After 24 to 72 h of starvation, cells in fruiting bodies convert to resistant, resting myxospores; each fruiting body contains 105 to 106 spores. Sporulation in M. xanthus differs from endospore formation in Bacillus spp. in that the entire cell converts to a myxospore, which is also the case during Rhodospirillum sp. bacterial cyst formation (6). During these developmental transitions, gene expression and the pattern of cell movements are highly regulated (112). When a food source becomes available, the myxospores germinate and resume vegetative growth. Spores are viable for long periods of time and provide a strong survival benefit to cells during periods of starvation and desiccation.
Nonaggregated cells (peripheral rods) surround the fruiting bodies as a monolayer of aligned cells. Lysed M. xanthus cells induce the peripheral rods to ripple as they feed on the nutrients that are released. It has been suggested that the function of the peripheral rods is to provide hunting parties that can attack and lyse microorganisms that approach the fruiting bodies (94). Lysed cells can provide a food source for the peripheral rods and, when sufficient, allow for myxospore germination and movement of cells away from the fruiting bodies.
M. xanthus Contains Two Motility Systems
The first breakthrough in our understanding of M. xanthus motility occurred 30 years ago, when Hodgkin and Kaiser used genetic studies to show that M. xanthus uses two different systems to move on surfaces (41). One motility system was called social (S) motility, because it was responsible for the movement of cells traveling in groups. Indeed, M. xanthus cells exhibiting only S motility (A−S+) were nonmotile when isolated but regained motility when placed near another cell. The other motility system was called adventurous (A) motility, because it was responsible for cell movements when cells were isolated from the group. The physical mechanisms for these two motility systems remained a mystery for many years, although some observations provided evidence for how the cells might move. For example, in 1979, Hodgkin and Kaiser (41) showed that S motility is correlated with the presence of pili at the cell poles, although the role for these pili was unclear at the time. Another intriguing observation was made by Shi and Zusman (111), who observed that the two motility systems of M. xanthus showed different selective advantages on different surfaces: A-motile cells (A+S−) moved best on relatively firm, dry surfaces (1.5% agar), but were virtually nonmotile on soft, wet surfaces (0.3% agar), whereas S-motile cells (A−S+) moved best on relatively soft, wet surfaces (0.3% agar) and moved much more slowly on drier surfaces such as 1.5% agar (Fig. 2).
The A- and S-motility systems are synergistic, as colony spreading by wild-type cells is faster than the sum of those of individual A+S− and A−S+ cells (53). In the absence of one or both motility systems, aggregation, fruiting body formation, and rippling are defective, indicating that motility is required for these social behaviors.
S MOTILITY
M. xanthus S motility is generally described as the coordinated movement of cells when present in groups; isolated A−S+ cells are nonmotile on agar. However, surprisingly, A−S+ cells can move as isolated cells when they are submerged in 1% methylcellulose (71, 121). Genetic and behavioral analyses show that S motility requires type IV pili (TFP) (126, 134-137) and extracellular matrix polysaccharide (EPS) (also referred to as fibrils) (72, 73, 100). Lipopolysaccharide (LPS) O antigen (16, 140, 144, 145) may also play an indirect role. Strains lacking genes required for the production of TFP or EPS fail to move by S motility. It is not known why S motility requires cell groups for movement. One possibility is that the EPS present on the surface of cells or in slime trails (see below) serves as a receptor that is required to trigger pilus retraction, pulling cells forward (see below) (72). This hypothesis can explain how cells maintain their cohesiveness and follow slime trails, but it cannot explain how colonies of M. xanthus can advance beyond slime trails. Alternatively, the pili might bind nonspecifically to surfaces, although if that was the case, cells should also be able to move when isolated. This is apparently the case when cells are placed in 1% methylcellulose, which might simulate EPS and trigger pilus retraction.
Type IV Pili Power S Motility
TFP are typically 5 to 7 nm in diameter and can extend to 5 μm or more in length (Fig. 4B). The mechanism of TFP-mediated motility is also known as twitching motility, a term that describes the “flagella-independent surface motility” (77) shared by several Gram-negative bacterial species, including Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Vibrio cholerae (40, 44, 115, 126). In this review, the reader will also find the term “swarming,” which we will use to describe the movement of cells in groups or colony spreading. In 1980, Bradley proposed that twitching motility was powered by pilus retraction (18). According to this hypothesis, the pili would bind to the surface of another cell or to the substrate, and then pilus depolymerization would pull the cell forward. He observed that only Pseudomonas aeruginosa strains with retractile pili were able to move, whereas those with no pili or nonretractile pili remained stationary (18). Twenty years later, evidence for this hypothesis was obtained by Skerker and Berg, who observed the extension and retraction of TFP in P. aeruginosa cells labeled with an amino-specific Cy3 fluorescent dye (115), and by Sun et al., who analyzed the motion of cells tethered by pili in M. xanthus (121). Apparently, the retraction of a single pilus provides sufficient force to pull cells forward. Indeed, the use of an optical trap to measure the force of retraction of pili in Neisseria gonorrhoeae showed that the force generated was very high, over 100 pN, one of the strongest molecular motors known (26, 72). The use of both polyclonal antibody and Cy3 staining showed that M. xanthus TFP localize at the leading pole of moving cells, similar to the case for P. aeruginosa (71, 87).
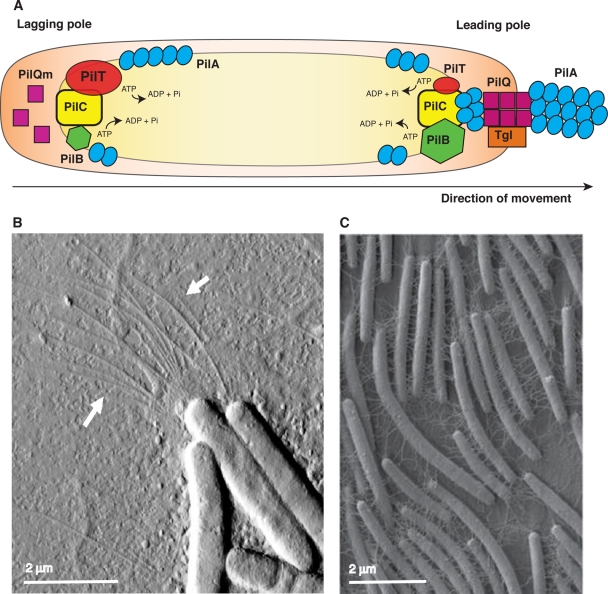
FIG. 4.
Components of the S-motility system. (A) A model showing the different components of the S-engine. The inner membrane PilC is present at both the leading pole and the lagging pole in equal amounts (19). The ATPase PilB is more abundant at the leading pole, where it catalyzes the polymerization of the TFP by hydrolyzing ATP. PilT is also an ATPase but with an opposing function: it catalyzes the disassembly of the TFP. PilT localizes mostly at the lagging pole. It is also present at the leading cell pole in smaller amounts to power the retraction of the TFP. The combined activities of PilB and PilT lead to the periodic assembly and retraction of TFP, which allow the cell to move forward. The lipoprotein Tgl is present only at the leading pole, and this causes the secretin PilQ to be assembled at the leading pole and disassembled at the lagging pole. The presence of PilQ channels at the leading pole allows the assembly of PilA filaments from the pool of monomers stored in the membrane or synthesized de novo. (B) Polar TFP. (The picture was obtained by atomic force microscopy and adapted from reference 97.) (C) Dried extracellular matrix material appears as fibrils that interconnect cells when viewed by scanning electron microscopy. (The picture of the M. xanthus fibril material was adapted from reference 60 with permission from Elsevier.)
Assembly and Retraction of Type IV Pili
The similarity between M. xanthus TFP and TFP from Pseudomonas and Neisseria has greatly facilitated the identification of many S-motility proteins. PilA, a 23-kDa protein, is the major subunit of the M. xanthus pilus filament (134). PilA monomers are anchored to the inner membrane through their 25 hydrophobic N-terminal residues. Prior to assembly, the PilA monomers are processed by the prepilin peptidase, PilD, which cleaves off the N-terminal leader sequence of the protein (49). The processed PilA monomers are polymerized by PilB, a cytosolic hexameric ATPase, and translocated through the outer membrane by the PilQ secretin and the Tgl lipoprotein (Fig. 4A). In contrast, TFP retraction and disassembly are mediated by PilT, a PilB homolog with an opposing function (49). The molecular mechanisms by which PilB and PilT extend and retract the pili are unknown. It is presumed that the binding of PilB to ATP followed by ATP hydrolysis provides the energy required for the insertion of PilA subunits into the growing pilus from a reservoir of pilin subunits in the inner membrane. Then, upon the stimulation of pilus retraction, PilT catalyzes the depolymerization of pili by inserting pilin subunits into the inner membrane. However, alternative models have been proposed based on the observation that certain biological fibers assemble and disassemble spontaneously (76). For example, hydrolysis of ATP might be required only for either assembly or disassembly, whereas the ATP binding of the second enzyme might be required only for regulatory functions to stimulate the switch to disassemble or assemble (49, 85).
It has recently been observed that the PilB and PilT ATPases show bipolar asymmetric localization patterns in M. xanthus cells (19). However, PilB localizes primarily at leading cell pole, whereas PilT forms a large cluster at the lagging cell pole with periodic localization at the leading cell pole (Fig. 4A). A model has been proposed in which PilB localizes primarily at the leading cell pole where TFP are assembled, while PilT localizes primarily at the lagging cell pole; the periodic accumulation of PilT at the leading pole would then be associated with TFP retractions, whereas the function of large PilT clusters at the lagging pole is not clear (19). The fact that cells lacking PilT show TFP only at the leading pole suggests that PilT does not simply function to inhibit the extension of TFP at the lagging pole (19). Interestingly, PilB and PilT localization depends on their ATPase activity and on the Frz pathway (see below) (19).
The Secretin PilQ and Its Cognate Lipoprotein Tgl
PilA is transported through the outer membrane via a channel composed of PilQ (Fig. 4A) (127). PilQ belongs to a large family of integral outer membrane secretin proteins that are essential for both pilus biogenesis and TFP-mediated motility (93, 126). Secretins often have cognate lipoproteins that facilitate their assembly in the outer membrane (92); Tgl is the lipoprotein necessary to assemble the PilQ secretin in M. xanthus. In the absence of Tgl, only the monomeric form of PilQ is observed and cells are not able to secrete PilA. Interestingly, cells with a tgl mutation can be stimulated to assemble PilQ channels and pili after being mixed with wild-type cells. The presence of Tgl in the tgl mutant cells after stimulation suggests that transient contact and fusion between outer membranes of Tgl+ donor cells and Tgl− recipient cells can result in the physical transfer of Tgl protein from one cell to another (92). Whether membrane fusion between neighbor M. xanthus cells occurs remains unproven. Recently, Palsdottir et al. showed that M. xanthus cells release large amounts of vesicles, and they suggest that such vesicles may function as vehicle for the transfer of Tgl between cells (96).
Tgl localizes at one pole, presumably the leading pole, since it is required for assembly of the PilQ channel and, consequently, pilus assembly (93). In contrast, PilQ is bipolar; its localization is Tgl independent. Thus, only the leading pole should contain the assembled (functional) form of PilQ (19, 93) (Fig. 4A).
EPS and Fibril Material
The M. xanthus extracellular matrix or fibril material was initially identified and characterized by Arnold and Shimkets (1). By using electron microscopy (EM), they observed thick “flaccid filaments” of 50 nm in diameter that they called fibrils. Later studies revealed that these fibrils formed a coat of extracellular matrix material around the cell surface (Fig. 4C). This material was composed of proteins and carbohydrate in a 1.0:1.2 ratio (4). The carbohydrate portion (extracellular exopolysaccharide [EPS]) was composed mostly of the monosaccharides galactose, glucosamine, glucose, rhamnose, and xylose (4), but its macromolecular structure was never determined. The protein fraction of isolated fibrils is composed of 21 proteins (28). The most abundant protein associated with the extracellular matrix is FibA, a zinc metalloprotease of the elastase family (5, 58). It was shown to have a function associated with fruiting body formation and response to membrane phospholipids containing the fatty acid 16:1ω5c (14, 15, 29, 61). The spore coat protein U (37), the protease B (another zinc metalloprotease) (99), and two amidohydrolases are also associated with the protein fraction of the extracellular matrix (28). The remaining identified proteins have unknown functions (28).
Genes responsible for encoding and assembling this matrix material were identified by screening for calcofluor white binding activity (73, 100). The biogenesis of extracellular fibril material was shown to require a DnaK homolog encoded by sglK (130); the eps operon, which encodes many genes involved in polysaccharide biosynthesis (73); a set of chemotaxis homologs encoded by the dif genes (68); and the eas locus, which is of unknown function (73). Mutants lacking any of these genes are able to form TFP and LPS but do not produce EPS and are incapable of performing S motility.
The carbohydrate portion of the extracellular fibril material appears to be the receptor or anchor for TFP, triggering pilus retraction and enabling S motility (72). Evidence to support this hypothesis came from the study of fibril-defective mutants, which are hyperpiliated, suggesting that in the absence of fibrils, TFP extend but cannot retract. However, retraction could be stimulated by mixing the mutant cells with wild-type cells, by adding protein-free fibril material, or by adding chitin (a GlcNAc polymer) (72). Interestingly, the polysaccharide requirement for TFP-mediated motility is not restricted to M. xanthus. For example, P. aeruginosa TFP bind specifically to the carbohydrate sequence βGalNAc(1-4)βGal found on eukaryotic cell surfaces (108).
A MOTILITY
In 1979, Hodgkin and Kaiser showed that M. xanthus cells lacking a functional S-motility system were still able to move as isolated cells at the edges of colonies (Fig. 2), indicating the presence of an additional motility system which they called A motility (41). Extensive genetic screens, especially by the Hartzell and Kaiser laboratories, have identified numerous genes involved in A motility (143, 144, 146). Specifically, over 35 genes were identified as being involved in A motility, although none of these genes encoded obvious motility structures or machinery (143). Indeed, A motility best fits the definition of “gliding” motility, defined as “a translocation along solid bodies … during which no wriggling, contraction or peristaltic alterations are visible, the change of shape being restricted to bending …” (20). Over the years, many models have been suggested to explain A motility, but despite several decades of research, the molecular basis of A motility remains elusive. Readers interested in the history of A-motility models may refer to reference 86. In this review, we will discuss two recent and opposing hypotheses that have become a matter of controversy. Evidence supporting each mechanism will be discussed.
Analysis of A-Motility Mutants
The first screen of M. xanthus motility mutants identified strains lacking motility as isolated cells but still capable of moving in groups (A−S+). Surprisingly, some of these mutants showed restored motility when mixed with wild-type cells, while others did not. Therefore, these mutants were classified in two major subclasses: agl and cgl (41, 42). If the defect in single-cell gliding of an A-motility mutant could be complemented by mixing with wild-type cells or mutant cells of another motility class, then the gene was designated cgl (contact-transient gliding) (42, 103). A-motility mutants that could not be rescued were named agl (adventurous gliding) mutants (41, 132, 143). Examples of each subclass are the cglB and aglU genes, which encode lipoproteins. Interestingly, although an aglU mutant cannot be complemented for motility extracellularly, it can be a donor to complement the defect of a cglB mutant (132).
More recently, the genetic screens conducted by the Hartzell and Kaiser laboratories using the transposable element mariner allowed identification of sites of insertional mutagenesis and sequencing of the genes involved. These screens identified over 35 genes involved in A motility (143). The genes fell into several classes: (i) genes that encode membrane proteins with a large representation of Tol/Ton-like protein complexes, which are known to be involved in macromolecule transport, membrane integrity (Tol), and import of ferric-siderophore complexes (Ton) in Gram-negative bacteria (143); (ii) genes predicted to encode enzymes involved in the biosynthesis of polysaccharides (143); and (iii) genes with unknown functions. Unfortunately, none of the mutants in these screens identified a set of genes encoding an obvious molecular engine. Thus, the mechanism of A motility remains open to different hypotheses and subject to different models, which are discussed below.
The “slime gun” model.
The “slime gun” model originated from the observation that A motility is associated with the deposition of visible trails of extracellular polysaccharide material, referred to as “slime.” What is slime? We actually know very little about this extracellular material because of the difficulty of separating it from other secreted polymers released and deposited by actively gliding cells. Furthermore, the search for mutants defective in slime production and/or secretion has not been successful (146). One reason for this failure might be that redundant enzymatic activities are required for the synthesis of slime. Alternatively, it has been suggested that slime production might be essential, shielding cells from their own extracellular degradative enzymes (55).
Time-lapse microscopy of moving cells revealed that slime becomes visible at the lagging ends of cells and that the trails elongate as the cells move forward. When cells reverse their direction of movement, they usually follow previously laid trails, showing that cells glide preferentially over slime (21). Early work suggested that molecular machineries at the lagging pole might push cells forward by actively secreting slime (48). This motility model was recently expanded by Wolgemuth et al. (133), who proposed that the extruded polymer is a polysaccharide with the chemical properties of a polyelectrolyte gel (133). As the polymer is synthesized from precursors at the inner membrane, it is transported across the cell wall in a dehydrated form within a secretion organelle located in the outer membrane. Within the secretion apparatus, the polymer swells as it progressively becomes hydrated. According to this model, the hydration process creates a compressive force that propels cells forward upon exit of the chamber (Fig. 5) (133).
FIG. 5.
Slime secretion model for A motility. A slime polymer (yellow) is synthesized within a putative inner membrane-localized biosynthetic machinery (pink), dehydrated, and introduced into a nozzle (blue) embedded within the peptidoglycan. Water flowing from the extracellular space creates a gradient of hydration leading to the swelling of the hydrogel within the nozzle chamber. Release of the hydrogel at the outer membrane creates a force (black arrow) that pushes the cell forward. (A) EM image of ribbons at a M. xanthus cell pole. (B) Average of side-view projection images of the Phormidium nozzle, a 40-nm-long symmetric open complex with a central hole of variable diameter (8 to 14 nm). (C) Rings observed in the M. xanthus outer membrane. (All panels were adapted from reference 133 with permission from Elsevier.)
The slime gun model was also invoked to explain gliding motility by the cyanobacteria Phormidium and Anabaena spp. (43). In these organisms, multicellular filaments glide coordinately over solid substrates and leave trails of slime that appear to be secreted at cellular junctions within the filaments, where circumferential rings of electron-dense pores (termed nozzles or junctional pores) were also observed (43). India ink staining of the mucus secreted by live cells appeared to originate at the cross walls, suggesting that the junctional pores were the actual sites of slime extrusion (43). Outer membrane complexes could be partially purified, and EM images showed both side and end-on views. The complexes had the shape of an “amphora” opened at both ends with 2-fold mirror symmetry (Fig. 5B). Importantly, the dimensions of the nozzles are comparable to those of outer membrane channels assembled by proteins belonging to the “secretin” family (23, 27). Thus, it cannot be excluded that the observed pores are in fact multiple secretin complexes.
The cyanobacterial junctional pores inspired the search for comparable structures in M. xanthus. M. xanthus cell envelopes also contained electron-dense pores that seemed to be enriched at the cell poles (Fig. 5C) (133). On EM grids, “ribbons” seemed to form at the cell poles, which led to the hypothesis that they were secreted through the observed pores, as suggested for Phormidium and Anabaena (Fig. 5A) (133). On the assumption that the secretory machineries in Phormidium and M. xanthus were structurally similar, it was calculated that the slime secretion produces a thrust that would account for the measured velocities of myxobacterial and cyanobacterial movements (133). Unfortunately, M. xanthus mutants that lack nozzles or that have defective nozzles have never been isolated, nor have the nozzle proteins been identified. Thus, it is still not clear whether the myxobacterial slime nozzles and the cyanobacterial septal pores are indeed similar structures or whether they share similar motility functions.
Another objection to the slime gun hypothesis is that it predicts that the engine for A motility, slime secretion, should be focused at the lagging cell pole. Sliusarenko et al. (118) tested this prediction by treating cells with cephalexin, thereby blocking cell division and causing cells to form long flexible filaments or “snakes.” These snakes continued to move by A motility, with motion from the leading cell pole preceding that of the lagging cell pole. Time-lapse videos of these cells were consistent with distributed motors rather than motors limited to the lagging cell pole (Fig. 6A).
FIG. 6.
Focal adhesion model for A motility. Focal adhesion complexes (green) are assembled at the leading cell pole along the MreB cytoskeleton (red). The complexes appear to retain fixed positions as an associated motor pulls on the MreB filament. The blow up shows a cartoon of the proposed machinery. An unidirectional engine (green) pulls the MreB filament on one side and is connected to an envelope-spanning protein system (pink) that ends with an adhesin (blue). Movement is produced because the inner and outer semifluidic membranes “flow” through the complex and a machinery-bound peptidoglycan hydrolase (purple) digests the cell wall locally. (A) Gliding motility of a cephalexin-treated cell. The cell unfolds as if it is pulled by its front part while the back part remains inert. The cell is stained with the membrane FM4-64 dye and shows no obvious septa. (The picture of the M. xanthus cephalexin-treated cell was adapted from reference 118.) (B) Fixed AglZ-YFP clusters in a moving cell. Frames of a movie (30-s intervals) showing a moving M. xanthus cell expressing AglZ-YFP (artificially colored in magenta) are shown. Black arrow, direction of movement. Scale bar = 2 μm. (The picture of the AglZ-YFP-labeled M. xanthus cell was adapted from reference 89.)
The “focal adhesion complex” model.
Recently, an alternative model, resulting from cytological observations of AglZ, an A-motility protein (143), was proposed to explain A motility. AglZ, discovered by the Hartzell laboratory, is a protein important for A motility (143). AglZ is structurally similar to a protein important for S motility swarming, FrzS (128) (see below), in that it contains an N-terminal pseudoreceiver domain and a long C-terminal coiled-coil domain (88, 89, 143). It has been recently shown to have a regulatory function during A motility (see below) (80). Fully motile cells showed that AglZ fused to yellow fluorescent protein (AglZ-YFP) was present in clusters distributed in an ordered array spanning the length of cells (89). As cells moved forward, these clusters maintained fixed positions with respect to the agar surface rather than to their relative positions in the cell (Fig. 6B). Interestingly, the only AglZ-YFP cluster that moved relative to the cell body was the one located at the leading pole, suggesting that new clusters were assembled at the leading pole. When cells reversed, AglZ localized rapidly to the new leading pole, after which it was distributed in clusters.
The dynamics of AglZ-YFP localization suggested an alternative mechanism for A motility in which intracellular motor complexes that connect to membrane-spanning adhesion sites and to the cytoskeleton power motility by pushing against the substratum and moving the cell body forward (Fig. 6). According to this model, the AglZ clusters only appear to be stationary but are actually moving in the direction opposite that of the cell body. The movement of clusters must consume energy that is then converted into locomotion. Cluster dynamics are believed to reflect the activity of as-yet-uncharacterized protein motors that are anchored to envelope-spanning substrate adhesion systems that pull on an internal cytoskeleton (Fig. 6).
Several lines of evidence suggest that adhesion occurs at clusters sites. (i) First, when moving cells bend, AglZ clusters are found to be localized at the sites where bending occurs (89). (ii) Furthermore, when a cell is blocked from moving forward, for example, because it confronts an obstacle, it undergoes characteristic “flailing” movements as the A engine keeps pushing against the obstacle. In these blocked cells, the AglZ clusters form between the bends; following these changes in cell shape, the clusters are dispersed at the back of the cell. These movements are consistent with tight anchor points binding the substrate (89). Evidence that the force powering cell movements is applied at the focal adhesion complexes comes from studies performed using 30-μm-long cephalexin-induced filaments (snakes). In these cells, the number of clusters is largely independent of the cell length but is proportional to the cell velocity (more exactly, the “drag force overcome” or the force necessary to power locomotion of a cell of given length and velocity) (89). These experiments further suggest that in order to produce movement in the snakes, the A motor cannot be localized at the rear of the cell but must be distributed along the cell body (118, 120).
Evidence that the cytoskeleton participates directly in A motility was recently provided by analysis of M. xanthus MreB. MreB is an actin-like protein that is important for cell division and cell shape determination in rod-shaped bacteria (36, 52, 66, 79). It is a dynamic cytoskeletal protein that forms helices with a pitch of about 0.5 μm (31, 66, 79). Indeed, deconvolution microscopy revealed that M. xanthus contains a double-helical MreB filament that spans the length of the cells (79). The pitch of the M. xanthus MreB helices was measured to be 0.47 ± 0.1 μm (79). This spacing is almost identical to the distribution of the AglZ-YFP complexes, suggesting that the MreB cytoskeleton might provide a scaffold to anchor the proposed adhesion sites (79, 89).
The involvement of the MreB cytoskeleton in A motility was demonstrated by treating gliding cells with the MreB-perturbing agent A22 (79). A22 is a small molecule that prevents MreB polymerization by occupying the ATP binding sites of MreB monomers (36). Since MreB filaments are very dynamic, this treatment effectively causes the MreB cytoskeleton to depolymerize. Interestingly, treating moving M. xanthus cells with A22 also caused the rapid arrest of motility and the dispersal of AglZ clusters (79). AglZ and MreB were shown to interact in vitro, confirming a direct link between these proteins (79). These experiments establish that the cytoskeleton is required for A motility and is essential for the positioning of the focal adhesion complexes. In the future, it will be important to test whether active traction occurs on the MreB filament.
Hybrid models.
Published data support the hypothesis that both slime and focal adhesion complexes have a function in A motility. The controversy is, therefore, about which component actively produces motility. Since definitive evidence for either model is still lacking, could slime secretion and focal adhesion complexes operate together to generate A motility? Three hybrid models to explain the data can be considered. (i) A motility is powered by distributed focal adhesion complexes; the complexes require slime which is secreted underneath the cell body to provide specific adhesions and limit viscous forces at the surface of the substrate. In our opinion, this model best fits the current data. (ii) Slime jets propel the cells forward, and focal adhesion points help dissipate friction forces that accumulate along the cell body as the cell is pushed forward. The focal adhesions could glue the cell surface to the substrate, thus acting as molecular ratchets that dissipate the stress that would oppose forward motion. However, this model predicts that if slime jets were still working in mutants defective in focal adhesion complexes, abnormal yet detectable movements would still occur. These movements are not observed in cells with absent or nonfunctional focal adhesion complexes (79, 89, 143). (iii) Slime jets and focal adhesions both actively promote locomotion. Genetic evidence does not favor this hypothesis because it predicts that a complete block in A motility necessitates mutations in both motor systems, whereas single mutations that abolish A motility can be readily isolated (143, 146).
GLIDING MOTILITY IN OTHER BACTERIAL SYSTEMS
Surface motility is found in many distantly related bacterial species (20), but it is not clear whether these organisms simply utilize different motility mechanisms or whether gliding motility has a common origin involving a universal mechanism which then diverged in different motility systems (50, 86). Clearly, the mollicutes, a class of bacteria distinguished by the absence of a cell wall, have evolved distinct motility mechanisms to accommodate their unusual surface architecture (86, 90). What about other bacterial species? The most commonly studied and perhaps most frequently encountered gliding systems are similar to M. xanthus S motility, as they utilize TFP binding and retraction to move over solid surfaces. This “twitching motility” has been studied in Gram-negative species, such as P. aeruginosa, N. gonorrhoeae, and V. cholerae, and is also found in Gram-positive species such as Clostridium perfringens (40, 108, 123, 126), probably reflecting a widespread mechanism in nature. One advantage of this motility system, which is frequently found in pathogens, is that it provides a mechanism for cell movement over solid surfaces and specific attachment to host cells.
Whether A-motility-like surface translocation is a conserved process remains an open question. Genetic studies do not favor the existence of an universal A-motility machinery; M. xanthus A-motility genes do not seem to be very conserved between gliding systems. For example, there is relatively little overlap between the motility genes that are found in M. xanthus and those that are found in Flavobacterium johnsoniae (81, 143). Also, F. johnsoniae moves 60-fold faster than M. xanthus (2 to 4 μm/s versus 2 to 4 μm/min), suggesting the presence of a more efficient motility apparatus. Analyses of mutants defective in motility revealed the presence of unique proteins of extremely high molecular weight on the surface of F. johnsoniae cells. One of these, SprB (669 kDa), seems to be of special interest, as anti-SprB antibodies completely blocked cell movement. Furthermore, latex beads carrying anti-SprB antibodies were observed to adhere to and be propelled along the cell surface at approximately the same rate as cell velocity on agar (69, 91). Jarrell and McBride published a recent review of these systems (50).
Bacterial motility over solid surfaces may have evolved multiple times, but there still may be core components that are conserved. Unfortunately, except for M. xanthus and Flavobacterium/Cytophaga, most gliding organisms are currently genetically intractable, limiting research with these organisms to microscopy. These studies showed helical ribbons at the surface of several gliding bacterial species, including M. xanthus, and, as already discussed, slime secretion (74, 97). The role of these structures in motility remains to be established. In M. xanthus, the finding that MreB is critical for A motility (79) may also suggest that there are universal features common to several motility systems.
DIRECTIONAL CONTROL AND CELL POLARITY
Reichenbach (101), in his pioneering time-lapse films of moving myxobacteria, showed that individual M. xanthus cells not only move slowly at a speed of approximately 2 μm/min in slime trails (51) but periodically reverse. Wild-type cells usually reverse about once every 7 to 14 min, depending on the culture conditions and the strain used. Cell reversals in M. xanthus are complex and need to be finely regulated, as they require a complete inversion of cell polarity: the leading cell pole becomes the lagging cell pole, and the lagging cell pole becomes the leading cell pole. Thus, all of the proteins and structures that show unipolar localization need to be relocated to the opposite cell pole. In the case of S motility, many components are fixed at both cell poles, whereas the pili and other S-motility components must be periodically disassembled and reassembled at the new leading pole (19). For example, FrzS, a regulator of S motility, appears to be actively translocated from pole to pole along a helical trajectory that may involve movement along a cytoskeletal filament (see below) (70, 79, 87). In contrast, the A-motility engine is distributed along the cell body. However, the A-motility engine also functions in a polar manner, as the focal adhesion complexes are always generated at the leading cell pole and A-motility proteins are translocated from pole to pole in order to regulate directional motility (89).
Cell reversals and the coordination of the two motility systems are needed to achieve directional movements, for example, during cellular aggregation, when cells form fruiting bodies, or during rippling behavior, when thousands of cells align and periodically reverse (9). Control of cell reversals and the coordination of the two motility systems are achieved by a set of chemotaxis-like proteins encoded by the frz operon (Fig. 7). The ability of cells to reverse direction (and polarity) is thought to be required for cells to reorient themselves as part of a biased random walk, in much the same way that changing the rotation of flagella in enteric bacteria causes tumbles, allowing cellular reorientation (17, 148). However, while cells traveling in groups show directional (chemotactic) movements toward nutrients, such as a mix of peptides found in Casitone medium, the biased movement of isolated cells toward common nutritional “chemoattractants” has never been observed (109). Thus, the Frz system mediates positive chemotactic responses in M. xanthus that differ significantly from the enteric paradigm, and these responses are limited to when individual cells are in contact with each other or when cells interact in swarms (8, 33).
FIG. 7.
Schematic diagram of the Mcp7, Dif, Frz, and Che4 chemotaxis systems of M. xanthus. These chemosensory systems are important in the regulation of M. xanthus motility. Chemosensory proteins might form complexes analogously to their enteric counterparts. Dashed arrows indicate the cross talk between the different chemosensory systems.
The Frz Chemosensory System
The frz (frizzy) genes constitute one of eight chemotaxis-like operons of M. xanthus. The frz genes were first discovered in a screen for mutants defective in aggregation during development because cells failed to form discrete mounds and instead aggregated into “frizzy” filaments that never matured into fruiting bodies (147). The formation of “frizzy” aggregates was not due to defects in the two motility systems but rather was due to the inability of cells to control their reversal frequency during gliding motility (13). For example, while wild-type cells reversed their direction of gliding about every 7 to 14 min on low-nutrient agar, frz null mutants reversed very infrequently, about once every 1 to 2 h (13); in contrast, some constitutively signaling frzCD mutants reversed very frequently, about every 2 min. Although the A- and S-motility systems are functional in frz mutants and isolated frz cells move at the same rate as wild-type cells (13), the loss of the ability to control the cell reversal frequency results in the loss of cell-cell coordination and directed group movements. Thus, vegetative swarming and developmental aggregation are defective in the frz mutants (13).
Sequence analysis of the frz genes performed by McBride et al. (82) showed that the frz genes encoded proteins that are highly similar to the chemotaxis proteins of enteric bacteria (Fig. 7 and Table 1). For example, the frz operon encodes FrzCD, a cytoplasmic methyl-accepting chemotaxis protein (MCP), and the CheW-like coupling proteins FrzA and FrzB (82) (Table 1). Further sequencing showed that the frz operon also contains genes encoding FrzE, a CheA (histidine kinase)-CheY (response regulator) fusion protein (84); FrzF, a CheR (methyltransferase); FrzG, a CheB (methylesterase); and FrzZ, a protein with two CheY domains (22, 47, 83) (Table 1). All of these genes, when mutated, caused aberrant cellular reversal frequencies.
TABLE 1.
Similarity and identity between E. coli Che protein and M. xanthus Frz proteins
| M. xanthus Frz protein | E. coli Che protein | Similarity (%)a | Identity (%)a |
|---|---|---|---|
| FrzCDMA | TarMA | 50 | 30 |
| FrzECheA | CheA | 53 | 33 |
| FrzECheY | CheY | 60 | 36 |
| FrzZCheY1 | CheY | 49 | 27 |
| FrzZCheY2 | CheY | 47 | 26 |
| FrzA | CheW | 48 | 32 |
| FrzB | CheW | NDb | ND |
| FrzB | FrzA (M. xanthus) | 52 | 34 |
| FrzF | CheR | 49 | 30 |
| FrzG | CheB | 52 | 30 |
Determined by using BLAST (www.blast.ncbi.nlm.nih.gov).
ND, not determined.
Signaling through the Frz Pathway
FrzCD is the MCP receptor associated with the Frz chemosensory system. It is encoded by a single gene, frzCD. Prior to DNA sequencing, it was thought that the frzCD locus consisted of two genes because transposon insertions in the first part of the locus, frzC, caused hyporeversing mutants while insertions in the second part of the locus, frzD, resulted in hyperreversing cells. FrzCD is an unusual chemoreceptor because it lacks the transmembrane and periplasmic domains typical of enteric MCPs. It contains two major domains: a unique N-terminal domain that lacks homology to other protein domains and a C-terminal domain that has good homology with the methylation and signaling domains of the enteric MCPs. While the unique N-terminal region of FrzCD has been shown to be important for the regulation of A motility (see below), its deletion results in only minor defects in S motility and development (22, 80). However, the methylation of the conserved C-terminal domain of FrzCD has proven to play a central role in regulating the cell reversal frequency, as well as S motility and development (2, 107).
Reversible methylation of chemoreceptors on glutamate residues enables bacteria to sense subtle changes in stimuli and is important for adaptation. To determine the roles of these sites in vivo, E-to-D and E-to-A mutations were created in the methylation sites of FrzCD and their impact on single cell reversals, swarming, and fruiting body formation determined. Analyses of single, double, and triple methylation site mutants revealed that each site plays a unique role in M. xanthus behavior and that the pattern of receptor methylation determines receptor activity. While certain methylation site mutants cause stimulation of the Frz pathway, leading to hyperreversing cells, other mutants inhibit Frz signaling and result in cells that rarely reverse (2, 107).
FrzF is the enzyme that methylates FrzCD. It also contains two domains: an N-terminal domain that is very similar to CheR from E. coli and a C-terminal domain containing three tetratricopeptide repeats (TPRs). These TPR domains are thought to play a significant role in regulating the methyltransferase activity of FrzF. In fact, in vitro assays showed that while full-length FrzF can methylate FrzCD at only one residue (E182), FrzF lacking the TPR domain could methylate FrzCD at three residues (E168, E175, and E182) (107). This shows that the TPR domains of FrzF have a regulatory role and modulate methylation at specific sites. How the methylation of FrzCD affects the level of activation of the downstream histidine kinase, FrzE, has yet to be determined.
FrzE is a two-domain protein containing an N-terminal histidine protein kinase similar to CheA and a C-terminal receiver domain. As shown in Fig. 7, when FrzCD is stimulated (in conjunction with the coupling protein FrzA), it causes FrzE to autophosphorylate at H49 and then transfer the phosphate to FrzZ, a protein consisting of two response regulator domains, at positions D52 and D220 (46, 47). Interestingly, the receiver domain of FrzE strongly inhibits the autophosphorylation of the CheA domain. This suggests that the receiver domain of FrzE is regulated by other, still-uncharacterized proteins (46). A genetic screen indicated that FrzZ may be the only direct output of the Frz system (47). However, how the Frz system communicates with the two motility motors is still unclear. Based on the enteric paradigm, we predict that FrzZ-P should be critical for this function. The target of the enteric counterpart, CheY-P, has been shown to be the switch component of the flagellar motor, FliM. However, M. xanthus does not contain a FliM homolog. Identifying the FrzZ-P interacting partners will supply an important missing component in the signaling pathway.
FrzS, an S-Motility Protein with Pole-Specific Localization
FrzS was discovered by Ward et al. (128) while they were examining proteins encoded by genes linked to the frz operon. frzS mutants are impaired in S-motility swarming because they are defective in regulating pilus-mediated directional movements. Mignot et al. (87), while investigating the in vivo localization of FrzS fused to green fluorescent protein (FrzS-GFP), showed that the protein was localized in polar fluorescent patches but that most of the fluorescence was concentrated at the leading, piliated cell pole. During a reversal cycle, fluorescence at the leading pole slowly decreased while gradually increasing at the trailing pole. When fluorescence at the leading pole dispersed, the cells reversed direction, and the new leading pole contained the main fluorescent focus (Fig. 8). Since FrzS is localized at the piliated pole, its oscillation from one pole to the other is thought to be related to the disassembly of TFP from the old leading pole and reassembly of pili at the new leading pole (87).
FIG. 8.
The localizations of A- and S-motility proteins change synchronously as cells reverse. Time-lapse fluorescence microscopy of a moving M. xanthus cell expressing FrzS-GFP and RomR-mCherry is shown. The top panels show fluorescence images. The bottom panels show the same cell in bright field. The cartoon shows schematically the asymmetric bipolar localization of FrzS (green) and RomR (red). The FrzS larger cluster is at the cell leading pole while the RomR larger cluster is at the lagging pole. During a reversal, proteins redistribute and appear in equal amount at both poles. As soon as the cell starts moving again, polarity is reestablished and FrzS appears at the new leading pole, whereas RomR localizes at the lagging end of the cell. (The picture of the FrzS-GFP RomR-mCherry-labeled M. xanthus cell was adapted from reference 70 by permission from Macmillan Publishers Ltd.)
The mechanism by which FrzS shifts from one pole to another has been clarified by a detailed analysis of the major domains present in the protein. FrzS consists of an N-terminal pseudoreceiver domain and a long coiled-coil C-terminal domain (128). An in-frame deletion of the receiver domain in a strain that expressed FrzS-GFP showed an S-motility defect as severe as that seen in a frzS deletion mutant (88). However, this strain showed that the truncated GFP fusion protein was now preferentially localized to the lagging cell pole rather than the leading cell pole. More precisely, in the absence of the receiver domain, the truncated FrzS was still targeted to the new leading pole during a reversal; however, without the receiver domain, the protein rapidly redistributed to the lagging pole (88).
What is the role of the Frz pathway in FrzS oscillations? FrzS oscillations seem to reflect the signaling activity of the Frz pathway, since frz hyporeversing strains rarely show FrzS switching from one cell pole to the other, whereas frz hyperreversing strains show extremely frequent FrzS oscillations (87). Genetic analyses and construction of double mutants suggest that FrzS is downstream of the Frz pathway (Fig. 7). However, it is very unlikely that FrzE∼P is the source of phosphate for FrzS phosphorylation, as FrzS lacks the conserved aspartate residue that is phosphorylated in canonic receiver domains, and moreover, the S motility of a strain mutated in what would be the most likely phosphorylation site (D55) is indistinguishable from that of the wild type (35, 88). The exact relationship between the Frz pathway and FrzS remains to be determined. However, it is clear that the protein plays an important function in the regulation of the reversals of the S-motility system.
The A-motility protein AglZ is similar to FrzS: AglZ also has an N-terminal pseudoreceiver domain and a C-terminal coiled-coil domain. Furthermore, the protein switches polarity coordinately with cell reversals. However, a recent genetic study showed that AglZ is more likely to act upstream of the Frz pathway in the regulation of A motility (see below), while FrzS is likely to act downstream (80).
RomR, an A-Motility Protein with Pole-Specific Localization
The discovery of the A-motility protein RomR occurred as part of an analysis of genes required for fruiting body formation (70). The romR mutant was defective in A motility but also showed a small S-motility defect. Cytological studies revealed that RomR and FrzS show divergent localization patterns and dynamics within cells. For example, RomR-mDsRed showed a bipolar asymmetric localization. However, in contrast to the case for FrzS, the brighter clusters of RomR localized to the lagging cell pole (Fig. 8) (70). Like FrzS, RomR clusters switch polarity with cell reversals. RomR consists of an N-terminal response regulator and a C-terminal proline-rich output domain. However, unlike that of FrzS, the RomR response regulator domain likely undergoes cycles of phosphorylation and dephosphorylation. Indeed, mutations in the conserved phosphorylation site caused cells to show altered cell reversal frequencies. This result suggests that the phosphorylation of RomR plays a direct role in the regulation of reversal frequency during A motility (70).
What is the role of the Frz pathway in the RomR oscillations? Like for FrzS, RomR oscillations seem to reflect the signaling activity of the Frz pathway in that frz hyporeversing strains show infrequent RomR oscillations. RomR may act downstream of the Frz pathway to induce reversals in the A-motility system, as RomR mutants locked in the phosphorylated state are able to bypass the reversal frequency defect of frz mutants (70). It has been proposed that RomR controls slime secretion at the rear of the cell (70). However, it is also possible that RomR acts to inhibit the assembly of the distributed focal adhesion motors at the lagging pole. In a strain coexpressing FrzS-GFP and RomR-mDsRed, these proteins showed coordinated shifts in their localizations, suggesting that the switch in polarity of the A- and S-motility systems is controlled by a common regulator (70) (Fig. 8). However, RomR and FrzS localize and relocate between the cell poles independently of each other (70). This finding rules out the possibility that the polarity of one system drives the switching of the other system.
MglA, a Small GTPase, May Be an Output of the Frz Pathway
In 1979, Hodgkin and Kaiser (41) reported finding a gene, mglA (mutual gliding A), required for the functioning of both A- and S-motility systems (41). This was the only gene that they found that was required for both motility systems. mglA is cotranscribed with mglB and encodes a 22-kDa protein similar to monomeric GTPases of the Ras superfamily, a class of eukaryotic proteins that perform a broad range of functions, including transport, signal transduction, and cell migration (24, 38, 39). While mglA mutant cells appear at first to be completely nonmotile because they show no net translocation, time-lapse films show that they actually make spastic movements with extremely frequent uncontrolled reversals (119). These “nonmotile” cells were unable to sporulate, but recombinant clones that expressed the yeast SAR1 protein, a member of the Ras/Rab/Rho superfamily of GTPases, were able to rescue sporulation. This surprising result and the fact that MglA hydrolyzes GTP in vitro indicate that MglA may indeed be a member of the Ras superfamily (39, 79).
MglA may be a good candidate for an output that links the Frz pathway to both the A and S motors, especially since genetic analyses show that MglA is downstream from the Frz pathway (119). Additionally, small GTPases in eukaryotic cells often act as regulatory proteins by recruiting factors to their sites of action. Therefore, it is possible that upon activation by the Frz pathway, MglA recruits molecules that are involved in the reversal of the two motility systems. For example, MglA is essential for the localization of both FrzS and AglZ: in an mglA mutant, FrzS remains localized to only one cell pole, while AglZ is no longer found in clusters but is diffused in the cytoplasm. Furthermore, protein interaction studies show that MglA can directly interact with both FrzS and AglZ (79). MglA is also important for RomR localization, since in mglA mutant cells, RomR appears as a single cluster at the piliated pole, rather than asymmetrically bipolar with the larger cluster at the lagging pole (70).
REGULATION OF MOTILITY
The Dif Chemosensory System and Its Role in Controlling S Motility
The dif mutants were first identified in a genetic screen for mutations that caused defects in fruiting (139). difA was found to be clustered on the chromosome with difC, difD, difE, and difG, all of which encode homologs of bacterial chemotaxis proteins (139). DifA is similar to bacterial MCP receptors, DifE to CheA kinases, DifD to CheY response regulators, DifC to CheW coupling proteins, and DifG to CheC phosphatases. The Dif system does not contain CheR (methyltransferase) or CheB (methylesterase) homologs. Motility assays showed that difA, difC, and difE mutants displayed wild-type A motility but were deficient in S motility (A+S−); difD and difG mutants still possessed S motility, but their swarming was different from that of the wild type on soft agar (12). These studies indicate that the Dif proteins play critical roles in S motility.
Subsequent work showed that the Dif chemosensory proteins regulate EPS (fibril) production, which is essential for TFP-mediated motility in M. xanthus. For example, difA, difC, and difE mutants were found to produce no EPS (142). In contrast, difD and difG mutants showed increased EPS production (12). However, difD and difG mutations, either alone or in combination, failed to restore EPS production or S motility in a difE mutant (12). These results suggest that in this pathway, DifD (CheY-like) does not function downstream of DifE (CheA-like), as observed in most chemotaxis pathways. Despite this difference, yeast two- and three-hybrid experiments indicated interaction patterns among Dif proteins similar to the ones observed for other chemotaxis proteins (141). Based on these results and other bacterial chemotaxis paradigms, a model in which DifA, DifC, and DifE form a membrane ternary signaling complex was proposed (Fig. 7). The DifE kinase is proposed to transmit signals downstream to an unknown protein (DifX). In this model, DifD negatively regulates EPS production, possibly acting as a phosphate sink for DifE. DifG stimulates the function of DifD or DifD-phosphate (DifD-P). Results from preliminary in vitro phosphorylation studies with Dif proteins are consistent with this model, as DifE can autophosphorylate and then serve as the source of phosphate for DifD, while DifG is able to accelerate the dephosphorylation of DifD-P (Z. Yang, unpublished data).
Interestingly, mutants with defective pili are also defective in EPS production (11, 30). Moreover, the hyperpiliated pilT mutant was found to overproduce EPS in a quantitative assay (11). Genetic epistasis experiments with pil and dif mutations established that Dif proteins function downstream of TFP in the regulation of EPS production. It has been proposed that TFP filaments act as the sensor for the Dif pathway to stimulate EPS production. EPS on neighboring cells is the signal that is perceived and relayed by TFP and its membrane complex to the Dif system (11). This model presents a positive feedback loop regulated by intercellular signals in M. xanthus, which is known to display many different social behaviors resulting from cell-cell communication.
Recent genetic studies indicate that the Dif system interacts with the Che7 pathway in the regulation of EPS production (Fig. 7). For example, a genetic screen showed that the defects in EPS production and development of a difA mutant were suppressed by a mutation in cheW7, which encodes a coupling protein homolog of the che7 gene cluster (10). This suppression required DifC and DifE from the Dif system and Mcp7 from the Che7 system (10). Evidence indicates that Mcp7 competes with DifA (also an Mcp) for interactions with DifC (CheW-like) under certain conditions.
Interactions between Dif and Frz Chemosensory Pathways
Besides its role in EPS regulation, the Dif system has been shown to modulate the reversal frequency of starved M. xanthus cells (59). Starved M. xanthus cells are known to respond to certain phosphatidylethanolamine (PE) species as chemoattractants (15, 60); that is, the reversal frequency of single gliding cells is transiently suppressed by these PE molecules. This transient suppression of reversal, known as the excitation response, requires an intact Dif system. Analysis of M. xanthus cell movement in response to PE was performed by tracking isolated cells. Since isolated cells are presumably moving by A motility, the observed excitation response indicates that the reversal of the A-motility motor is responsive to the Dif chemosensory system (Fig. 7).
The response to PE also helped to uncover a more intimate interaction between the Dif and Frz systems. In typical bacterial chemotaxis, excitation is followed by adaptation (34). During adaptation, the motility behavior of stimulated cells returns to the prestimulus pattern when there is no further change in the concentration of the stimulus. In the M. xanthus response to PE, temporal adaptation occurs about 80 min after PE exposure (59). Although excitation by PE depends on the Dif pathway, adaptation relies on Frz, as frz mutants never adapt after excitation but are still able to suppress reversal, suggesting that the frz pathway is not important for excitation (59). These results indicate that the Dif and Frz pathways must interact physically or biochemically in order to coordinate their functions in the regulation of the M. xanthus gliding motors.
The evidence thus far is supportive of a model in which FrzCD methylation is responsible for adaptation to PE in M. xanthus. Indeed, exposure to PE has been shown to increase FrzCD methylation (82), and frzCD mutant cells show no temporal adaptation to PE in single-cell motility assays (59). The Dif and Frz systems can probably sense the presence of PE independently. While the signal from Dif promotes excitation, the signal from Frz leads to adaptation by increasing FrzCD methylation. These signals from two pathways are antagonistic at the level of FrzCD modification: FrzCD shows increased methylation in dif mutants relative to the wild type. It is suggested that the signals from the Dif pathway suppress adaptation by modulating FrzCD methylation and this suppression functions to prolong excitation to ensure a meaningful response on the spatial scale of the slow-moving M. xanthus (137).
AglZ Controls A Motility through the Frz Pathway
The Frz chemosensory pathway controls reversals for both A and S motility. To identify proteins that might provide a regulatory input to the pathway, FrzCD, the receptor for the pathway, was examined for interacting proteins. Unexpectedly, AglZ and AgmU, previously described A-motility proteins (138, 143), were identified as interacting with the unique N-terminal domain of FrzCD (80). These results were surprising since it was expected that the Frz pathway should provide an output to the motility systems rather than receive an input. aglZ and agmU mutants were clearly defective in A motility. However, aglZ frzCD and agmU frzCD double mutants showed restored A motility (80; unpublished data). These results show that AglZ and AgmU are not components of the motor powering A motility but rather are regulators of the A-motility system. However, this observation does not rule out the possibility that AglZ may be an important component of the A-motility focal adhesion complexes, which would be expected to include some motor proteins as well as regulatory factors. Therefore, AglZ provides us a valuable marker to follow the localization and dynamics of motor complexes in vivo. Moreover, these results indicate that the Frz pathway negatively regulates A motility, since motility is restored when this pathway is mutated in aglZ strains. Thus, AglZ (and probably AgmU) are upstream inhibitors that counteract this inhibitory activity (Fig. 7) (80).
To explore the interaction of these proteins in vivo, fluorescence microscopy was used to localize FrzCD-GFP and AglZ-mCherry in a strain coexpressing these labeled proteins. FrzCD-GFP was found to localize in dynamic clusters distributed throughout the cytoplasm except near the cell poles (78). In contrast, AglZ-mCherry clusters localized primarily at the leading cell pole or in regular distributed positions along the cell which were not occupied by FrzCD-GFP clusters (80). This suggests that any interactions between AglZ and FrzCD must be limited, perhaps only to cluster interfaces (80). Interestingly, fluorescence microscopy also indicated that when cells are placed under conditions that favor S motility, for example, in cell groups or isolated cells in methylcellulose, AglZ-mCherry appears diffuse or as a single polar cluster. This suggests that the putative AglZ-focal adhesion complexes are not assembled when cells are moving by S motility and, additionally, that the AglZ interactions with FrzCD might be reduced when cells are moving in groups. This suggests that the Frz pathway can inhibit A motility when the use of S motility is favored and predominant (80).
The Che4 Chemosensory System
M. xanthus has eight chemosensory systems, although only four have been described in print: Frz, Dif, Che3, Che4, and Che7 (10, 148). The Che3 system has been shown to be important for regulating gene expression during the transition between vegetative growth and development and does not appear to play a direct role in the control of motility (64). The Che4 system functions to control S motility (124). For example, in A−S+ strains, but not in a wild-type strain, a deletion of the entire che4 operon or cheY4 resulted in enhanced swarming on 1.5% agar surfaces. This suggests that the Che4 system negatively regulates swarm expansion under these conditions. In contrast, deleting only mcp4 caused reduced swarming. Single-cell analysis revealed a previously unknown inverse correlation between swarming rate and reversal frequency: cells that swarmed faster (cheY4) showed a reduced reversal frequency; cells with a reduced swarming rate (mpc4) showed an increased reversal frequency (124). Since the Frz system has been shown to be the master regulator of reversal frequency in M. xanthus, these results suggest that cross talk between the Frz and Che4 systems exists. This hypothesis is supported by yeast two-hybrid experiments showing that FrzCD interacts with CheA4 (H. C. Vlamakis and D. R. Zusman, unpublished data). Clearly more work needs to be done to elucidate the possible cross talk networks between the multiple M. xanthus chemosensory systems that may lead to regulation of motility.
COORDINATING CELL MOVEMENTS
In M. xanthus, the formation of organized multicellular aggregates, like ripples and fruiting bodies, is essential for social behaviors associated with predation and development (7, 116). These behaviors are all cell density dependent and must rely on the coordination of cell movements. Unfortunately, little is known about how cell-cell communication is achieved in M. xanthus.
In other bacterial systems, cell-cell communication usually involves the production and detection of secreted signaling molecules called autoinducers (129). These small-molecule signals are short peptides or acylhomoserine lactones, depending on the species. These molecules are diffusible, allowing bacteria to communicate over long distances, both within the bacterial species and between different species (3). In contrast, in addition to the response to certain soluble amino acids known as A signals during development (25, 57, 67, 112), M. xanthus appears to utilize signals requiring direct cell-cell contacts (63, 92). An example of a protein reported to be involved in contact-dependent signal transmission during fruiting body formation is the C signal, a 17-kDa peptide (63). Mutants that lack C signaling show a strong defect in fruiting body formation but no defects in vegetative swarming (62, 63). Another example of a molecule that can be exchanged by cell contact is the lipoprotein Tgl, which can be transferred from one cell to another when cells are moving by social motility (Fig. 4A) (92). M. xanthus chemotaxis might also require cell-cell contacts and coordinated motility, since isolated cells exhibit only nonvectorial movements in the presence of nutrient gradients (122). Additionally, during the tactic behavior termed predataxis, M. xanthus cells will attack prey microorganisms only after making physical contact with the prey cells (8, 9).
Cell Movement during Development
Organized cell movements are very important during fruiting body formation. Specifically, changes in reversal frequency and cell velocities are important for directing cell movements (51, 110). For example, during the course of development, cells in large groups reverse much less frequently than isolated cells or cells in small groups (110). Cells were also shown to double their velocities on starvation medium (51, 110). These changes are probably due to the accumulation of substances such as C signal, EPS, and PE (51, 65, 98). Indeed, mutant cells lacking the C signal (csgA) were not observed to decrease their reversal frequency under starvation conditions (51). Additionally, cells lacking a functional S-motility system, of which EPS is a part, were not observed to decrease their reversal frequency in response to increased cell density (110). Decreased reversal frequency with increased cell density during development was associated with increased FrzCD methylation (110). These observations suggest that a complex signaling network mediates the regulation of cell movements during development.
A different model was proposed by Sliusarenko et al. (117). They found that during cellular aggregation, reductions in cell velocity rather than cell reversals were principally responsible for the accumulation of cells in aggregates or fruiting bodies. On the basis of their data and computer simulations, they proposed that decreased cell velocities but not cell reversals were required for aggregation. It is not clear why different observations were made by different workers, although it is possible that the use of different culture conditions (submerged cultures rather than agar pads) might account for some observed differences.
FrzCD and Cell-Cell Communication
A recent study of the localization of the cytoplasmic receptor FrzCD suggested that this protein might have an important function during cell-cell communication (78). FrzCD shows a localization pattern that is very different from that observed for chemosensors in other bacterial species. For example, FrzCD does not form membrane-bound polar clusters that are typical of most bacterial MCPs (75, 125) but rather is found in cytoplasmic clusters that appear helically arranged and span the cell length. The involvement of cytoskeletal elements in FrzCD localization is still under investigation. The distribution of FrzCD in living cells was found to be dynamic: FrzCD was localized in clusters that continuously changed their size, number, and position. Interestingly, the number of FrzCD clusters was correlated with cellular reversal frequency: fewer clusters were observed in hyporeversing mutants, and additional clusters were observed in hyperreversing mutants. Perhaps the most unexpected finding in this study was that M. xanthus cells making side-to-side contacts show transient FrzCD cluster alignments (Fig. 3D) (78). Detailed statistical analyses show that the alignment of FrzCD clusters occurs only at the moment of the side-to-side contact and that before and after that moment the distributions of the FrzCD clusters in two different cells are not correlated. Similar analyses showed that cluster alignments are not observed in strains lacking the histidine kinase FrzE, indicating the requirement of feedback regulation mechanisms from the Frz pathway.
What is the function of the FrzCD cluster alignments? Interestingly, in the same study, side-to-side cell contacts were shown to influence not only the organization of the clusters but also the timing between cell reversals (78). In fact, converging cells making side-to-side contacts exhibit increased cellular reversals (78). These reversals are not seen in frzCD mutants (22). Contact-dependent cellular reversals may be important for coordinated cell movements, in particular “rippling,” the wave-like periodic movements associated with predation and fruiting body formation (7, 114, 116).
Rippling
Rippling is the coordinated rhythmic waves of cells observed when myxobacteria feed on macromolecules or lysing cells (7, 8, 106, 114, 131). Because the waves reflect off each other, Sliusarenko et al. (116) named them “accordion waves.” Several lines of evidence support the hypothesis that ripples represent a predatory behavior. First, rippling is induced by many different macromolecules, such as peptidoglycan, DNA, or protein, but not by the monomeric constituents of these macromolecules (7, 114). Also, rippling is observed during incubation with different prey substrates such as phages, Gram-negative bacteria, Gram-positive bacteria, and yeast (8). The occurrence of rippling in some strains of fruiting M. xanthus has been shown to be caused by autolysis in these strains. Indeed, strains with low level of autolysis, such as the wild-type strain DZ2, rarely show rippling unless they are mixed with a strain that does show autolysis (7, 8).
Several models have been proposed to account for rippling behavior, but a clear understanding of this phenomenon requires more experimentation.
(i) The first model is based on the hypothesis that M. xanthus cells exchange signals through head-to-head collisions (45, 106). This model proposes that when two cells approach each other from opposite directions, collisions lead to the exchange of the C signal, which in turn primes an internal response that results in cell reversals through the Frz pathway. The C signal is a 17-kDa protein that results from the proteolysis of the larger 25-kDa protein CsgA (104). Evidence for this model is based on the observation that csgA is required for rippling as well as for development (62, 106). However, this model cannot explain why purified C signal could not rescue the rippling defect of a csgA mutant, even though it can rescue the developmental defects (62). Additionally, cells do not make head-to-head contacts but rather make side-to-side contacts during rippling (116). Unfortunately, since the receptor for the C signal has never been identified, the CsgA signaling pathway with respect to rippling remains speculative.
(ii) A second hypothesis, proposed by Berleman and Kirby (8), is that M. xanthus cells do not directly signal to each other during the rippling behavior. Instead, the signal that drives rippling behavior may come solely from prey macromolecules. In this model, each M. xanthus cell responds to the presence of prey autonomously, and the ripple structures that arise are a consequence of the shifting movements of individuals reaching a tenuous state of equilibrium. As the local density of M. xanthus cells increases, each cell will be more likely to trigger a reversal as it becomes surrounded by inedible sister cells. A reversal under this circumstance has the potential to move a cell away from an area crowded with predators and back toward an area with more prey contacts available (8).
(iii) A third hypothesis is that side-to-side contacts between leading cells in opposing waves mediate the exchange of chemical or physical signals that are perceived directly by FrzCD, the receptor of the Frz pathway. This model is supported by observations that side-to-side contacts stimulate both FrzCD relocalization and cell reversals (see above) (78) and by the observation that colliding rippling waves penetrate each other by about one cell length before triggering reversals (116). Analyses of FrzCD localization during rippling might provide more insights into the function of FrzCD clusters during the biogenesis of accordion waves.
CONCLUSIONS
While many recent discoveries have provided important insights into the nature of A and S gliding motility in M. xanthus, many questions remain. For example, while the S-motility engine has been elucidated, the A-motility engine and its constituent proteins and mechanism of action have still not been identified. Is it a unique engine designed to power gliding motility in the myxobacteria, or is it a general motility system found in other bacteria but never identified? Why do M. xanthus cells need two engines for gliding motility? How are they coordinated? Why are both motility systems so slow, 2 to 4 μm/min (one cell body length per minute)? Studies of bacterial gliding motility have long been frustrated by the lack of appropriate tools, but the use of M. xanthus as a genetically tractable system and the emergence of high-resolution fluorescence microscopy now presents exciting opportunities for new discoveries. Indeed, the very slowness of myxobacterial motility provides a special advantage in studies involving protein localization and dynamics, as these experiments would be difficult to perform in bacteria that move at rates of 10 to 100 μm/s.
Clearly, the complexities of motility and its regulation in M. xanthus are still far from being well understood. However, the novel nature of the M. xanthus motility systems promises to provide numerous insights toward our understanding of the cell biology of these bacteria and their adaptation to their soil habitat. Indeed, M. xanthus is sufficiently different from the other well-characterized bacterial systems that important discoveries and surprises are sure to follow in the not-too-distant future.
Acknowledgments
We thank Eva Campodonico, Juan Jesus Vicente, Beiyan Nan, James Berleman, and Mireille Ansaldi for helpful criticisms and discussions.
Research in our laboratories is funded by grants from the National Institutes of Health to D.R.Z. (GM020509) and Z.Y. (GM071601) and by an ANR “Jeunes Chercheurs-Jeunes Chercheuses” to T.M. (ANR-07-JCJC-0131).
Biography
 Emilia Mauriello received her Ph.D. in Applied Microbiology in 2006 from the Universita' degli Studi di Napoli Federico II (Italy). During her Ph.D. studies, she worked on the regulation of type I fimbriation in the mouse pathogen Citrobacter rodentium. She also worked, during a master program in microbiology, on the development of vaccines using Bacillus subtilis spores as a vehicle for various antigens. Emilia Mauriello joined the Zusman lab as a postdoctoral fellow in 2006. Since then, she has been working on uncovering the mechanisms by which a cytoplasmic chemosensory protein regulates cell-cell communication and motility in Myxococcus xanthus.
Emilia Mauriello received her Ph.D. in Applied Microbiology in 2006 from the Universita' degli Studi di Napoli Federico II (Italy). During her Ph.D. studies, she worked on the regulation of type I fimbriation in the mouse pathogen Citrobacter rodentium. She also worked, during a master program in microbiology, on the development of vaccines using Bacillus subtilis spores as a vehicle for various antigens. Emilia Mauriello joined the Zusman lab as a postdoctoral fellow in 2006. Since then, she has been working on uncovering the mechanisms by which a cytoplasmic chemosensory protein regulates cell-cell communication and motility in Myxococcus xanthus.
 Tâm Mignot is CNRS CR scientist at the Institut de Microbiologie de la Méditerranée in Marseille, France. He received his Ph.D. from the Université Paris 7-Pasteur Institute in 2002 and his M.S. in Biochemistry at the University of Nice in 1998. His Ph.D. work was on anthrax toxin regulation. He has been working on motility in Myxococcus xanthus since 2002, when he started his postdoctoral training at the University of California, Berkeley. He joined CNRS in Marseille in 2007 to start a project on bacterial motility.
Tâm Mignot is CNRS CR scientist at the Institut de Microbiologie de la Méditerranée in Marseille, France. He received his Ph.D. from the Université Paris 7-Pasteur Institute in 2002 and his M.S. in Biochemistry at the University of Nice in 1998. His Ph.D. work was on anthrax toxin regulation. He has been working on motility in Myxococcus xanthus since 2002, when he started his postdoctoral training at the University of California, Berkeley. He joined CNRS in Marseille in 2007 to start a project on bacterial motility.
 Zhaomin Yang is an Associate Professor of Microbiology in the Department of Biological Sciences at Virginia Tech. He received his Ph.D. in Microbiology in 1996 and his M.S. in Food Science in 1993 from the University of California at Davis. His B.S. in Biology was from Beijing University in China. His M.S. and Ph.D. work was on the regulation of glucose transport in Saccharomyces cerevisiae. He has been working on motility and signal transduction in myxobacteria since 1996, when he started his postdoctoral training at the University of California, Los Angeles. He joined Virginia Tech in 2002 and was an Assistant Professor of Biological Sciences at Auburn University from 1999 to 2002.
Zhaomin Yang is an Associate Professor of Microbiology in the Department of Biological Sciences at Virginia Tech. He received his Ph.D. in Microbiology in 1996 and his M.S. in Food Science in 1993 from the University of California at Davis. His B.S. in Biology was from Beijing University in China. His M.S. and Ph.D. work was on the regulation of glucose transport in Saccharomyces cerevisiae. He has been working on motility and signal transduction in myxobacteria since 1996, when he started his postdoctoral training at the University of California, Los Angeles. He joined Virginia Tech in 2002 and was an Assistant Professor of Biological Sciences at Auburn University from 1999 to 2002.
 David Zusman is currently Professor of Biochemistry and Molecular Biology at the University of California, Berkeley. He began his studies of the myxobacteria in 1967, when he joined the inspirational laboratory of Eugene Rosenberg at the University of California, Los Angeles. After receiving his Ph.D. in 1970, Dr. Zusman did postdoctoral work at Princeton University with Arthur B. Pardee, working on the regulation of cell division in Escherichia coli. In 1973, Dr. Zusman joined the Department of Bacteriology and Immunology at the University of California, Berkeley, where he returned to his long-standing interest in Myxococcus xanthus. Dr. Zusman's principal focus has been on the role of the frizzy (Frz) chemosensory system in regulating cellular movements. The Frz system has turned out to be a key regulator for many of the behaviors of M. xanthus, including swarming, rippling, predation, and fruiting body formation.
David Zusman is currently Professor of Biochemistry and Molecular Biology at the University of California, Berkeley. He began his studies of the myxobacteria in 1967, when he joined the inspirational laboratory of Eugene Rosenberg at the University of California, Los Angeles. After receiving his Ph.D. in 1970, Dr. Zusman did postdoctoral work at Princeton University with Arthur B. Pardee, working on the regulation of cell division in Escherichia coli. In 1973, Dr. Zusman joined the Department of Bacteriology and Immunology at the University of California, Berkeley, where he returned to his long-standing interest in Myxococcus xanthus. Dr. Zusman's principal focus has been on the role of the frizzy (Frz) chemosensory system in regulating cellular movements. The Frz system has turned out to be a key regulator for many of the behaviors of M. xanthus, including swarming, rippling, predation, and fruiting body formation.
REFERENCES
- 1.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astling, D. P., J. Y. Lee, and D. R. Zusman. 2006. Differential effects of chemoreceptor methylation-domain mutations on swarming and development in the social bacterium Myxococcus xanthus. Mol. Microbiol. 59:45-55. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125:237-246. [DOI] [PubMed] [Google Scholar]
- 4.Behmlander, R. M., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behmlander, R. M., and M. Dworkin. 1994. Integral proteins of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6304-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berleman, J. E., and C. E. Bauer. 2004. Characterization of cyst cell formation in the purple photosynthetic bacterium Rhodospirillum centenum. Microbiology 150:383-390. [DOI] [PubMed] [Google Scholar]
- 7.Berleman, J. E., T. Chumley, P. Cheung, and J. R. Kirby. 2006. Rippling is a predatory behavior in Myxococcus xanthus. J. Bacteriol. 188:5888-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berleman, J. E., and J. R. Kirby. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 33:942-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berleman, J. E., J. Scott, T. Chumley, and J. R. Kirby. 2008. Predataxis behavior in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 105:17127-17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, W. P., Q. Xu, C. L. Cadieux, S. J. Suh, W. Shi, and Z. Yang. 2009. Isolation and characterization of a suppressor mutation that restores Myxococcus xanthus exopolysaccharide production. Microbiology 155:3599-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black, W. P., Q. Xu, and Z. Yang. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447-456. [DOI] [PubMed] [Google Scholar]
- 12.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackhart, B. D., and D. R. Zusman. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. U. S. A. 82:8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonner, P. J., W. P. Black, Z. Yang, and L. J. Shimkets. 2006. FibA and PilA act cooperatively during fruiting body formation of Myxococcus xanthus. Mol. Microbiol. 61:1283-1293. [DOI] [PubMed] [Google Scholar]
- 15.Bonner, P. J., and L. J. Shimkets. 2006. Phospholipid directed motility of surface-motile bacteria. Mol. Microbiol. 61:1101-1109. [DOI] [PubMed] [Google Scholar]
- 16.Bowden, M. G., and H. B. Kaplan. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30:275-284. [DOI] [PubMed] [Google Scholar]
- 17.Boyd, A., and M. Simon. 1982. Bacterial chemotaxis. Annu. Rev. Physiol. 44:501-517. [DOI] [PubMed] [Google Scholar]
- 18.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 19.Bulyha, I., C. Schmidt, P. Lenz, V. Jakovljevic, A. Hone, B. Maier, M. Hoppert, and L. Sogaard-Andersen. 2009. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol. 74:691-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burchard, R. P. 1981. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu. Rev. Microbiol. 35:497-529. [DOI] [PubMed] [Google Scholar]
- 21.Burchard, R. P. 1982. Trail following by gliding bacteria. J. Bacteriol. 152:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustamante, V. H., I. Martinez-Flores, H. C. Vlamakis, and D. R. Zusman. 2004. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol. Microbiol. 53:1501-1513. [DOI] [PubMed] [Google Scholar]
- 23.Chami, M., I. Guilvout, M. Gregorini, H. W. Remigy, S. A. Muller, M. Valerio, A. Engel, A. P. Pugsley, and N. Bayan. 2005. Structural insights into the secretin PulD and its trypsin-resistant core. J. Biol. Chem. 280:37732-37741. [DOI] [PubMed] [Google Scholar]
- 24.Charest, P. G., and R. A. Firtel. 2007. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 401:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268-281. [DOI] [PubMed] [Google Scholar]
- 26.Clausen, M., V. Jakovljevic, L. Sogaard-Andersen, and B. Maier. 2009. High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191:4633-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins, R. F., S. A. Frye, A. Kitmitto, R. C. Ford, T. Tonjum, and J. P. Derrick. 2004. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J. Biol. Chem. 279:39750-39756. [DOI] [PubMed] [Google Scholar]
- 28.Curtis, P. D., J. Atwood III, R. Orlando, and L. J. Shimkets. 2007. Proteins associated with the Myxococcus xanthus extracellular matrix. J. Bacteriol. 189:7634-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis, P. D., R. Geyer, D. C. White, and L. J. Shimkets. 2006. Novel lipids in Myxococcus xanthus and their role in chemotaxis. Environ. Microbiol. 8:1935-1949. [DOI] [PubMed] [Google Scholar]
- 30.Dana, J. R., and L. J. Shimkets. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 175:3636-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Defeu Soufo, H. J., and P. L. Graumann. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dworkin, M. 1966. Biology of the myxobacteria. Annu. Rev. Microbiol. 20:75-106. [DOI] [PubMed] [Google Scholar]
- 33.Dworkin, M. 1983. Tactic behavior of Myxococcus xanthus. J. Bacteriol. 154:452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenbach, M. 1996. Control of bacterial chemotaxis. Mol. Microbiol. 20:903-910. [DOI] [PubMed] [Google Scholar]
- 35.Fraser, J. S., J. P. Merlie, Jr., N. Echols, S. R. Weisfield, T. Mignot, D. E. Wemmer, D. R. Zusman, and T. Alber. 2007. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol. Microbiol. 65:319-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 37.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartzell, P., and D. Kaiser. 1991. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J. Bacteriol. 173:7615-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartzell, P. L. 1997. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc. Natl. Acad. Sci. U. S. A. 94:9881-9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 42.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 8:1161-1168. [DOI] [PubMed] [Google Scholar]
- 44.Howie, H. L., M. Glogauer, and M. So. 2005. The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism. PloS Biol. 3:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Igoshin, O. A., A. Mogilner, R. D. Welch, D. Kaiser, and G. Oster. 2001. Pattern formation and traveling waves in myxobacteria: theory and modeling. Proc. Natl. Acad. Sci. U. S. A. 98:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inclan, Y. F., S. Laurent, and D. R. Zusman. 2008. The receiver domain of FrzE, a CheA-CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 68:1328-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inclan, Y. F., H. C. Vlamakis, and D. R. Zusman. 2007. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol. Microbiol. 65:90-102. [DOI] [PubMed] [Google Scholar]
- 48.Jahn, E. 1924. Beiträge zur botanischen Protistologie. Gebrüder Borntraeger, Leipzig, Germany.
- 49.Jakovljevic, V., S. Leonardy, M. Hoppert, and L. Sogaard-Andersen. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarrell, K. F., and M. J. McBride. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466-476. [DOI] [PubMed] [Google Scholar]
- 51.Jelsbak, L., and L. Sogaard-Andersen. 2002. Pattern formation by a cell surface-associated morphogen in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 99:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 53.Kaiser, A. D., and C. Crosby. 1983. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 3:227-245. [Google Scholar]
- 54.Kaiser, D. 2006. A microbial genetic journey. Annu. Rev. Microbiol. 60:1-25. [DOI] [PubMed] [Google Scholar]
- 55.Kaiser, D. 2009. Are there lateral as well as polar engines for A-motile gliding in myxobacteria? J. Bacteriol. 191:5336-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 57.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 58.Kearns, D. B., P. J. Bonner, D. R. Smith, and L. J. Shimkets. 2002. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 184:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kearns, D. B., and L. J. Shimkets. 1998. Chemotaxis in a gliding bacterium. Proc. Natl. Acad. Sci. U. S. A. 95:11957-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearns, D. B., and L. J. Shimkets. 2001. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 9:126-129. [DOI] [PubMed] [Google Scholar]
- 61.Kearns, D. B., A. Venot, P. J. Bonner, B. Stevens, G. J. Boons, and L. J. Shimkets. 2001. Identification of a developmental chemoattractant in Myxococcus xanthus through metabolic engineering. Proc. Natl. Acad. Sci. U. S. A. 98:13990-13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 63.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 64.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroos, L., P. Hartzell, K. Stephens, and D. Kaiser. 1988. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 2:1677-1685. [DOI] [PubMed] [Google Scholar]
- 66.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lancero, H., J. E. Brofft, J. Downard, B. W. Birren, C. Nusbaum, J. Naylor, W. Shi, and L. J. Shimkets. 2002. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 184:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lapidus, I. R., and H. C. Berg. 1982. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 151:384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leonardy, S., G. Freymark, S. Hebener, E. Ellehauge, and L. Sogaard-Andersen. 2007. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 26:4433-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li, Y., R. Lux, A. E. Pelling, J. K. Gimzewski, and W. Shi. 2005. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology 151:353-360. [DOI] [PubMed] [Google Scholar]
- 72.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu, A., K. Cho, W. P. Black, X. Y. Duan, R. Lux, Z. Yang, H. B. Kaplan, D. R. Zusman, and W. Shi. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55:206-220. [DOI] [PubMed] [Google Scholar]
- 74.Lunsdorf, H., and H. U. Schairer. 2001. Frozen motion of gliding bacteria outlines inherent features of the motility apparatus. Microbiology 147:939-947. [DOI] [PubMed] [Google Scholar]
- 75.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 76.Mahadevan, L., and P. Matsudaira. 2000. Motility powered by supramolecular springs and ratchets. Science 288:95-100. [DOI] [PubMed] [Google Scholar]
- 77.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Cell Dev. Biol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 78.Mauriello, E. M., D. P. Astling, O. Sliusarenko, and D. R. Zusman. 2009. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc. Natl. Acad. Sci. U. S. A. 106:4852-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mauriello, E. M., F. Mouhamar, B. Nan, A. Ducret, D. Dai, D. R. Zusman, and T. Mignot. 2010. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 29:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mauriello, E. M., B. Nan, and D. R. Zusman. 2009. AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol. Microbiol. 72:964-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McBride, M. J. 2004. Cytophaga-Flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 7:63-71. [DOI] [PubMed] [Google Scholar]
- 82.McBride, M. J., T. Kohler, and D. R. Zusman. 1992. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J. Bacteriol. 174:4246-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McBride, M. J., R. A. Weinberg, and D. R. Zusman. 1989. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc. Natl. Acad. Sci. U. S. A. 86:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCleary, W. R., and D. R. Zusman. 1990. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 87:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Merz, A. J., and K. T. Forest. 2002. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr. Biol. 12:R297-R303. [DOI] [PubMed] [Google Scholar]
- 86.Mignot, T. 2007. The elusive engine in Myxococcus xanthus gliding motility. Cell Mol. Life Sci. 64:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mignot, T., J. P. Merlie, Jr., and D. R. Zusman. 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310:855-857. [DOI] [PubMed] [Google Scholar]
- 88.Mignot, T., J. P. Merlie, Jr., and D. R. Zusman. 2007. Two localization motifs mediate polar residence of FrzS during cell movement and reversals of Myxococcus xanthus. Mol. Microbiol. 65:363-372. [DOI] [PubMed] [Google Scholar]
- 89.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyata, M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6-12. [DOI] [PubMed] [Google Scholar]
- 91.Nelson, S. S., S. Bollampalli, and M. J. McBride. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 190:2851-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nudleman, E., D. Wall, and D. Kaiser. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125-127. [DOI] [PubMed] [Google Scholar]
- 93.Nudleman, E., D. Wall, and D. Kaiser. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60:16-29. [DOI] [PubMed] [Google Scholar]
- 94.O'Connor, K. A., and D. R. Zusman. 1991. Behavior of peripheral rods and their role in the life cycle of Myxococcus xanthus. J. Bacteriol. 173:3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palsdottir, H., J. P. Remis, C. Schaudinn, E. O'Toole, R. Lux, W. Shi, K. L. McDonald, J. W. Costerton, and M. Auer. 2009. Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by ectron microscopic tomography. J. Bacteriol. 191:2077-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pelling, A. E., Y. Li, W. Shi, and J. K. Gimzewski. 2005. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc. Natl. Acad. Sci. U. S. A. 102:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pham, V. D., C. W. Shebelut, B. Mukherjee, and M. Singer. 2005. RasA is required for Myxococcus xanthus development and social motility. J. Bacteriol. 187:6845-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quillet, L., L. Bensmail, S. Barray, and J. Guespin-Michel. 1997. Cloning and sequencing of two genes, prtA and prtB, from Myxococcus xanthus, encoding PrtA and PrtB proteases, both of which are required for the protease activity. Gene 198:135-140. [DOI] [PubMed] [Google Scholar]
- 100.Ramaswamy, S., M. Dworkin, and J. Downard. 1997. Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J. Bacteriol. 179:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reichenbach, H. 1966. Myxococcus spp. (Myxobacterales) schwarmentwicklung und Bildung von protocysten., vol. 1A. Publikationen zu wissenschaftlichen Filmen, Gottingen, Germany.
- 102.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 103.Rodriguez, A. M., and A. M. Spormann. 1999. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 181:4381-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rolbetzki, A., M. Ammon, V. Jakovljevic, A. Konovalova, and L. Sogaard-Andersen. 2008. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev. Cell 15:627-634. [DOI] [PubMed] [Google Scholar]
- 105.Rosenberg, E., B. Vaks, and A. Zuckerberg. 1973. Bactericidal action of an antibiotic produced by Myxococcus xanthus. Antimicrob. Agents Chemother. 4:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 107.Scott, A. E., E. Simon, S. K. Park, P. Andrews, and D. R. Zusman. 2008. Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase. Mol. Microbiol. 69:724-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheth, H. B., K. K. Lee, W. Y. Wong, G. Srivastava, O. Hindsgaul, R. S. Hodges, W. Paranchych, and R. T. Irvin. 1994. The pili of Pseudomonas aeruginosa strains PAK and PAO bind specifically to the carbohydrate sequence beta GalNAc(1-4)beta Gal found in glycosphingolipids asialo-GM1 and asialo-GM2. Mol. Microbiol. 11:715-723. [DOI] [PubMed] [Google Scholar]
- 109.Shi, W., T. Kohler, and D. R. Zusman. 1993. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 9:601-611. [DOI] [PubMed] [Google Scholar]
- 110.Shi, W., F. K. Ngok, and D. R. Zusman. 1996. Cell density regulates cellular reversal frequency in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 93:4142-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. U. S. A. 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 113.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shimkets, L. J., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sliusarenko, O., J. Neu, D. R. Zusman, and G. Oster. 2006. Accordion waves in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 103:1534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sliusarenko, O., D. R. Zusman, and G. Oster. 2007. Aggregation during fruiting body formation in Myxococcus xanthus is driven by reducing cell movement J. Bacteriol. 189:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sliusarenko, O., D. R. Zusman, and G. Oster. 2007. The motors powering A-motility in Myxococcus xanthus are distributed along the cell body. J. Bacteriol. 189:7920-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spormann, A. M., and D. Kaiser. 1999. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 181:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun, H., Z. Yang, and W. Shi. 1999. Effect of cellular filamentation on adventurous and social gliding motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 96:15178-15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 122.Taylor, R. G., and R. D. Welch. 2008. Chemotaxis as an emergent property of a swarm. J. Bacteriol. 190:6811-6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varga, J. J., V. Nguyen, D. K. O'Brien, K. Rodgers, R. A. Walker, and S. B. Melville. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol. Microbiol. 62:680-694. [DOI] [PubMed] [Google Scholar]
- 124.Vlamakis, H. C., J. R. Kirby, and D. R. Zusman. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol. Microbiol. 52:1799-1811. [DOI] [PubMed] [Google Scholar]
- 125.Wadhams, G. H., A. C. Martin, S. L. Porter, J. R. Maddock, J. C. Mantotta, H. M. King, and J. P. Armitage. 2002. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol. Microbiol. 46:1211-1221. [DOI] [PubMed] [Google Scholar]
- 126.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 127.Wall, D., P. E. Kolenbrander, and D. Kaiser. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ward, M. J., H. Lew, and D. R. Zusman. 2000. Social motility in Myxococcus xanthus requires FrzS, a protein with an extensive coiled-coil domain. Mol. Microbiol. 37:1357-1371. [DOI] [PubMed] [Google Scholar]
- 129.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 130.Weimer, R. M., C. Creighton, A. Stassinopoulos, P. Youderian, and P. L. Hartzell. 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 180:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Welch, R., and D. Kaiser. 2001. Cell behavior in traveling wave patterns of myxobacteria. Proc. Natl. Acad. Sci. U. S. A. 98:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.White, D. J., and P. L. Hartzell. 2000. AglU, a protein required for gliding motility and spore maturation of Myxococcus xanthus, is related to WD-repeat proteins. Mol. Microbiol. 36:662-678. [DOI] [PubMed] [Google Scholar]
- 133.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 134.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 135.Wu, S. S., and D. Kaiser. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu, S. S., J. Wu, Y. L. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]
- 137.Xu, Q., W. P. Black, C. L. Cadieux, and Z. Yang. 2008. Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol. Microbiol. 69:714-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang, R., S. Bartle, R. Otto, A. Stassinopoulos, M. Rogers, L. Plamann, and P. Hartzell. 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 186:6168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123-1130. [DOI] [PubMed] [Google Scholar]
- 140.Yang, Z., D. Guo, M. G. Bowden, H. Sun, L. Tong, Z. Li, A. E. Brown, H. B. Kaplan, and W. Shi. 2000. The Myxococcus xanthus wbgB gene encodes a glycosyltransferase homologue required for lipopolysaccharide O-antigen biosynthesis. Arch. Microbiol. 174:399-405. [DOI] [PubMed] [Google Scholar]
- 141.Yang, Z., and Z. Li. 2005. Demonstration of interactions among Myxococcus xanthus Dif chemotaxis-like proteins by the yeast two-hybrid system. Arch. Microbiol. 183:243-252. [DOI] [PubMed] [Google Scholar]
- 142.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Youderian, P., N. Burke, D. J. White, and P. L. Hartzell. 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49:555-570. [DOI] [PubMed] [Google Scholar]
- 144.Youderian, P., and P. L. Hartzell. 2006. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics 172:1397-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Youderian, P., and P. L. Hartzell. 2007. Triple mutants uncover three new genes required for social motility in Myxococcus xanthus. Genetics 177:557-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu, R., and D. Kaiser. 2007. Gliding motility and polarized slime secretion. Mol. Microbiol. 63:454-467. [DOI] [PubMed] [Google Scholar]
- 147.Zusman, D. R. 1982. “Frizzy” mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J. Bacteriol. 150:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862-872. [DOI] [PubMed] [Google Scholar]