Abstract
Neisseria meningitidis is naturally competent for transformation throughout its growth cycle. Transformation in neisserial species is coupled to the expression of type IV pili, which are present on the cell surface as bundled filamentous appendages, and are assembled, extruded and retracted by the pilus biogenesis components. During the initial phase of the transformation process, binding and uptake of DNA takes place with entry through a presumed outer-membrane channel into the periplasm. This study showed that DNA associates only weakly with purified pili, but binds significantly to the PilQ complex isolated directly from meningococcal membranes. By assessing the DNA-binding activity of the native complex PilQ, as well as recombinant truncated PilQ monomers, it was shown that the N-terminal region of PilQ is involved in the interaction with DNA. It was evident that the binding of ssDNA to PilQ had a higher affinity than the binding of dsDNA. The binding of DNA to PilQ did not, however, depend on the presence of the neisserial DNA-uptake sequence. It is suggested that transforming DNA is introduced into the cell through the outer-membrane channel formed by the PilQ complex, and that DNA uptake occurs by non-specific introduction of DNA coupled to pilus retraction, followed by presentation to DNA-binding component(s), including PilQ.
INTRODUCTION
Neisseria meningitidis (the meningococcus) is constitutively competent for transformation throughout its growth cycle. Natural transformation is a process unique to bacteria and involves active uptake of exogenous DNA from the environment, which can result in the acquisition of new genetic information in a heritable form, and is particularly important for genetic exchange and diversity (Chen & Dubnau, 2004). Transformation in Gram-negative bacteria differs from that in Gram-positive bacteria in that DNA has to pass through an additional layer, the outer membrane. Transformation is coupled to the expression of type IV pili in a number of Gram-negative bacteria, including N. meningitidis (Frøholm et al., 1973), Neisseria gonorrhoeae (Mathis & Scocca, 1984; Swanson et al., 1971), Eikenella corrodens (Tønjum et al., 1985), Moraxella nonliquefaciens (Bøvre et al., 1970; Bøvre, 1964), Legionella pneumophila (Stone & Kwaik, 1999), Pseudomonas stutzeri (Lorenz & Wackernagel, 1990) and Thermus thermophilus (Friedrich et al., 2001). Type IV pili are long fibrous structures emanating from the bacterial surface, and as multifunctional organelles, they are involved in a variety of bacterial processes. In addition to their role in competence, the type IV pili of many species also play a role in adherence (Swanson et al., 1971), twitching motility (Bradley, 1974; Mattick, 2002), biofilm formation (O'Toole & Kolter, 1998) and bacteriophage infection (Bradley, 1974). Twitching motility is driven by pilus retraction.
Efficient neisserial transformation further requires the presence of the frequently occurring 10 bp long signature sequence 5′-GCCGTCTGAA-3′, termed the DNA uptake sequence (DUS), in the transforming DNA (Goodman & Scocca, 1988). Homologous recombination of the incoming DNA with the chromosome is mediated by RecA (Koomey & Falkow, 1987), and is partially dependent upon other recombination components such as the RecBCD complex (Mehr & Seifert, 1998) and RecN (Skaar et al., 2002). Current knowledge on the N. gonorrhoeae transformation process has recently been reviewed by Hamilton and Dillard (2006).
Neisserial competence for transformation is dependent on the expression of pilus biogenesis components (Carbonnelle et al., 2006; Tønjum & Koomey, 1997) and several pilus-related components, including the minor pilin ComP (Wolfgang et al., 1999; Aas et al., 2002). Among the pilus biogenesis components are secretins, which belong to a large family of bacterial complexes associated with translocation of single proteins and macromolecules across the outer membrane. A subset of this family, termed PilQ proteins, is required for type IV pilus extrusion and retraction in N. meningitidis, N. gonorrhoeae and Pseudomonas aeruginosa (Drake & Koomey, 1995; Tønjum et al., 1995). Neisserial PilQ null mutants are not piliated and are non-competent for natural transformation (Drake & Koomey, 1995; Tønjum et al., 1998). Meningococcal PilQ is unique among secretins because of its abundance in the outer membrane and its N-terminally located polymorphic region containing small basic repeat (SBR) elements. We have previously shown that the native PilQ complex from meningococcal outer membranes adopts a cage-like structure of ∼900 kDa in total mass, consisting of 12 identical subunits (Collins et al., 2001, 2003, 2004). Furthermore, we have provided evidence that PilQ and the pilus fibre interact, identified the region of PilQ that is involved in the PilQ–pilus interaction, and shown that this interaction induces a conformational change in the structure of the PilQ oligomer. These findings indicate that the PilQ complex is the channel through which its substrate, the moving pilus fibre (polymerized PilE), is directed to the bacterial surface (Collins et al., 2005).
Transformation of neisserial DNA is unequivocally coupled to pilus expression, although the molecular basis for this correlation has not been elucidated. Mutations that affect the expression of type IV pili in N. meningitidis and N. gonorrhoeae greatly reduce the amount of DNA taken up during transformation (Koomey, 1998; Tønjum et al., 1998; Tønjum & Koomey, 1997; Aas et al., 2002). It is not clear, however, whether this phenomenon is solely due to the lack of pili per se, or to the direct involvement of pilus biogenesis components themselves in introducing DNA into the meningococcal cell. On the other hand, gonococcal pilin variants that do not express observable assembled pili, but express pilin subunits, can be competent for transformation to some extent (Long et al., 2003). Due to its strong preference for DUS-containing DNA in transformation, the neisserial cell is able to distinguish between self DNA (containing DUS) and non-self DNA (lacking DUS). The DNA-binding events during transformation can thus be classified into two types: specific binding mediated by a putative DUS-specific receptor, and non-specific affinity mediated by non-specific DNA-binding component(s) that are required to support the entry of DNA into the cell. Among the components contributing to non-specific DNA binding during transformation, the periplasmatic protein ComE has been identified (Chen & Gotschlich, 2001).
In P. aeruginosa, evidence for direct binding of DNA to purified type IV pili has been obtained (van Schaik et al., 2005), although this bacterial species is found to be non-competent for transformation. Here, we have addressed the question whether of the neisserial pilus itself and/or its corresponding secretin PilQ exhibit DNA-binding activity. The DNA-binding behaviour of purified pili and PilQ from meningococcal outer membranes was assessed by DNA band-shift, solution and solid-phase overlay assays, as well as by electron microscopy. We showed that purified pili from N. meningitidis and P. aeruginosa exhibit weak DNA binding, while the PilQ complex binds DNA to a significant extent. The PilQ-mediated binding of DNA was also assessed with regard to the structural location of the PilQ DNA-binding site, DUS specificity, and the relative preference for binding of ssDNA versus dsDNA. This work is important for the understanding of how DNA is introduced into the neisserial cell during the early phase of natural transformation.
METHODS
Bacterial strains and media.
The bacterial strains used in this study were N. meningitidis M1080 (Frasch & Chapman, 1972), M1080-ΔpilQ and TT31-221 (Tønjum et al., 1998), P. aeruginosa PAK (Bradley, 1974), and Escherichia coli ER2566 (New England Biolabs). Strain TT31-221 contains chimeric PilQ, in which the N-terminal SBR region is replaced by the corresponding but non-homologous region in P. aeruginosa PilQ. Meningococcal and E. coli strains were grown as previously described (Tønjum et al., 1995). Meningococcal strain M1080 was employed for PilQ complex purification. P. aeruginosa strain PAK was grown on Luria–Bertani (LB) or lactose medium. The M1080-ΔpilQ mutant has a transposon insertion in the pilQ start codon (Tønjum et al., 1998).
Bioinformatics analysis and prediction of secondary structure and electrostatic charge.
The deduced amino acid sequence of the pilQ of strain M1080 (GenBank accession no. AJ564200) was searched for the presence of recognized DNA-binding motifs and the electrostatic charge was calculated by using the charge program from the emboss package (Rice et al., 2000).
Cloning of the full-length and partial pilQ.
All standard methods of DNA manipulation were performed as previously described (Sambrook et al., 1989; Tønjum et al., 1995). The partial pilQ gene was PCR-amplified from genomic M1080 DNA (Frye et al., 2006). For the generation of recombinant PilQ with an N-terminal His-tag, the vector pQE30 was used (Qiagen), and for the C-terminal His-tag, the pET28b(+) vector (Novagen) was used. The 5′ part of pilQ was amplified by using primers QBamHI and QEcoRI, generating plasmid pPilQ-1. The central portion of pilQ was amplified with the primers SF21 and SF22, resulting in plasmid pPilQ-3. The 3′ part of pilQ was amplified with the primers SF20 and SF19 and subcloned into pET28b(+), resulting in plasmid pPilQ-4 (Table 1).
Table 1.
Primers used in this study
Restriction sites are underlined.
| Primer | Orientation in relation to pilQ | Sequence (5′–3′) | Restriction site |
|---|---|---|---|
| QBamHI | + | CGGGATCCGGAAACATTACAGACATCAAAG | BamHI |
| PilQ4B | − | GCGGATCCGCATCAATAGCGCAGGCTGTTGC | BamHI |
| SF20 | + | GCCCATGGCCATCCTGCAGATTTTGGC | NcoI |
| SF19 | − | GCCTCGAGATAGCGCAGGCTGTTGCCGG | XhoI |
| SF21 | + | GCGGATCCACCAATATCGATTTCCGCAAAG | BamHI |
| SF22 | − | GCAAGCTTGGCGGGATCGATCAGCACGCTG | HindIII |
| SF13 | + | ATCGGACGATACCGTGTCC | − |
| SF14 | − | TCGATATTGGTTTGTTTTGCTG | − |
| SF7 | + | GACCGTCAAAATCAACAAGGA | − |
| SF8 | − | AATACCGCCGACAATCAATG | − |
| SF1-for | + | AAGAGGCCAAAATCGAATCC | − |
| SF1-rev | − | CAGGCGAGTCCTTGTTGATT | − |
| SF9 | + | GCGAATTCCATCGCCTTGGACTTTGAAC | EcoRI |
| SF12b | − | GCGGATCCTGAGGGAGAGGGTCATTTTG | BamHI |
| PilQ1 | + | GCGAATTCGCGTGCTGATCGATCCCGCC | EcoRI |
| PilQ4c | − | GCGAATTCGCATCAATAGCGCAGGCTGTTGC | EcoRI |
Recombinant PilQ and MutY expression and purification.
For recombinant PilQ and MutY production, the plasmids were transformed into E. coli ER2566 (New England Biolabs), overexpressed, and the recombinant proteins were purified to homogeneity. For this purpose, cells were grown at 37 °C in LB broth until they reached OD600 0.5, then protein overexpression was induced with 1 mM IPTG. Cells were grown for another 4 h, harvested by centrifugation and lysed by means of a French Press (Thermo Electron) at 20 000 p.s.i. (138 MPa). The His-tagged proteins were purified by immobilized metal affinity chromatography with nickel–nitrilotriacetic acid agarose (Qiagen), under native or denaturing conditions as recommended by the manufacturer. Protein concentrations were determined by Bradford analysis (Bradford, 1976), and the purity was determined by SDS-PAGE and Coomassie blue staining. Protein specificity was confirmed by immunoblotting using a polyclonal rabbit antiserum raised against the purified PilQ complex.
Purification of native PilQ complex.
Purification of the PilQ complex from meningococcal outer membranes was performed as previously described (Collins et al., 2001, 2003, 2004).
Purification of type IV pilus fibres.
Type IV pili were purified from the N. meningitidis and P. aeruginosa cell surface using ammonium sulphate precipitation of a shearing fraction (Brinton et al., 1978). Briefly, bacterial cells were vortexed for 1 min in 0.15 M ethanolamine buffer (pH 10.5), and cellular debris was removed by centrifugation. Pilus fibres were precipitated at room temperature for 30 min by addition of one-tenth volume ammonium sulphate-saturated 0.15 M ethanolamine buffer, and collected by centrifugation. Pili were subsequently washed twice with 50 mM Tris-buffered saline, pH 7.5, and treated with DNase (Sigma), prior to DNA-binding experiments to ensure that they were not naturally saturated with DNA.
Rabbit immunization and antibody production.
Rabbit polyclonal antibodies were raised against the native PilQ complex, purified recombinant His-tagged PilQ fragments and purified pili, as previously described (Table 2, Fig. 2) (Frye et al., 2006). Procedures for SDS-PAGE and immunoblotting have been described previously (Tønjum et al., 1995, 1998).
Table 2.
Oligonucleotide substrates used in this study
SUD, DNA uptake sequence 3′–5′ (DUS is 5′–3′). DUS and SUD sequences are shown in bold type.
| Oligonucleotide | Sequence (5′–3′) | Pyrimidine content (%) | DUS |
|---|---|---|---|
| T1 | CAACAACAACAACAGCCGTCTGAACCAAATTCAGACGGCAACAACAACAACA | 40 | DUS+SUD |
| T2 | TGTTGTTGTTGTTGCCGTCTGAATTTGGTTCAGACGGCTGTTGTTGTTGTTG | 60 | DUS+SUD |
| T3 | CAACAACAACAACAGGCCTGTCATCCAAAATGACAGGCCAACAACAACAACA | 40 | None |
| T4 | TGTTGTTGTTGTTGGCCTGTCATTTTGGATGACAGGCCTGTTGTTGTTGTTG | 60 | None |
| T7 | CCGCCGCCTGCCGTCTGAAAGATATTCAGACGGCATCGGC | 52 | DUS+SUD |
| T8 | GCCGATGCCGTCTGAATATCTTTCAGACGGCAGGCGGCGG | 48 | DUS+SUD |
| T9 | CAACAACAACAACAGCCGTCTGAACCAAAGCCGTCTGAAAACAACAACAACA | 40 | DUS+DUS |
| T10 | TGTTGTTGTTGTTTTCAGACGGCTTTGGTTCAGACGGCTGTTGTTGTTGTTG | 60 | SUD+SUD |
| T11 | CAACAACAACAACACTGTCAGTTGCCAAAGACAGTCAACAACAACAACAACA | 40 | None |
| T12 | GTTGTTGTTGTTGTGACAGTCAACGGTTTCTGTCAGTTGTTGTTGTTGTTGT | 60 | None |
Fig. 2.
Gene organization and pilQ constructs. (a) Schematic representation of the N. meningitidis strain M1080 pilQ gene and the predicted gene product. The positions of the N-terminal SBRs and the PstI restriction site are indicated. The locations of positively charged areas within the predicted sequence are marked in black. (b) Schematic representation of the PilQ truncated proteins used in this study, depicted relative to Fig. 2(a). The position of the hexa-histidine tag is indicated with an ‘H’ and the sizes of the recombinant proteins are given on the right.
Pilus–DNA-binding assay in solution.
The DNA-binding assay was based on a method previously described by van Schaik et al. (2005), and used the same buffers and volumes. Briefly, microtitre plates were coated with poly-l-lysine, but not blocked with BSA (Sigma), and coated with salmon sperm DNA (200 μg ml−1). Blocking with 4 % (w/v) BSA was then performed. Pili, purified by the shearing and ammonium-sulfate precipitation method of Brinton et al. (1978), were biotinylated with sulfosuccinimidyl-6-(biotin-amido)hexonoate (Pierce), following the manufacturer's instructions, and allowed to bind for 2–3 h. Binding was detected with avidin–alkaline phosphatase (Sigma), using 4-nitrophenyl phosphate (Fluka) as substrate in developing buffer (50 mM Tris/HCl, 3 mM MgCl2, pH 9.6). Absorption was measured at 405 nm, with 490 nm as background. Purified DNA glycosylase MutY (Davidsen et al., 2005) was used as a positive control for assessing DNA binding.
DNA substrates.
The sequences of the oligonucleotide substrates, with and without DUSs, employed in DNA-binding assays are listed in Table 2. Salmon sperm DNA (Roche Diagnostics), N. meningitidis chromosomal DNA prepared in 0.01 M phosphate buffer (pH 7.4), as well as non-labelled oligonucleotides were used as competitive DNA.
Labelling of DNA substrates.
Synthesized oligomers (3.5 pmol) (Table 2) were end-labelled with [γ-32P]ATP (GE Healthcare) or biotin (Invitrogen) using T4 polynucleotide kinase (New England Biolabs), as described by Sambrook et al. (1989). Double-stranded substrates were generated by mixing equal amounts of complementary oligomers, before heating to 95 °C for 5 min and slowly cooling to room temperature, which allowed the oligomers to hybridize. Labelled substrates were separated on 20 % non-denaturing PAGE and extracted by diffusion in water.
Band-shift analysis.
For electromobility shift assays, 200 ng protein was mixed with 2 μl 5× gel shift buffer (250 mM MOPS, pH 7.5, 5 mM EDTA, 5 mM DTT, 25 mM MgCl2, 25 %, v/v, glycerol) in a final volume of 10 μl. Where indicated, competing DNA was added at 1–1400-fold excess 10 min prior to the addition of labelled substrate. Labelled DNA (2000 c.p.m.) substrate was added to the sample on ice, and the mixture was incubated at room temperature for 20 min. Electrophoresis was carried out on 6 % polyacrylamide gels in Tris/glycine/EDTA buffer. Gels were dried, exposed to a PhosphorImager cassette, and scanned in a PhosphorImager 445 SI or Typhoon scanner (both from GE Healthcare).
Southwestern analysis.
For the solid-phase overlay assays, protein samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were equilibrated and incubated overnight in renaturing buffer (0.5 % BSA, 0.25 % gelatin, 0.2 % Triton X-100, 10 mM Tris/HCl, pH 7.5, 5 mM β-mercaptoethanol, 100 mM NaCl). Probing with biotinylated DNA substrates (Invitrogen) (Table 2) was done overnight in renaturing buffer. The membranes were washed three times with washing buffer (10 mM Tris/HCl, pH 7.5, 100 mM NaCl) before incubation with alkaline phosphatase-conjugated streptavidin (Chemicon), dilution in renaturing buffer, and subsequent detection using 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NBT) as substrates (Life Technologies).
Electron microscope imaging of PilQ bound to biotin-labelled DNA.
PilQ complex was purified from N. meningitidis strain M1080 membranes as previously described (Collins et al., 2001, 2003). Ten-base-pair oligonucleotides for both strands corresponding to the DUS (5′-GCCGTCTGAA-3′) and a DUS-negative sequence with five residues changed by 5′ biotinylation (5′-GAAGTACGAC-3′) were purchased from MWG. For gold-labelling experiments, 10 μl streptavidin with 5 nm gold label (Sigma) was mixed with equimolar amounts of biotin-labelled DNA and incubated on ice for 12 h. These DNA–gold conjugates were then added to 10 μl purified PilQ at a protein concentration of 50 μg ml−1, and incubated for 24 h at 4 °C with gentle agitation. Samples were then centrifuged at 13 000 r.p.m. in a bench-top centrifuge for 5 min, before the supernatant was extracted and prepared for negative staining. Aliquots of the PilQ oligomer–DNA incubation mixture were adsorbed to freshly glow-discharged carbon-coated copper grids, prior to application of negative stain (4 %, w/v, uranyl acetate) for 30 s, and briefly blotted onto double-layered Whatman 50 filter paper. Grids were analysed using a Philips CM100 transmission electron microscope (Philips Electron Optics) operating at an accelerating voltage of 100 keV.
RESULTS
Purified pili from N. meningitidis and P. aeruginosa exhibit weak binding of DNA
Interactions between purified N. meningitidis and P. aeruginosa pili and DNA were assessed in solution (Fig. 1). Various concentrations of protein and DNA were tested for binding under several different conditions, and the protein–DNA interaction was monitored using DNA glycosylase MutY as a positive control (Fig. 1a). This analysis showed that assembled N. meningitidis and P. aeruginosa fimbrial structures weakly bound DNA under the conditions employed (Fig. 1b), and that binding could be inhibited by competing DNA- and pilus-specific antibodies (Fig. 1c, d). Purified N. meningitidis and P. aeruginosa pili did not bind DNA in a band-shift assay (data not shown).
Fig. 1.
Detection of pilus–DNA interaction in solution. DNA was immobilized in the well and overlaid with protein in solution. (a) Binding of DNA glycosylase MutY to DNA (positive control protein). Saturation was reached at 1 μg MutY. One representative experiment out of four is shown. (b) Binding of N. meningitidis M1080 and P. aeruginosa PAK pili to DNA. The mean of four independent measurements using each purified pilus preparation is shown. Error bars indicate sd. (c) Competition of pili binding to immobilized DNA by preincubation with DNA in solution. The average of two independent measurements using pilus protein is shown, as above. Error bars indicate minimum and maximum values. (d) Competition of pili binding to immobilized DNA by preincubation with pilus-specific antibodies. The average of two independent measurements using 0.125 μl pilus proteins is shown, as above. Error bars indicate minimum and maximum values.
Distribution of positively charged sequences within PilQ
The weak affinity of pili for DNA suggested that other pilus biogenesis components could contribute to DNA binding. The search for DNA-binding signatures in the predicted PilQ secondary structure, using various algorithms, did not reveal any recognized DNA-binding motifs such as helix–turn–helix, helix–loop–helix, leucine zipper or zinc finger motifs (data not shown). An analysis of the distribution of positively charged residues within the PilQ sequence showed that several stretches of such sequences were distributed throughout the reading frame (Fig. 2a). In common with many DNA-binding proteins, some of these regions could be involved in mediating nucleic acid recognition (Misra & Honig, 1995). The N-terminal octameric repeat region (SBR) (Tønjum et al., 1998) was one of the major positively charged regions identified.
Observation of DNA binding to native PilQ complex using a band-shift assay
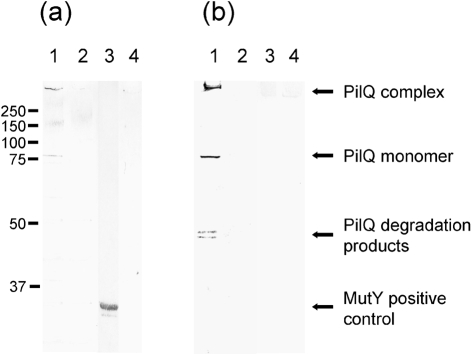
The PilQ complex, purified directly from meningococcal outer membranes, was assessed for DNA binding using band-shift assays. PilQ was mixed with radioactively labelled DNA substrates, some with and some without the DUS recognition sequence. The native PilQ complex showed a preference for binding to ssDNA, although some binding to dsDNA was also discernable (Fig. 3a). In a competition assay, ssDNA was more effective at competing for binding than dsDNA (Fig. 3b). To evaluate the possibility that DNA binding is dependent on the presence of the DUS or base composition, short oligonucleotides with various DUS contents were employed (Table 2). No sequence or DUS specificity in PilQ complex-mediated DNA binding was observed (Fig. 3b). To validate the DNA-binding properties of PilQ, a solid-phase overlay assay in the form of Southwestern analysis was also employed (Fig. 4). The binding of ssDNA to the PilQ multimer, which runs at the top of the stacking gel, was readily apparent using samples from whole-cell lysates (Fig. 4a). Similar results were obtained using dsDNA (data not shown), although binding of ssDNA was observed to be of higher affinity than that of dsDNA, irrespective of the presence or absence of DUS in the DNA substrate. The PilQ degradation product, routinely observed in secretin preparations and representing the C-terminal part of the monomer, did not bind DNA. This indicates that the C-terminal part of the PilQ monomer, which makes up the degradation product (Tønjum et al., 1998), is not the region of PilQ that promotes binding of DNA.
Fig. 3.
DNA-binding characteristics of the native PilQ complex measured by band-shift analysis. The interaction of native PilQ complex with ssDNA and dsDNA substrates was studied. (a) Band-shift analysis of PilQ complex binding to ssDNA (T7) or dsDNA (T7+T8 hybrid) substrates. Lane 1, free labelled DNA substrates (no PilQ); lane 2, DNA and PilQ complex; lane 3, DNA and Taq polymerase (positive control); lane 4, DNA and BSA (negative control). (b) Competition band-shift experiments, showing the ability of unlabelled ss- or dsDNA ligands to compete with the binding of labelled ligands. Results are compared for oligonucleotides containing (T1/T2) or not containing (T3/T4) DUSs. Lane 1, free labelled DNA substrates; lanes 2–4, PilQ complex.
Fig. 4.
Binding of the PilQ complex to DNA assessed by a solid-phase overlay assay in the form of Southwestern analysis. Southwestern analysis of the proteins reacted with ssDNA substrate (T1) (a) and immunoblotting with PilQ-specific antibody (b) is shown. Lanes 1, M1080 whole-cell lysate; lanes 2, M1080-ΔPilQ whole-cell lysate; lanes 3, MutY (positive control); lanes 4, BSA (negative control). Markers on the left give the molecular mass in kDa; arrows on the right indicate the migration positions of the PilQ complex, PilQ monomer, PilQ degradation product typical for secretins, and DNA glycosylase MutY (positive control protein for DNA binding).
Investigation of DNA binding using recombinant PilQ fragments
The results obtained in the band-shift and Southwestern assays established that native PilQ, purified directly from meningococcal membranes in an assembled state, was able to bind various DNA oligomers. When optimizing the protein–DNA ratio, it was found that 200 ng protein was adequate, which is the amount of protein regularly employed in band-shift assessments of, for example, DNA repair proteins (Davidsen et al., 2005). In order to determine the PilQ region responsible for mediating DNA binding, a range of recombinant fragments of PilQ was prepared (Fig. 2b). ssDNA- and dsDNA-binding capability was detected for some of the recombinant PilQ fragments (Fig. 5, Table 4). The PilQ25–354 fragment exhibited the most evident DNA-binding capability in band-shift analysis. Interestingly, increasing protein concentrations produced increasing band-shift patterns for ssDNA, as well as for dsDNA (Fig. 5). The partial band shifts observed might have been due to protein degradation during non-saturated binding of DNA, in combination with the effect of the electrostatic charge of this protein on its migration pattern.
Fig. 5.

Interaction of recombinant PilQ with DNA substrates in band-shift analysis: dependence on protein concentration and DNA substrate. Free labelled (2000 c.p.m.) ssDNA T7 and dsDNA (T7+T8 hybrid) ligands were incubated with the indicated amount of recombinant N-terminal PilQ protein PilQ25–354, and run under standard conditions. The migration of the shifted DNA substrates appeared to be affected by the PilQ protein concentration and/or charge. Filled arrow, free oligonucleotide; open arrow, partial shift, typical of PilQ25–354; filled arrowhead, full band shift. At high protein concentrations, the sticking of oligonucleotides to protein in the well was also observed.
Table 4.
Competitive DNA band-shift analysis with recombinant N-terminal PilQ
The interaction of recombinant N-terminal PilQ25–354 with DNA with and without the DUS is shown. DNA substrates were single stranded (T1, T3) and double stranded (T1+T2 hybrid, T3+T4 hybrid). Competition with non-labelled competitor ss- or dsDNA is indicated. +, Reactivity in the band shift; (+), weak binding of DNA; −, no visible band shift.
| DNA substrate | Competing DNA | ||
|---|---|---|---|
| None | ssDNA | dsDNA | |
| T1/DUS+ | ++ | − | + |
| T1+T2/DUS+ | ++ | − | − |
| T3/DUS− | ++ | − | (+) |
| T3+T4/DUS− | ++ | − | − |
The determination of the binding of all four recombinant PilQ fragments and control proteins to ssDNA and dsDNA by band-shift analysis is shown in Table 3. Among the PilQ recombinants, the PilQ25–354 protein bound DNA much more strongly than did the other proteins, mapping the region mediating DNA binding to N-terminal PilQ, and corroborating the results from the solid-phase overlay analysis (Fig. 4). Further testing of chimeric meningococcal PilQ complex from strain TT31-221, in which the N-terminal positively charged SBR region is replaced by the corresponding but non-homologous region in P. aeruginosa PilQ (Tønjum et al., 1998), demonstrated that this macromolecule exhibited reduced DNA binding only (data not shown). As described above, the wild-type PilQ complex bound more readily to ssDNA than to dsDNA, and dsDNA was less competitive than ssDNA. One exception to this finding was the N-terminal PilQ25–354 fragment, which bound both dsDNA and ssDNA (Table 3, Fig. 5). Analysis of other recombinant truncated PilQ proteins showed that they and the neisserial outer-membrane proteins (HmbR and OpcA) employed as negative controls did not bind DNA (Table 3). Competitive band-shift analysis with the N-terminal PilQ25–354 fragment confirmed that both dsDNA and ssDNA compete for binding to this particular protein, although ssDNA was a more effective competitor than dsDNA (Table 4). The more potent effect of ssDNA in competition for DNA-binding affinity was verified by adding a surplus of competing non-labelled salmon sperm or meningococcal chromosomal DNA (data not shown).
Table 3.
Interaction of recombinant truncated PilQ proteins in band-shift analysis with ss- and dsDNA
ssDNA comprised primers T1, T2, T3, T7 and T8, and dsDNA comprised T1+T2, T3+T4 and T7+T8 hybrids (see Table 2). +, Positive reactivity in the band shift; (+), weak binding of DNA; −, no visible band shift.
| PilQ recombinant | DNA substrates | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ssDNA | dsDNA | ||||||||
| T1 | T2 | T3 | T4 | T7 | T8 | T1+T2 hybrid | T3+T4 hybrid | T7+T8 hybrid | |
| DUS+ | DUS+ | DUS− | DUS− | DUS+ | DUS+ | DUS+ | DUS− | DUS+ | |
| PilQ25–132 | − | − | − | − | − | (+) | (+) | + | (+) |
| PilQ25–354 | + | + | + | + | + | + | + | + | + |
| PilQ218–478 | − | (+) | − | − | (+) | − | − | (+) | (+) |
| PilQ352–761 | − | − | − | − | − | − | − | − | − |
| OpcA* | − | − | − | − | − | − | − | − | − |
| HmbR* | − | − | − | − | − | − | − | − | − |
| MutY† | (+) | (+) | (+) | (+) | (+) | (+) | + | + | + |
*Negative control.
†Positive control.
Examination of PilQ complex–DNA interaction by electron microscopy
The binding of DNA to the PilQ complex was studied by transmission electron microscopy. Biotinylated DNA was incubated with the native PilQ complex, and binding detected with an avidin–nanogold conjugate (Fig. 6). Control incubation, which contained just PilQ and the avidin–nanogold reagent, showed evidence for separate PilQ (Fig. 6c, black arrow) and nanogold particles (white arrows). Pre-incubation of the avidin–gold with either ds- or ss-biotinylated DNA resulted in labelling of the PilQ complex in a highly specific manner (Fig. 6a, b, arrows). This was apparent from the stain exclusion visible around the circumference of the nanogold particles (Fig. 6d, compare left with right column). High electron density was consistently found at the centre of the projection of the complex, rather than at the periphery. The results suggested that DNA binding to PilQ is mediated by a specific site or sites within the PilQ complex.
Fig. 6.
Interaction between DNA and the PilQ complex visualized by transmission electron microscopy. (a) PilQ complex incubated with avidin–nanogold plus biotinylated dsDNA; complexes are indicated with arrows. Bar, 50 nm. (b) PilQ complex incubated with avidin–nanogold plus biotinylated ssDNA; complexes are indicated with arrows. Bar, 50 nm. (c) PilQ complex incubated with avidin–nanogold; nanogold particles alone are indicated by white arrows, and an unliganded PilQ particle is indicated by the black arrow (negative control). Bar, 50 nm. (d) Panel of example particles in each class: left column, PilQ–avidin–nanogold plus biotinylated ssDNA; middle column, unlabelled PilQ; right column, avidin–nanogold label alone. The perimeter of each particle is indicated. Bar, 16.5 nm. For clarity, the contrast in all the figures has been enhanced.
DISCUSSION
N. meningitidis is naturally and constitutively competent for transformation, and the process is coupled to the expression of type IV pili. However, the exact relationship between the transformation process, pili and pilus biogenesis components is poorly understood. We asked whether neisserial pili or PilQ binds DNA, facilitating the entry of DNA during transformation. Therefore, we investigated the ability of these components to bind DNA by band-shift analysis, in solution and Southwestern analysis, as well as by electron microscopy. We demonstrated that neisserial and P. aeruginosa type IV pili bind DNA only weakly in solution. However, recombinant PilQ monomers and native PilQ complex purified from meningococcal outer membranes bound DNA from several sources. PilQ-mediated DNA binding was not DUS or sequence specific. Interestingly, binding of ssDNA to PilQ was preferred over the binding of dsDNA. The binding appeared to take place in the centre of the PilQ ring-like projection, rather than at the periphery, suggesting that the DNA-binding sites are located within the central portions of the PilQ oligomer. The band-shift analysis of truncated recombinant PilQ monomers showed that the N-terminal PilQ25–354 was the region predominantly involved in DNA binding. Analysis of the PilQ complex made up of chimeric subunits lacking the N-terminal SBR region exhibited much-reduced DNA binding, compared to that of the wild-type PilQ complex. We have previously shown that N-terminal PilQ is predominantly located in the periplasm (Frye et al., 2006), and most likely also contributes to forming the central channel of the PilQ complex. Electrostatic charge prediction of the deduced PilQ amino acid sequence suggested that there is a region of increased positive charge in the N-terminal portion of this secretin (Fig. 2a).
Electron microscopy studies have shown that secretins form stable doughnut-like structures in projection, with the diameter of the central cavity ranging from 6 to 8.8 nm (Bitter et al., 1998; Collins et al., 2003; Nouwen et al., 2000). Electrophysiological measurements have shown that secretins can indeed form aqueous channels (Nouwen et al., 1999). Clearly, such large channels must be gated to preserve the integrity of the outer membrane and periplasmic compartment. Among these secretins, the best characterized is PilQ from N. meningitidis, which also functions in pilus biogenesis (Tønjum et al., 1998). The diameter of the central cavity in the PilQ 12-mer (Collins et al., 2003) fits the proposed pilus fibre model (∼6 nm diameter) (Collins et al., 2003, 2004), and could therefore easily accommodate the DNA double helix (∼2.4 nm). DNA uptake in N. meningitidis requires the presence of a DUS, although no DUS specificity in PilQ DNA binding was observed. This suggests that DUS specificity is conferred at another level. The conundrum that competence for transformation is dependent on pilus expression precludes some of the biological testing one would like to perform to validate the biological significance of the DNA-binding properties detected.
The binding and uptake of transforming DNA into the meningococcal cell can be divided into four stages: entry through the outer membrane, transit of the periplasm, transport across the inner membrane, and integration of the new DNA into the chromosome. We propose that the early part of meningococcal transformation is coupled to pilus retraction, and that transforming DNA is introduced into the cell through the transiently opened PilQ channel in the wake of the retracting pili. A similar model has been suggested for P. stutzeri (Graupner et al., 2001). We further suggest that DNA is introduced to the inner membrane through the positively charged channel formed by the PilQ complex, and that other DNA-binding components exert sequence specificity and process DNA. In such a scenario, ComP (as a composite of the pilus structure or as a separate structure) (Aas et al., 2002) could contribute by catching DNA and presenting it to DNA-binding components such as PilQ and ComE (Chen & Gotschlich, 2001). During these events, one strand of the DNA must be degraded and the ssDNA transported through the inner membrane. The presence of ssDNA in the cytoplasm and periplasm has been detected by Hill and co-workers (Chaussee & Hill, 1998). Degradation of one strand of the transforming DNA has until now been suggested to take place in the periplasm (Chaussee & Hill, 1998), although no recognized nuclease has so far been found. The fact that ssDNA binding to PilQ predominates over dsDNA binding indicates that during transformation either one strand of the transforming DNA is degraded or the DNA-binding epitopes of PilQ are located at the periplasmic surface of the outer membrane, which agrees with recent data on PilQ topology (Frye et al., 2006). On the other hand, there might be a substantial amount of ssDNA in the natural environment of N. meningitidis at the mucosal surface. Wackernagel and co-workers have demonstrated that P. stutzeri can be transformed by ssDNA (Meier et al., 2002), and an abundance of ssDNA has been demonstrated in the N. gonorrhoeae periplasm during transformation (Chaussee & Hill, 1998). Yet another explanation might be that the PilQ-mediated binding of ssDNA is attributable to functions other than transformation, such as phage transduction and/or conjugation. PilQ has recently been described as the secretin used by a filamentous phage (Bille et al., 2005). The genomes of these phages are ssDNA, and the DNA has to interact with PilQ from the periplasm. In addition, no DUS is required for phage production or conjugation.
No pilus–DNA interaction could be detected by band-shift analysis, in either N. meningitidis or P. aeruginosa, probably due to steric hindrance. The lack of DNA binding by neisserial pili in a solid-phase overlay has been demonstrated by Mathis and Scocca (1984). We could, however, detect weak N. meningitidis and P. aeruginosa pilus-mediated DNA binding when employing a solution-based assay, corroborating the findings of van Schaik et al. (2005) for P. aeruginosa pili. An extended positively charged surface patch has been proposed for P. aeruginosa pili, and this patch is suggested to be responsible for binding DNA without sequence specificity (van Schaik et al., 2005). Tainer and co-workers have, based on their structural analysis, predicted similar positively charged patches or grooves along the assembled neisserial pilus (Parge et al., 1995). If electrostatic charge is important in promoting protein–DNA interactions, the neisserial pilin subunit PilE (theoretical pI 9.19) should be even more prone to charge-mediated interaction with the negatively charged DNA than the P. aeruginosa pilin subunit PilA (theoretical pI 6.24). Furthermore, it has been demonstrated that the pilA gene of P. aeruginosa can complement a pilE null mutant in N. gonorrhoeae to regain competence, implying that, in neisserial transformation, the DUS specificity is not imparted by the neisserial pilus subunit (Graupner et al., 2001). Based on these collective findings, we propose that binding of neisserial and P. aeruginosa pili to DNA takes place only to a minor extent compared to the DNA binding exerted by PilQ.
We believe that we have described for the first time that the outer-membrane protein PilQ binds DNA in a non-DUS-specific manner. Based on these findings, we suggest that the PilQ complex is involved in non-sequence-specific and DUS-independent DNA binding in the meningococcus during the transformation process. The ability of the PilQ complex, and to some extent pili, of N. meningitidis to bind DNA could therefore contribute to the presentation of DNA to the meningococcal cell during the early part of natural transformation. In addition, the interactions among macromolecules such as PilQ, type IV pili and DNA could be involved in biofilm formation during mucosal surface colonization, or in the course of infection. The ultimate goal is to define how DNA-binding components are involved in the dynamic multi-site targeting, entry and processing of DNA during natural transformation.
Acknowledgments
This work received financial support from the Research Council of Norway (RCN), the FUGE (National Program for Research in Functional Genomics in Norway) platform CAMST (Consortium for Advanced Microbial Sciences and Technologies) supported by RCN, EU fifth framework grant QLRT CT 1999-00359, and the Wellcome Trust, UK. We are indebted to H. K. Tuven for excellent technical assistance.
Abbreviations
DUS, DNA uptake sequence
SBR, small basic repeat
References
- Aas, F. E., Wolfgang, M., Frye, S., Dunham, S., Lovold, C. & Koomey, M. (2002). Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol 46, 749–760. [DOI] [PubMed] [Google Scholar]
- Bille, E., Zahar, J. R., Perrin, A., Morelle, S., Kriz, P., Jolley, K. A., Maiden, M. C., Dervin, C., Nassif, X. & Tinsley, C. R. (2005). A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med 201, 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter, W., Koster, M., Latijnhouwers, M., de Cock, H. & Tommassen, J. (1998). Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Bøvre, K., Bergan, T. & Froholm, L. O. (1970). Electron microscopical and serological characteristics associated with colony type in Moraxella nonliquefaciens. Acta Pathol Microbiol Scand [B] Microbiol Immunol 78, 765–779. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bradley, D. E. (1974). The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58, 149–163. [DOI] [PubMed] [Google Scholar]
- Brinton, C. C., Bryan, J., Dillon, J.-A., Guerina, N., Jacobson, L. J., Labik, A., Lee, S., Levine, A., Lim, S. & other authors (1978). Uses of pili in gonorrhea control: role of bacterial pili in disease, purification and properties of gonococcal pili, and progress in the development of a gonococcal pilus vaccine for gonorrhoeae. In Immunobiology of Neisseria gonorrhoeae, pp. 155–178. Edited by G. E. Brooks, E. C. Gotschlich, K. H. Homes, W. D. Sawyer & F. E. Young. Washington, DC: American Society for Microbiology.
- Bøvre, K. (1964). Studies on transformation in moraxella and organisms assumed to be related to moraxella. 1. A method for quantitative transformation in moraxella and neisseria, with streplomycin resistance as the genetic marker. Acta Pathol Microbiol Scand 61, 457–473. [DOI] [PubMed] [Google Scholar]
- Carbonnelle, E., Helaine, S., Nassif, X. & Pelicic, V. (2006). A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol 61, 1510–1522. [DOI] [PubMed] [Google Scholar]
- Chaussee, M. S. & Hill, S. A. (1998). Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J Bacteriol 180, 5117–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I. & Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat Rev Microbiol 2, 241–249. [DOI] [PubMed] [Google Scholar]
- Chen, I. & Gotschlich, E. C. (2001). ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J Bacteriol 183, 3160–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. F., Davidsen, L., Derrick, J. P., Ford, R. C. & Tønjum, T. (2001). Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J Bacteriol 183, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. F., Ford, R. C., Kitmitto, A., Olsen, R. O., Tønjum, T. & Derrick, J. P. (2003). Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J Bacteriol 185, 2611–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R. F., Frye, S. A., Kitmitto, A., Ford, R. C., Tønjum, T. & Derrick, J. P. (2004). Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 Å resolution. J Biol Chem 279, 39750–39756. [DOI] [PubMed] [Google Scholar]
- Collins, R. F., Frye, S. A., Balasingham, S., Ford, R. C., Tønjum, T. & Derrick, J. P. (2005). Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J Biol Chem 280, 18923–18930. [DOI] [PubMed] [Google Scholar]
- Davidsen, T., Bjørås, M., Seeberg, E. C. & Tønjum, T. (2005). Antimutator role of DNA glycosylase MutY in pathogenic Neisseria species. J Bacteriol 187, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, S. L. & Koomey, M. (1995). The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol 18, 975–986. [DOI] [PubMed] [Google Scholar]
- Frasch, C. E. & Chapman, S. S. (1972). Classification of Neisseria meningitidis group B into distinct serotypes. I. Serological typing by a microbactericidal method. Infect Immun 5, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, A., Hartsch, T. & Averhoff, B. (2001). Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl Environ Microbiol 67, 3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøholm, L. O., Jyssum, K. & Bøvre, K. (1973). Electron microscopical and cultural features of Neisseria meningitidis competence variants. Acta Pathol Microbiol Scand [B] Microbiol Immunol 81, 525–537. [DOI] [PubMed] [Google Scholar]
- Frye, S. A., Assalkhou, R., Collins, R. F., Ford, R. C., Petersson, C., Derrick, J. P. & Tonjum, T. (2006). Topology of the outer-membrane secretin PilQ from Neisseria meningitidis. Microbiology 152, 3751–3764. [DOI] [PubMed] [Google Scholar]
- Goodman, S. D. & Scocca, J. J. (1988). Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 85, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graupner, S., Weger, N., Sohni, M. & Wackernagel, W. (2001). Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J Bacteriol 183, 4694–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, H. L. & Dillard, J. P. (2006). Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol 59, 376–385. [DOI] [PubMed] [Google Scholar]
- Koomey, M. (1998). Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl 84, 56–61. [DOI] [PubMed] [Google Scholar]
- Koomey, J. M. & Falkow, S. (1987). Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol 169, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, C. D., Tobiason, D. M., Lazio, M. P., Kline, K. A. & Seifert, H. S. (2003). Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect Immun 71, 6279–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. G. & Wackernagel, W. (1990). Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol 154, 380–385. [DOI] [PubMed] [Google Scholar]
- Mathis, L. S. & Scocca, J. J. (1984). On the role of pili in transformation of Neisseria gonorrhoeae. J Gen Microbiol 130, 3165–3173. [DOI] [PubMed] [Google Scholar]
- Mattick, J. S. (2002). Type IV pili and twitching motility. Annu Rev Microbiol 56, 289–314. [DOI] [PubMed] [Google Scholar]
- Mehr, I. J. & Seifert, H. S. (1998). Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol 30, 697–710. [DOI] [PubMed] [Google Scholar]
- Meier, P., Berndt, C., Weger, N. & Wackernagel, W. (2002). Natural transformation of Pseudomonas stutzeri by single-stranded DNA requires type IV pili, competence state and comA. FEMS Microbiol Lett 207, 75–80. [DOI] [PubMed] [Google Scholar]
- Misra, V. K. & Honig, B. (1995). On the magnitude of the electrostatic contribution to ligand–DNA interactions. Proc Natl Acad Sci U S A 92, 4691–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen, N., Ranson, N., Saibil, H., Wolpensinger, B., Engel, A., Ghazi, A. & Pugsley, A. P. (1999). Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A 96, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen, N., Stahlberg, H., Pugsley, A. P. & Engel, A. (2000). Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J 19, 2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G. A. & Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Parge, H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D. & Tainer, J. A. (1995). Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378, 32–38. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I. & Bleasby, A. (2000). emboss: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Skaar, E. P., Lazio, M. P. & Seifert, H. S. (2002). Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol 184, 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B. J. & Kwaik, Y. A. (1999). Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol 181, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, J., Kraus, S. J. & Gotschlich, E. C. (1971). Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J Exp Med 134, 886–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønjum, T. & Koomey, M. (1997). The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships – a review. Gene 192, 155–163. [DOI] [PubMed] [Google Scholar]
- Tønjum, T., Hagen, N. & Bøvre, K. (1985). Identification of Eikenella corrodens and Cardiobacterium hominis by genetic transformation. Acta Pathol Microbiol Immunol Scand [B] 93, 389–394. [DOI] [PubMed] [Google Scholar]
- Tønjum, T., Freitag, N. E., Namork, E. & Koomey, M. (1995). Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol 16, 451–464. [DOI] [PubMed] [Google Scholar]
- Tønjum, T., Caugant, D. A., Dunham, S. A. & Koomey, M. (1998). Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol Microbiol 29, 111–124. [DOI] [PubMed] [Google Scholar]
- van Schaik, E. J., Giltner, C. L., Audette, G. F., Keizer, D. W., Bautista, D. L., Slupsky, C. M., Sykes, B. D. & Irvin, R. T. (2005). DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J Bacteriol 187, 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang, M., van Putten, J. P., Hayes, S. F. & Koomey, M. (1999). The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol 31, 1345–1357. [DOI] [PubMed] [Google Scholar]