Abstract
Generation of reprogrammed induced pluripotent stem cells (iPSC) from patients with defined genetic disorders promises important avenues to understand the etiologies of complex diseases, and the development of novel therapeutic interventions. We have generated iPSC from patients with LEOPARD syndrome (LS; acronym of its main features: Lentigines, Electrocardiographic abnormalities, Ocular hypertelorism, Pulmonary valve stenosis, Abnormal genitalia, Retardation of growth and Deafness), an autosomal dominant developmental disorder belonging to a relatively prevalent class of inherited RAS-MAPK signaling diseases, which also includes Noonan syndrome (NS), with pleiomorphic effects on several tissues and organ systems1,2. The patient-derived cells have a mutation in the PTPN11 gene, which encodes the SHP2 phosphatase. The iPSC have been extensively characterized and produce multiple differentiated cell lineages. A major disease phenotype in patients with LEOPARD syndrome is hypertrophic cardiomyopathy. We show that in vitro-derived cardiomyocytes from LS-iPSC are larger, have a higher degree of sarcomeric organization and preferential localization of NFATc4 in the nucleus when compared to cardiomyocytes derived from human embryonic stem cells (HESC) or wild type (wt) iPSC derived from a healthy brother of one of the LS patients. These features correlate with a potential hypertrophic state. We also provide molecular insights into signaling pathways that may promote the disease phenotype.
Approximately 90% of LS cases, and 45% of NS, are caused by missense mutations in the PTPN11 gene that encodes the protein tyrosine phosphatase SHP2. PTPN11 is ubiquitously expressed, essential for normal development, and somatic mutations in this gene contribute to leukemogenesis in children3,4. For LS, two mutations, T468M and Y279C, are most recurrent5. Hipertrophic cardiomyopathy is the most common life–threatening cardiac anomaly in LS2. Animal models of LS have been generated in Drosophila and zebrafish6,7, but the molecular pathogenesis of LS remains obscure.
Ectopic expression of four transcription factors (OCT4, SOX2, KLF4 and c-MYC) in adult human dermal fibroblasts can generate pluripotent iPSC8–10. Together with defined in vitro differentiation protocols, this suggests the possibility of developing reliable disease models11–14. We have established iPSC lines from two LS patients, a 25-year-old female (L1), and a 34-year-old male (L2). A heterozygous T468M substitution mutation in PTPN11 is present in both.
Fibroblasts were transduced with OCT4-, SOX2-, KLF4- and c-MYC-encoding VSV-pseudotyped Moloney-based retroviral vectors. Compact ESC-like colonies emerged after two weeks and TRA-1-81-positive colonies were clonally expanded to create stable LS-iPSC lines (Supplementary Fig. 1)8. Three iPSC lines per patient were used for preliminary characterization: L1-iPS1, L1-iPS6, L1-iPS13, L2-iPS6, L2-iPS16 and L2-iPS18.
To verify that the iPSC originated from patient-derived fibroblasts, we performed DNA fingerprinting analysis (Supplementary Fig. 2a). All iPSC had normal karyotypes of 46,XX (L1) and 46,XY (L2) (Supplementary Fig. 2b and data not shown). In addition, they carried the expected T468M mutation (Supplementary Fig. 3a). Restriction fragment length polymorphism analysis of an RT-PCR amplimer containing the mutation with BsmFI showed biallelic expression of PTPN11 (Supplementary Fig. 3b). PCR and Southern blots indicated the presence of all four transgene proviruses in the LS-iPSC (Supplementary Fig. 4) and quantitative RT-PCR (qRT-PCR) results confirmed efficient transgene silencing (Supplementary Fig. 5).
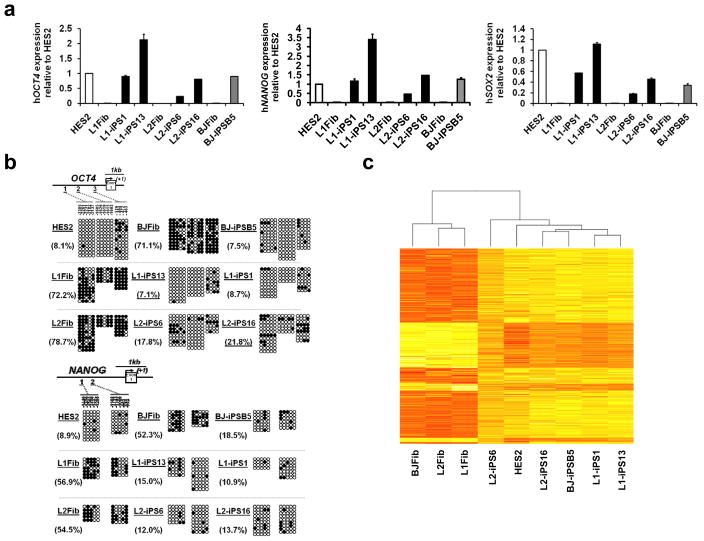
To further characterize the LS-iPSC clones, expression of several HESC markers in two LS-iPSC lines from each patient (L1-iPS1, L1-iPS13, L2-iPS6 and L2-iPS16) was analyzed and compared to the HES2 HESC and a wt-iPSC line, BJ-iPSB5, derived in our lab from a normal human fibroblast line (BJ). The BJ-iPSB5 cell line was also karyotypically normal (46,XY), contained all four transgene proviruses, which were silenced (Supplementary Fig. 2b, 4b and 5b). All LS and control iPSC lines exhibited high alkaline phosphatase activity, and expressed pluripotency markers, including surface antigens TRA-1-81, TRA-1-60, and SSEA-4, as well as the nuclear transcription factors OCT4 and NANOG (Supplementary Fig. 6). Activation of a series of endogenous stemness genes (OCT4, NANOG, SOX2, GDF3, DPPA4, REX1 and TERT) in iPSC was confirmed by qRT-PCR (Fig. 1a and Supplementary Fig. 7a). Extensive demethylation of CpG dinucleotides in the OCT4 and NANOG promoters compared to their parental fibroblasts was confirmed by bisufite sequencing (Fig. 1b).
Figure 1. Gene expression profile in LS-iPSC is similar to HESC.
a, Quantitative real-time PCR assay for the expression of endogenous hOCT4, hNANOG and hSOX2 in iPSC and parental fibroblasts (Fib). PCR reactions were normalized against β-ACTIN and plotted relative to expression levels in HES2. Error bars indicate ± s.d. of triplicates. b, Bisulfite sequencing analyses of the OCT4 and NANOG promoters. The cell line and the percentage of methylation is indicated to the left of each cluster. c, Heat map showing hierarchical clustering of 3657 genes with at least two-fold expression change between the average of the three fibroblast cell lines versus all the iPSC lines/HES samples. Expression levels are represented by color; red indicates lower and yellow higher expression.
We next examined genome-wide mRNA expression profiles of two LS-iPSC lines from each patient, the BJ-iPSB5 cell line, parental fibroblasts and HES2 cells. The resulting heat map and scatter-plot analyses indicated that iPSC lines shared a higher degree of similarity with HES2 cells than with their parental fibroblast cell lines (Fig. 1c and Supplementary Fig. 7b).
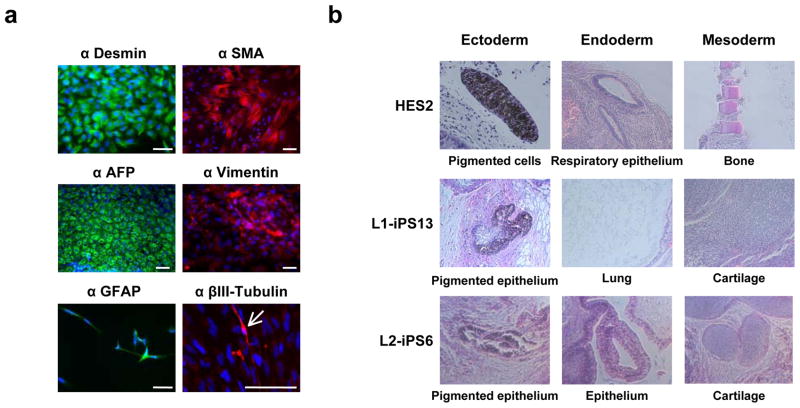
Pluripotent HESC can differentiate into cell types representative of all three germ layers. We tested the differentiation abilities of our iPSC using an in vitro floating embryoid body (EB) system, followed by replating on gelatin-coated dishes10,15. Immunocytochemistry analyses detected expression of α-smooth muscle actin (α-SMA, mesoderm), desmin (mesoderm), α-fetoprotein (AFP, endoderm), vimentin (mesoderm), glial fibrillary acidic protein (GFAP, ectoderm) and βIII-tubulin (ectoderm) markers (Fig. 2a and Supplementary Fig. 8). In order to determine pluripotency in vivo, we injected LS-iPS, BJ-iPSB5 and HES2 cells into immune-compromised NOD-SCID mice. Histological analyses of the resulting teratomas showed cell types representative of the three germ layers, including pigmented cells (ectoderm), lung, respiratory and gut-like epithelia (endoderm), and mesenchyme, adipose tissue and cartilage (mesoderm) (Fig. 2b and data not shown).
Figure 2. LS-iPSC differentiate in vitro and in vivo into all three germ layers.
a, L2-iPS6 cells were differentiated as floating EBs for eight days and then plated onto gelatin-coated dishes and allowed to differentiate for another eight days. Immunocytochemistry showed cell types positively stained for differentiation markers including Desmin/SMA (mesoderm), AFP (endoderm), vimentin (mesoderm), and GFAP/βIII-Tubulin (ectoderm). The arrow indicates a βIII-tubulin-positive cell. Scale bar, 100 μm. b, HES2, L1-iPSC and L2-iPSC were injected subcutaneously into the right hindleg of immuno-compromised NOD-SCID mice. The resulting teratomas were stained with hematoxylin and eosin and tissues representative of all three germ layers were observed.
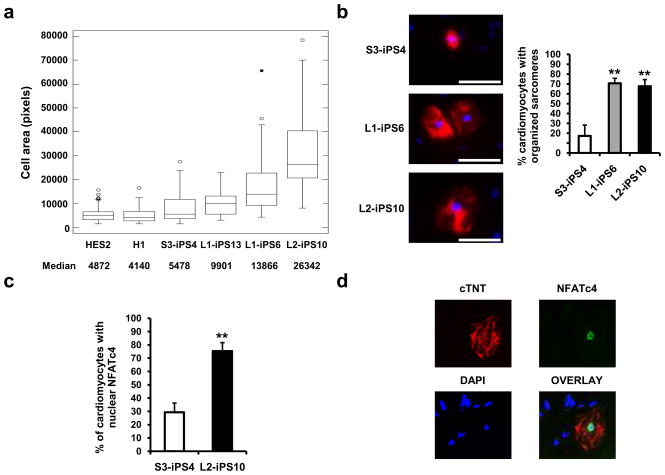
As mentioned previously, hypertrophic cardiomyopathy is one of the major features of LS, affecting 80% of the patients. In addition, affected individuals occasionally manifest hematologic complications such as myelodysplasia and leukemia16,17. Therefore, we asked if LS-iPSC were able to differentiate into hematopoietic and cardiac lineages. LS-iPSC from both patients differentiated into a variety of hematopoietic cell types including early hematopoietic progenitors (CD41+)18, early erythroblasts (CD71+/CD235a+)19, and macrophages (CD11b+)20 (Supplementary Fig. 9 and data not shown). The cardiac hypertrophic response includes induction of immediate-early genes (such as c-jun, c-fos and c-myc), an increase in cell size, and organization of contractile proteins into sarcomeric units21,22. To have an appropriate control cell line to analyze some of these parameters, besides HESC, we generated a wt-iPSC line (S3-iPS4) from fibroblasts obtained from an unaffected brother of L1 without the T468M mutation (Supplementary Fig. 10 and Supplementary Fig. 11). Using a well-established cardiac differentiation protocol23, we observed contracting EBs emerging around day 11 of differentiation (Supplementary Movies 1–7). In order to monitor cardiac development, we analyzed cardiac troponin T (cTNT) expression on day 18 of differentiation by flow cytometry (data not shown). Replated cells from beating EBs were processed as described in Material and Methods. Briefly, cells were fixed, immunostained for cTNT (Fig. 3b), and 50 cardiomyocytes were randomly chosen from each sample for surface area measurement using a computerized morphometric system (ImageJ software, NIH). Cardiomyocytes derived from LS-iPSC lines: L1-iPS13, L1-iPS6 and L2-iPS10 (Supplementary Fig. 10 and Supplementary Fig. 11), had a significantly increased median surface area compared to wt-iPSC cardiomyocytes; 1.8 times, 2.5 times and 4.8 times larger, respectively, whereas the area median of the cardiomyocytes obtained from HESC was similar to wt-iPSC cardiomyocytes (Fig. 3a). We also observed increased sarcomere assembly in L1-iPS6 and L2-iPS10 cells when compared to wt S3-iPS4 cells (Fig. 3b). Recently, the calcineurin-NFAT pathway has been shown to be an important regulator of cardiac hypertrophy. Active calcineurin dephosphorylates NFAT transcription factors, resulting in their nuclear translocation22,24. We analyzed the localization of NFATc4 using immunocytochemistry in 50 cTNT-positive cardiomyoctes derived from the L2-iPS10 cell line, which produced the largest cardiomyocytes, and wt S3-iPS4. We observed a significantly higher proportion of LS cardiomyocytes with nuclear NFATc4, (~80% versus ~30%, respectively) (Fig. 3c and 3d).
Figure 3. Cardiomyocytes derived from LS-iPSC show hypertrophic features.
a, HES2, H1, wt S3-iPS4 and three LS-iPS clones, were differentiated into cardiac lineage. Cell areas of 50 random cTNT-positive cardiomyocytes of each cell line were measured using ImageJ. Boxes show the span from the median (50th percentile) to the first and third quartiles. The lines represent the largest/smallest sizes that are no more than 1.5 times the median to quartile distance. Additional points drawn represent extreme values. b, Sarcomeric organization was assessed in 50 cTNT positive (red) cardiomyocytes. Data are presented as mean ± s.d. n = 3; **P < 0.01 (Student’s t-test). c, S3-iPS4 and L2-iPS10 cells-derived cardiomyocytes were restained with NFATc4 antibody, and the nuclear versus cytosolic expression was analyzed. n = 3; **P < 0.01 (Student’s t-test). d, Nuclear localization of NFATc protein in a cTNT-positive cell from L2-iPS10 is shown.
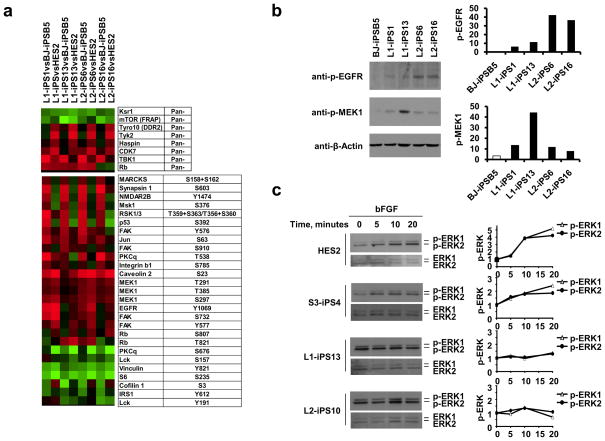
In order to identify potential molecular targets that could be affected by the T468M PTPN11 mutation, protein extracts from LS-iPS, wt BJ-iPSB5 and HES2 cells were analyzed using a phosphoproteomic microarray chip containing approximately 600 pan and phospho-specific antibodies (Kinexus Bioinformatics Corporation). We established eight groups for comparison, each of the LS-iPSC lines versus one control cell line, either HES2 or wt-iPSC. Proteins with a 1.5 fold change were filtered, and those that were conserved in most of the groups were represented in a heat map (Fig. 4a). Some of the proteins were more abundantly present in LS-iPS when compared to either HES2 (Tyro10, Tyk2 and Haspin) or wt-iPSC (p-MARCKs, p-Synapsin1, p-NMDAR2B, p-MSK1, p-RSK1/3 and p-p53). The phosphorylation of other proteins was increased (p-Caveolin2, p-MEK1, p-EGFR and p-FAK) or decreased (p-Vinculin, p-S6 and p-Lck) in LS-iPSC when compared to control cell lines. In order to eliminate false positives, we verified the phosphoproteomic results by Western blot (WB) for three of the most altered proteins (p-S6, p-EGFR and p-MEK1) in four LS-iPSC lines, in comparison to wt-iPSC. While we did not confirm a major change in the phosphorylation status of S6 protein (data not shown), WB confirmed that the phosphorylation of EGFR and MEK1 proteins was considerably increased in the LS-iPSC samples (Fig. 4b).
Figure 4. Phosphoproteomic and MAPK activation analyses.
a, Protein extracts of two iPSC from each LS patient (L1 and L2), wt iPSC (BJ-iPSB5) and HES2 were hybridized to an antibody microarray. The heat map represents the most significant protein changes preserved in all the comparison groups. b, pMEK1 and pEGFR expression was confirmed by Western blot using phospho-specific antibodies. Band density was measured (ImageJ software), and normalized to β-Actin. c, HES2, wt S3-iPS4, and LS-iPSC were serum- and bFGF-starved for 6 hours and then treated with bFGF (20 ng/ml) for the indicated time. Phosphorylated ERK1/2 (p-ERK1/2) and total ERK were assessed by immunoblotting and quantitated. p-ERK1/2 levels were compared to the untreated p-ERK1/2 level in each sample, normalized to the total ERK1/2 and represented graphically at the right of each panel.
RAS-MAPK represents the major signaling pathway deregulated by SHP2 mutants. NS mutants increase basal and stimulated phosphatase activity, whereas LS mutants are catalytically impaired and have dominant-negative effects, inhibiting growth factor-evoked ERK1/2 activation25. We analyzed the ability of LS-iPSC to respond to external growth factors. We used bFGF (basic Fibroblast Growth Factor), the main growth factor in the maintenance of HESC, to induce the stimulation of the MAPK signaling pathway. bFGF treatment increased the phosphorylation of ERK1/2 (p-ERK) levels over time in HES2 and wt S3-iPS4 cells (Fig. 4c). Although the LS-iPSC expressed the four FGF receptor (FGFR) family members (Supplementary Fig. 12a), bFGF stimulation did not cause any substantial change in p-ERK levels (Fig. 4c and Supplementary Fig. 12b-c). However, the LS-iPSC lines had higher basal p-ERK levels compared to HES2 and S3-iPS4 cells (Supplementary Fig. 12b-c), in accordance with the increased pMEK1, ERK upstream kinase, levels found in LS-iPSC samples in phosphoproteomic array results.
In summary, we have generated and characterized LS patient-specific iPSC, providing a new system for the study of disease pathogenesis. Some of the standard procedures to analyze cardiomyocyte hypertrophy (e.g., protein synthesis rate, re-activation of the fetal gene program) could not be reliably assessed due to the variably mixed population of cells obtained using this cardiac differentiation procedure (e.g. endothelial, cardiomyocytes), and the lack of a reliable cell surface marker for cardyomyocyte purification. However, we observed increases in cell size, sarcomeric organization and nuclear NFATc4 localization in LS-iPSC-derived cardiomyocytes, when compared to HESC and wt-iPSC-derived cardiomyocytes. These results would be consistent with cardiac hypertrophy, a condition commonly found in LS patients2, and suggest that this abnormality occurs through a cell autonomous mechanism due to the PTPN11 mutation. Since many human cell types, such as cardiomyocytes, cannot be propagated readily in cell culture, iPS-derived cells exhibiting disease-relevant phenotypes provide the requisite resource for precisely elucidating pathogenesis and pursuing novel therapeutic strategies.
In our studies, we also attempted to provide insights into the molecular events that could be affected by the PTPN11 mutation in the pluripotent iPSC using antibody microarrays. We found that the phosphorylation of certain proteins was increased in LS-iPSC when compared to wild-type HESC and iPSC. Further analysis will be required to elucidate if these proteins/signaling pathways are involved in the development of the disease phenotype. Interestingly, one of the more upregulated phosphoproteins was MEK1, the upstream kinase of ERK1/2, whose gene is sometimes mutated in the related disorder, cardiofaciocutaneous syndrome. PTPN11 mutations underlie 45% and 90% of NS and LS, respectively. It is not well understood how mutations that provoke opposite effects on SHP2 phosphatase activity cause syndromes with similar features26. In concordance with observations in the Drosophila LS model7, basal p-ERK levels were increased in LS-iPSC. Of note, receptor tyrosine kinase stimulation with bFGF in LS-iPSC failed to elicit further activation of ERK, as previously observed in a different cellular model25. Interestingly, this result demonstrates for the first time that RAS-MAPK signal transduction is perturbed in LS as early as the pluripotent stem cell stage.
Taken together, this is the first described human model of an inherited RAS pathway disorder.
METHODS SUMMARY
Cell culture
Dermal fibroblast lines were obtained from skin biopsies, collected under an Institutional Review Board-approved protocol and with informed consent. Fibroblasts and GP2 cells were maintained in Dulbeccós modified Eagle medium (DMEM) containing 10% fetal bovine serum and penicillin/streptomycin (Invitrogen). HES2 and iPSC were maintained on irradiated Swiss Webster mouse embryonic feeder cells (MEFs), in a serum free HESC medium containing 20 ng/ml basic fibroblast growth factor (bFGF, R&D systems).
LEOPARD syndrome iPSC generation
OCT4, SOX2, KLF4 and c-MYC transcription factors were introduced in dermal fibroblasts derived from two patients with LEOPARD syndrome via the pMXs retroviral vector. In parallel, pMXs-EGFP vector was used to estimate the infection efficiency (data not shown). Six days after infection, fibroblasts were seeded onto MEFs. The following day the medium was replaced with the HESC medium and bFGF.
Microarray analysis
Gene level mRNA abundance measures were extracted using the Affymetrix GeneChip Exon 1.0 ST array, and Robust Multi-Array (RMA)-normalized using the Affymetrix Expression Console software. Subsequently, these genes were clustered and a heat map was generated against a background subset of genes showing at least two-fold change between sample averages of iPSC/HES2 cells and fibroblast samples.
In vitro differentiation
For non-lineage specific and hematopoietic differentiation we used previously described protocols10,15, with certain modifications (unpublished data, Kennedy M. and Keller G. et al.). For cardiomyocytes induction, we used a well-established assay23.
Phosphoproteomics
HES2 and iPSC were incubated overnight in HESC culture medium deprived of bFGF and knockout serum replacement (KSR). Protein lysates were quantified by Bradford assay and sent to Kinexus Bioinformatics Corporation (Vancouver, Canada) for antibody microarray screening. The proteins with at least 1.5 fold change between the LS-iPSC samples and control sample (either HES2 or wt-iPSC), and conserved in the majority of the comparison groups were represented in a heat map.
METHODS
Cell culture
LEOPARD syndrome patients derived fibroblasts, BJ fibroblasts (American Type Culture Collection) and GP-2 cells were maintained in DMEM 10% FBS medium. iPSC and HESC were maintained on mitotically inactivated MEFs in HESC medium composed of DMEM/F12 (Cellgro, Mediatech) containing 20% (vol/vol) KSR (Invitrogen), 5% (vol/vol) MEF-conditioned medium, Penicillin/Streptomycin, L-glutamine (L-Gln), non-essential amino acids (Invitrogen), β-mercaptoethanol (β-ME, Sigma-Aldrich) and bFGF (R&D Systems).
Plasmid construction
Full-length sequences of human OCT4, SOX2, KLF4 and c-MYC transcription factors were obtained from Open Biosystems. The coding sequences were PCR amplified using Pfu Turbo (Stratagene) and cloned into pMXs vector and verified by sequencing. pMXs-EGFP vector was constructed by introducing the BamHI/NotI EGFP fragment from FUGW (kindly provided by Dr Lois, MIT, Massachusetts) into pMXs vector. The latter vector was used to monitor the transfection and infection efficiency. Detailed primer and cloning information will be provided upon request.
Retroviral infection and human iPSC generation
GP-2 cells were plated at 8×106 cells per 10-cm dish and transfected with pMXs, VSV-G and Gag-Pol vectors using SuperFect transfection reagent on the following day. The same day, human fibroblasts were seeded at 8×105 cells per 10-cm dish. Twenty-four hours after transfection, the four retroviruses (hOCT4, hSOX2, hKLF-4 and hC-MYC) containing supernatants were collected, and equal amounts of each were mixed and filtered through a 0.45 μm pore-size filter, and supplemented with 4 μg/mL polybrene. Retroviruses containing medium was added to the fibroblasts plates. The following day, forty eight hours post-transfection, the fibroblasts were reinfected following the same procedure as the day before. Six days after transduction, fibroblasts were transferred into four dishes coated with MEFs, at 50,000 fibroblasts per plate.
DNA Fingerprinting analysis
In order to verify the genetic relatedness of the iPSC to their parental fibroblasts, we PCR amplified across three discrete genomic loci containing highly variable numbers of tandem repeats. Genomic DNA (gDNA) was isolated with Easy-DNA™ kit (Invitrogen). Fifty nanograms of genomic DNA was used per reaction. Primers are summarized in Supplementary Table 1 on line.
Karyotype analysis
HES2 and iPSC were grown on Matrigel-coated glass coverslip dishes (MatTek, Ma). The day of culture harvest, 20 μl of colcemid (5 μg/ml) was added to the in situ ESCl culture which was 30–50% confluent. The culture was re-incubated for 15 min at 37°C. A robotic harvester (Tecan) was utilized, which included automatic addition of 2cc of hypotonic solution (sodium citrate solution 0.8%) with incubation for 20 min at room temperature, prefixation with addition of 2cc of fixative (methanol: glacial acetic acid; 3:1), followed by addition of 4cc of fixative, twice. The coverslip was dried completely at 37°C with 45–50% humidity and mounted on a microscope slide and GTG-banded according to standard protocols. Metaphases were captured and karyotypes were prepared using the CytoVision software program (Version 3.92 Build 7, Applied Imaging).
qPCR and transgenes integration
For quantitative real-time PCR (qPCR) analyses, total RNA was extracted from cells using Trizol® Reagent (Invitrogen) and subsequently column-purified with RNeasy kit (Qiagen) and treated with RNase-free DNase (Qiagen). One microgram of total RNA was reverse transcribed into cDNA using random primers and Superscript II Reverse Transcriptase (Invitrogen). PCR for transgene silencing was performed with Expand High Fidelity Enzyme Taq Polymerase (Roche). Real-time qPCR was performed on a StepOne Plus Real-Time PCR System (Applied Biosystems) with Fast SYBR® Green Master Mix (Applied Biosystems). The results were analyzed with the StepOne Software v2.0, normalized to β-Actin gene expression, and compared to HES2 cell expression levels. To examine the presence of transgenes in the iPSC lines, gDNA was isolated with Easy-DNA Kit (Invitrogen). PCR reactions were carried out with the Expand High Fidelity Enzyme Taq Polymerase (Roche). Primer sequences are described in Supplementary Table 1 on line. Primers for FGF receptors expression analysis have been previously described27.
Southern blot analyses
gDNA (2μg) was completely digested with Bgl II, separated on a 0.8% agroase gel, transferred to a positively charged nylon membrane, and hybridized with DIG-labeled hOCT4, hSOX2, hKLF-4 and hC-MYC cDNA probes. After hybridization, membranes were washed, blocked with DIG blocking solution, and incubated with anti-DIG-AP Fab fragments (Roche). Probe-target hybrids were then incubated with chemiluminescent CDP-Star substrates (Roche) and detected via exposure to X-ray film.
Bisulfite sequencing
We treated 500 ng of purified gDNA with sodium bisulfite using the Zymo EZ-DNA Methylation Kit, following the manufacturer’s intructions. The sequences of primers used for amplification of genomic fragments were previously published28. PCR products were then size fractionated in 1% TAE-agarose, extracted using the Qiaquick gel extraction kit (Qiagen) and cloned into the pGEM-T Easy Vector system (Promega). Blue-white selection was applied to eliminate false positives, and twelve random clones were picked and sequenced. Bisulfite conversion efficiency of non-CpG cytosines was >90% for all individual clones for each sample.
Immunocytochemistry, AP staining and FACS analysis
For in vivo immunostaining, HES2 and iPSC were washed once with DMEM medium supplemented with 10% FBS and antibiotics (DMEM 10%), and incubated with biotin-TRA-1-81 antibody (1:100, eBiosciences) for 2 h. Cells were washed three times with DMEM 10% and they were incubated with the secondary antibody streptavidin-FITC (1:100, eBiosciences) and the phycoeritrin TRA-1-60 (PE-TRA-1-60) antibody (1:100, eBiosciences) where indicated. All the incubations were performed in a humidified incubator at 37 °C with 5% CO2. For intracellular staining, cells were fixed in 2% paraformaldehyde for 30 min, and blocked and permeabilized in PBS containing 10% donkey serum, 1% BSA and 0.1% Triton X-100 for 45 min. Cells were incubated with primary antibody in blocking solution overnight at 4 °C, washed and incubated with the corresponding Alexa donkey secondary antibody for 1 h at room temperature (RT). Then cells were washed and stained with DAPI (1 μg/ml) for 20 min. The primary antibodies used for intracellular immunostaining were OCT4 (1:100, BioVision), NANOG (1:100, R&D systems), desmin (1:100, Lab Vision), α-SMA (pre-diluted, DAKO), vimentin (1:100, Chemicon), AFP (1:500, DAKO), GFAP (1:1000, DAKO), βIII-Tubulin (1:100, Chemicon), or NFATc4 (1:100, Santa Cruz Biotechnology). All the secondary antibodies Alexa 488 Donkey anti-Rabbit (1:100), Alexa 546 donkey anti-goat (1:100) and Alexa 546 donkey anti-mouse (1:100) were obtained from Invitrogen. Alkaline phosphatase staining was detected following manufacturer’s recommendations (Millipore). SSEA-4, troponin T, and hematopoietic markers expression were evaluated on a BD Biosciences LSRII FACS machine analyzer. Primary antibodies SSEA-4-PE (R&D systems), cardiac troponin T (Lab Vision), CD11b-APC (Caltag), and CD45-APC, CD45-PE, CD71-PE, CD41-PE and CD235a-APC were purchased from BD Biosciences.
Teratoma formation
All animal procedures were performed in accordance with the Mount Sinai Medical Center’s Institutional Animal Care and Use Committee. Approximately 1–2×106 cells were injected subcutaneously into the right hindleg of immuno-compromised NOD-SCID mice (The Jackson Laboratory). Teratomas were excised 6–10 weeks post-injection, fixed overnight in formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin by the Morphology and Assessment Core of the Department of Gene and Cell Medicine. Histological evaluation was performed using a Nikon TE2000-U microscope and ACT-1 software.
In vitro differentiation
For embryoid body (EB) formation, HES2 and iPSC were treated with collagenase B (Roche) for 10 min, and collected by scraping. After centrifuging, cell pellets were resuspended in basic differentiation media, StemPro 34 (Invitrogen) containing 2 mM L-Gln, 4×10−4 monothioglycerol (MTG), 50 μg/ml ascorbic acid (Sigma) and 150 μg/ml transferrin (Sigma). EBs were grown in ultra low-binding plates (Costar) and medium was changed every three days. After eight days of differentiation, EBs were collected, resuspended in DMEM 10% and transferred to gelatin-coated dishes to allow them to attach and differentiate for eight additional days before processing for immunocytochemistry analyses. For hematopoietic differentiation we used a described protocol29 with certain modifications (unpublished data, Kennedy M. and Keller G. et al.). For cardiomyocyte induction, we used a well-established protocol23.
Microarray analysis
RNA probes were hybridized to Affymetrix GeneChip Exon 1.0ST array according to the manufacturer’s protocols by the Genomics Core Lab in The Institute for Personalized Medicine at Mount Sinai Medical Center. Microarrays were scanned and data were analysed using the Affymetrix Expression Console software.
Cytology/cardiomyocyte size determination
On D18 of differentiation, beating EBs were plated on gelatin coated dishes. Three days after plating, EBs outgrowths were trypsinized, filtered through a 40 μm size pore-size filter, and single cells were replated at low density on gelatin coated dishes. The following day, cells were fixed with 4% para-formaldehyde, permeablized, blocked in PBS/1% BSA/0.1% Triton/10% donkey serum, and Stained for cardiac Troponin T (1:200, Lab Vision), overnight at 4 °C. Stained cells were washed three times with PBS, and then incubated with the Alexa Fluor 547 donkey-anti-mouse antibody (Invitrogen) for 1 h. The areas of HESC- and LS-iPSC-derived cardiomyocytes were analyzed using ImageJ software (NIH).
Phosphoproteomics and Western blotting
We prepared a lysis buffer (pH 7.2) containing 20 mM MOPS pH 7.0, 2 mM EGTA, 5 mM EDTA, 30 mM sodium fluoride, 60 mM β-glycerophosphate, 20 mM sodium phyrophospate and 1% Triton X-100. Protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 3 mM benzamidine, 10 μg/ml aprotinin, 10 μM leupeptin, 5 μM pepstatin, 1mM dithiothreitol and 1 mM sodium orthovanadate) were added to the lysis buffer immediately before use. Protein extracts were sent to Kinexus Bioinformatics Corporation (Vancouver, Canada). The antibody microarray results were processed following the company recommendations. Western blot was carried out as previously described30. The primary antibodies used were: pS6 S235/236 (1:1000, Cell Signaling), pEGFR Y1086 (1:1000, Cell Signaling), pMEK1 S298 (1:1000, Cell Sinaling), β-Actin (1:5000, Abcam), p-ERK1/2 T202/Y204 (1:2000 Cell Signaling) and ERK1 (1:2500, Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments
We thank Taneisha James, Xiaohong Niu and Dayna York for their technical support and lab management, and Ben Macarthur for his support in microarray analysis. We also would like to thank Dr. Kateri Moore and her lab, and Dr. Sonia Mulero-Navarro from B.G.’s lab for their help, and Drs. Valentin Fuster and Antonio Bernad for their support. This research was funded by grants from the National Institutes of Health (NIH) to I.R.L (5R01GM078465), the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM) C024410 to I.R.L. and C.S., C024176 (HESC-SRF) to I.R.L. and S.L.D., and C024407 to B.G., American College of Cardiology/Pfizer Research Fellowship to E.D.A., and ERA-Net for research programmes on rare diseases 2009 to M.T. X.C.-V. is a recipient of a Postdoctoral Fellowship from the Ministerio de Ciencia e Innovacion/Instituto de Salud Carlos III, D.F.L. is a New York Stem Cell Foundation Stanley and Fiona Druckenmiller Fellow and S.P. is a recipient of a Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (T32). Opinions expressed here are solely those of the authors.
Footnotes
Author Contributions X.C.-V. (iPSC establishment, project planning, experimental work and preparation of manuscript), A.S., S.L.D., Y.-S.A., L.Y., A.D.K., E.D.A., D.-F.L., A.W., B.C., J.S., and S.P. (experimental work), R.R. and Y.G. (microarray analysis), N.C. and L.J.E. (karyotype analysis), K.D.L., and M.T. (obtaining of fibroblast samples from patients), C.S. (project planning, experimental work), B.G. and I.R.L. (project planning, preparation of manuscript).
Author Information Microarrray data have been deposited in NCBI-GEO under the accession number GSE20473.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Gorlin RJ, Anderson RC, Moller JH. The Leopard (multiple lentigines) syndrome revisited. Birth Defects Orig Artic Ser. 1971;07:110–115. [PubMed] [Google Scholar]
- 2.Sarkozy A, Digilio MC, Dallapiccola B. Leopard syndrome. Orphanet J Rare Dis. 2008;3:13. doi: 10.1186/1750-1172-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loh ML, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 4.Tartaglia M, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 5.Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jopling C, van Geemen D, den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oishi K, et al. Phosphatase-defective LEOPARD syndrome mutations in PTPN11 have gain-of-function effects during Drosophila development. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2008 doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Z, et al. Human induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009 doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 16.Laux D, Kratz C, Sauerbrey A. Common acute lymphoblastic leukemia in a girl with genetically confirmed LEOPARD syndrome. J Pediatr Hematol Oncol. 2008;30:602–604. doi: 10.1097/MPH.0b013e31817588fb. [DOI] [PubMed] [Google Scholar]
- 17.Ucar C, Calyskan U, Martini S, Heinritz W. Acute myelomonocytic leukemia in a boy with LEOPARD syndrome (PTPN11 gene mutation positive) J Pediatr Hematol Oncol. 2006;28:123–125. doi: 10.1097/01.mph.0000199590.21797.0b. [DOI] [PubMed] [Google Scholar]
- 18.Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 19.Wu CJ, et al. Evidence for ineffective erythropoiesis in severe sickle cell disease. Blood. 2005;106:3639–3645. doi: 10.1182/blood-2005-04-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan ST, Edgington TS. Coupling of the adhesive receptor CD11b/CD18 to functional enhancement of effector macrophage tissue factor response. J Clin Invest. 1991;87:50–57. doi: 10.1172/JCI115000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoki H, Sadoshima J, Izumo S. Myosin light chain kinase mediates sarcomere organization during cardiac hypertrophy in vitro. Nat Med. 2000;6:183–188. doi: 10.1038/72287. [DOI] [PubMed] [Google Scholar]
- 22.Buitrago M, et al. The transcriptional repressor Nab1 is a specific regulator of pathological cardiac hypertrophy. Nat Med. 2005;11:837–844. doi: 10.1038/nm1272. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 26.Edouard T, et al. How do Shp2 mutations that oppositely influence its biochemical activity result in syndromes with overlapping symptoms? Cell Mol Life Sci. 2007;64:1585–1590. doi: 10.1007/s00018-007-6509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dvorak P, et al. Expression and potential role of fibroblast growth factor 2 and its receptors in human embryonic stem cells. Stem Cells. 2005;23:1200–1211. doi: 10.1634/stemcells.2004-0303. [DOI] [PubMed] [Google Scholar]
- 28.Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–1553. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvajal-Vergara X, et al. Multifunctional role of Erk5 in multiple myeloma. Blood. 2005;105:4492–4499. doi: 10.1182/blood-2004-08-2985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.