Abstract
Porcine reproductive and respiratory syndrome is a major cause of economic loss for the swine industry worldwide. Porcine reproductive and respiratory syndrome virus (PRRSV) triggers weak and atypical innate immune responses, but key genes and mechanisms by which the virus interferes with the host innate immunity have not yet been elucidated. In this study, genes that control the response of the main target of PRRSV, porcine alveolar macrophages (PAMs), were profiled in vitro with a time-course experiment spanning the first round of virus replication. PAMs were obtained from six piglets and challenged with the Lelystad PRRSV strain, and gene expression was investigated using Affymetrix microarrays and real-time PCR. Of the 1409 differentially expressed transcripts identified by analysis of variance, two, five, 25, 16 and 100 differed from controls by a minimum of 1.5-fold at 1, 3, 6, 9 and 12 h post-infection (p.i.), respectively. A PRRSV infection effect was detectable between 3 and 6 h p.i., and was characterized by a consistent downregulation of gene expression, followed by the start of the host innate immune response at 9 h p.i. The expression of beta interferon 1 (IFN-β), but not of IFN-α, was strongly upregulated, whilst few genes commonly expressed in response to viral infections and/or induced by interferons were found to be differentially expressed. A predominance of anti-apoptotic transcripts (e.g. interleukin-10), a shift towards a T-helper cell type 2 response and a weak upregulation of tumour necrosis factor-α expression were observed within 12 h p.i., reinforcing the hypotheses that PRRSV has developed sophisticated mechanisms to escape the host defence.
INTRODUCTION
Porcine reproductive and respiratory syndrome is a major cause of economic loss for the swine industry worldwide (Neumann et al., 2005) and causes high mortality of nursery piglets, reproductive failure in sows, respiratory distress in pigs of all ages and influenza-like symptoms in grow/finish swine (Mengeling & Lager, 2000; Nodelijk, 2002). The aetiological agent is porcine reproductive and respiratory syndrome virus (PRRSV), belonging to the family Arteriviridae with an enveloped, positive-stranded RNA genome of about 14.5 kb (Snijder & Meulenberg, 1998).
A typical hallmark of PRRSV is that it causes an acute viraemic phase (up to 14 days post-inoculation) during which the virus can be detected in serum and all susceptible organs (Beyer et al., 2000; Duan et al., 1997b). This acute phase is followed by virus elimination from serum and most organs, and by persistent replication in tonsils, lungs and some lymph nodes (Allende et al., 2000; Rowland et al., 2003; Wills et al., 2003). This prolonged replication does not represent a true persistent infection, as all animals clear the virus by 6 months after inoculation, thus indirectly showing that the immune system is capable of finally dealing with the virus, although not efficiently. Because of this persistent nature of PRRSV infections, numerous studies have analysed the immune responses that may control PRRSV infections or that may be altered by PRRSV (reviewed by Lopez & Osorio, 2004; Mateu & Diaz, 2007; Murtaugh et al., 2002).
The PRRSV-specific humoral immunity is generally characterized by a strong, non-neutralizing antibody response, which is detected from 5–6 days post-infection (p.i.). In contrast, induction of neutralizing antibodies is severely delayed (starting at 3–4 weeks p.i.) and their levels remain low (Lopez & Osorio, 2004); antibodies were shown to be ineffective in eliminating PRRSV-infected macrophages in combination with complement (Costers et al., 2006). Cellular immune responses against PRRSV infection are characterized by a late onset of lymphocyte proliferative responses (4 weeks p.i.) and the late appearance of gamma interferon (IFN-γ)-secreting cells (Meier et al., 2003). Several studies have also shown weak and atypical innate immune responses, such as weak IFN-α responses and high induction of interleukin (IL)-10. This inadequate recognition of virus infection by the innate defence mechanisms could be responsible for the initially crippled immune response (Albina et al., 1998; Buddaert et al., 1998; Murtaugh et al., 2002; Royaee et al., 2004; Suradhat et al., 2003; van Reeth et al., 1999; Xiao et al., 2004). The mechanism by which PRRSV interferes with innate immune responses has yet to be elucidated.
PRRSV has a highly specific tropism for cells of the monocyte/macrophage lineage, cells that are essential for immune function. In vivo, the virus mainly infects a subpopulation of differentiated macrophages that are present in tonsils, lungs and other lymphoid tissues (Beyer et al., 2000; Duan et al., 1997a, b). Besides macrophages, in vitro analysis of susceptible cells has identified cultivated monocytes and dendritic cells as potential targets, but their role during PRRSV infections in vivo remains to be established (Delputte et al., 2007; Duan et al., 1997a; Loving et al., 2007; Teifke et al., 2001; Voicu et al., 1994; Wang et al., 2007). Lung pathogenesis is another feature of PRRSV infections, and porcine alveolar macrophages (PAMs) are generally considered to be a major target for PRRSV.
The aim of this study was to gain insight into the putative mechanisms by which PRRSV can evade innate immunity, and consequently the adaptive response, using a genome-wide approach. A time-course gene expression profiling of PAMs infected in vitro with a reference strain (Lelystad) was conducted by utilizing an Affymetrix 24K Porcine Chip microarray. Collection of samples at different times during the infection cycle, from 1 h p.i. (virus entry) up to 12 h p.i. (virus release and cell death) allowed us to discriminate between changes in early and late gene expression during infection. Times later than 12 h p.i. were not analysed, as by that time PRRSV infection of macrophages has typically resulted in cell death.
METHODS
Cells and treatments.
Six 3-week-old hybrid piglets from a PRRSV- and porcine circovirus 2-negative herd of the Rattlerow–Seghers genetic line (a cross-breed between English Landrace, Belgian Landrace, Large White and a synthetic company Landrace) were injected daily with 1 ml enrofloxacin (5 % solution) and 1 ml lincospectin/spectinomycin (5 or 10 % solution) for 3 days to eliminate eventual bacterial pathogens. Two weeks later, the piglets were sacrificed. PAMs were collected by bronchoalveolar lavage and frozen in liquid nitrogen as described by Wensvoort et al. (1991).
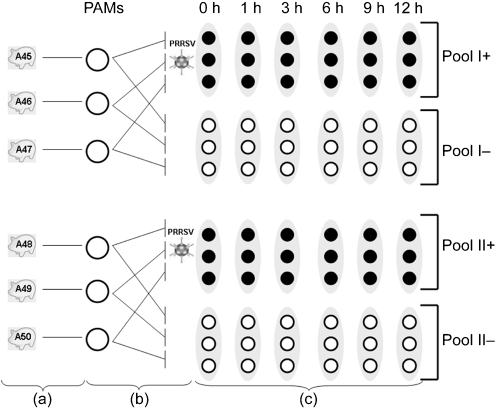
PAMs were thawed and cultured for 48 h before treatment as described previously by Delputte & Nauwynck (2004). One primary culture from each animal was split into two: one was infected at an m.o.i. of 10 with a 13th passage of PRRSV Lelystad virus (kindly provided by G. Wensvoort, Institute for Animal Science and Health, Lelystad, The Netherlands), which was semi-purified as described previously (Delputte & Nauwynck, 2004). The other culture was maintained as a control and was mock inoculated. The percentage of infected cells ranged between 60 and 70 % for all batches. Cells were collected at 1, 3, 6, 9 and 12 h p.i. in TRIzol (Invitrogen Life Technologies) for RNA extraction (Fig. 1).
Fig. 1.
Schematic representation of the experimental design used in this study to challenge PAMs with PRRSV in vitro. PAMs were obtained from six piglets (a). Each PAM culture was split into two and infected with PRRSV or mock infected as a control (b). The total RNA from PAMs of each piglet was extracted at different time points (0, 1, 3, 6, 9 and 12 h p.i.). The RNA of three piglets was pooled (pools I and II) for the subsequent microarray and real-time analyses (c).
RNA extraction, reverse transcription, RNA labelling and cRNA hybridization.
Total RNA extraction from PAMs was performed using TRIzol following standard instructions (Invitrogen) and a clean-up was carried out using RNeasy columns (Qiagen). RNA quality was assessed by microcapillary electrophoresis on an Agilent 2001 Bioanalyser (Agilent Technologies) with RNA 6000 Nanochips. RNA was quantified by spectrophotometry (ND-1000; NanoDrop Technologies). Reverse transcription of 20 μg total RNA and synthesis of biotin-labelled cRNA with one round of amplification were carried out following the standard Affymetrix one-cycle protocol according to the manufacturer's instructions.
Transcriptional profiles were assessed using Affymetrix 24K GeneChip Porcine Genome Arrays (http://www.affymetrix.com/products/arrays/specific/porcine.affx). Based on previous evidence that sample pooling does not significantly affect the results of Affymetrix chip analysis (see, for example, Han et al., 2004), three samples each from control and infected-cell cultures were pooled for each time point (Fig. 1), resulting in two control (pools I− and II−) and two infected pools (pools I+ and II+).
Hybridization and scanning of the arrays were carried out according to standard Affymetrix protocols (Shen et al., 2005) using a GeneChip Scanner 3000 7G.
Microarray data analysis.
Signal intensities were evaluated using the GeneChip Operating Software algorithm (gcos version 1.4; Affymetrix). Raw data and statistical analyses were performed with GeneSpring version 7.3.1 software (Agilent). Normalization was performed per chip (normalized to 50th percentile) and per gene (normalized to the median).
A statistical analysis of variance (ANOVA) model was applied to the data and significance was declared accepting a false discovery rate (FDR) of 0.05. Fixed effects of time point and status (infected−non-infected cells) were included in the ANOVA model. A further cut-off threshold was applied based on a fold change of 1.5 between infected and control PAMs. Hierarchical clustering of the conditions was performed using Pearson's correlation coefficient (r) as a measure of similarity and the average linkage method as the clustering algorithm.
In order to test for the presence of outliers in the two pools, the transcriptional profiles of infected animals were analysed separately at the 3 h p.i. time point. A paired t-test (paired across pools by gene) was performed using the range of minimum and maximum corrected expression values within each pool for each gene. The test was applied (i) to the whole set of genes and (ii) to the subset of genes that appeared to be significant for differential expression in the general analysis. No significant difference was observed either for the whole set of genes included in the study or for the subset of differentially expressed genes.
The Database for Annotation, Visualization and Integrated Discovery (DAVID 2006; http://david.abcc.ncifcrf.gov/), an expanded version of the original web-accessible programs described by Dennis et al. (2003), was used to allocate transcripts with similar biological questions into the three gene ontology (GO) categories.
Real-time PCR.
Quantitative real-time PCR analysis was conducted on ten selected swine transcripts and on the ORF7 gene of PRRSV. Hypoxanthine phosphoribosyltransferase (HPRT1) was chosen as the reference gene because the amplifications of all control and infected samples showed very similar threshold cycle (Ct) values (data not shown). The transcript-specific primers were designed using ProbeFinder software (version 2.3) on the Roche website (https://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp) using standard settings for the human Universal Probe Library Set catalogue (see Supplementary Table S1, available with the online version of this paper).
Two micrograms of total RNA from pools I and II were reverse-transcribed using the Superscript II RT-PCR System (Invitrogen Life Technologies) and standard procedures. The real-time reaction mixture (total 20 μl) included 5 μl cDNA as template (diluted 1 : 50), 200 nM of each of the two primers (forward and reverse), 100 nM Roche probe and 1× master mix (Applied Biosystems). Real-time PCR was performed in 384-well optical plates using a Tecan Freedom EVO-150 liquid handling workstation (Tecan Trading) and an ABI 7900HT real-time PCR machine (Applied Biosystems) with the GeneAmp 7900HT sequence detection system software (PerkinElmer).
A control cDNA dilution series (1 : 50, 1 : 100, 1 : 500 and 1 : 5000) was created for each transcript to establish a standard curve for each plate; real-time reactions of the same pools described for the microarray analysis were performed in triplicate. Briefly, the log input amount of the standard curve was plotted against the output Ct values; all amplifications had a slope of between −3.48 and −2.99 and were accepted as quantitative. The log input amount of each sample was then calculated according to the formula (Ct−b)/m, where b is the y-intercept and m is the slope. The log input amount was converted to input amount according to the formula 10log input amount and triplicate input amounts were averaged for each sample. The mean input amount of each gene was normalized to the mean input amount of HPRT1. A t-test (with thresholds for statistical significance set to 0.1 and 0.05) was applied to each gene to verify whether the difference between control and infected macrophages at each time point was significant.
Pearson's correlation coefficient (r) was calculated for each gene on the normalized data to quantify the consistency between microarray experiments and real-time PCR.
Microarray data.
The data of the microarray analysis were deposited in the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress) with ArrayExpress accession number ×10−MEXP-1350, following the guidelines of the rationale of minimum information about a microarray experiment (MIAME) (Brazma et al., 2001).
RESULTS
Microarray analysis
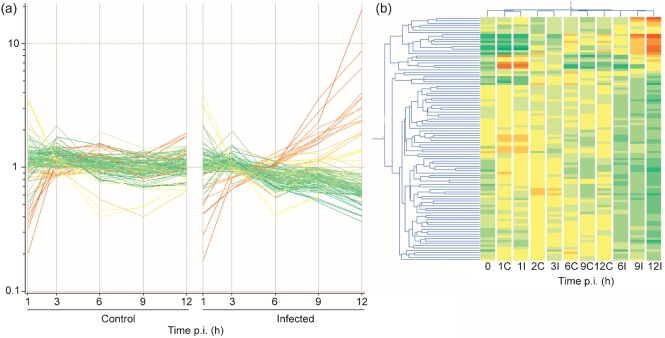
ANOVA analysis (FDR=0.05) showed that 1409 genes were differentially expressed in macrophages after PRRSV infection. After applying a further filter of 1.5-fold change in expression, two, five, 25, 16 and 100 transcripts were differentially expressed at 1, 3, 6, 9 and 12 h p.i., respectively, compared with the controls at the same time points. Overall, the effect of PRRSV on the host transcription machinery was one of downregulation (115/148 transcripts). The differentially expressed transcripts were annotated based on a previous work (Tsai et al., 2006) and are reported in Table 1. The distribution of signal intensities of the 100 differentially expressed transcripts at 12 h p.i. and the hierarchical clustering of controls and infected replicates for the five time conditions (plus the time 0) are shown in Fig. 2.
Table 1.
Transcripts differentially expressed in PAMs at 1, 3, 6, 9 and 12 h p.i. following PRRSV infection
A total of 148 transcripts showed differential expression and are listed from the highest to the lowest fold change at the different time points p.i. The Affymetrix probe set IDs are reported with fold changes, gene symbols and gene description (Tsai et al., 2006).
| Affymetrix probe set ID | Fold change | Gene symbol | Gene description |
|---|---|---|---|
| 1 h p.i. | |||
| Ssc.10997.1.S1_at | 1.699 | GRP58 | Protein disulfide-isomerase A3 precursor |
| Ssc.1313.1.A1_at | 0.661 | NP_077001 | XTP3-transactivated protein A (Homo sapiens) |
| 3 h p.i. | |||
| Ssc.20199.2.S1_at | 0.666 | HIVEP2 | Human immunodeficiency virus type I enhancer-binding protein 2 |
| Ssc.30752.1.S1_at | 0.659 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 |
| Ssc.23248.1.S1_at | 0.653 | PTPRC | Leukocyte common antigen precursor |
| AFFX-Ss_IRP_3_at | 0.648 | IRG6 | Sus scrofa inflammatory response protein 6 |
| Ssc.390.2.S1_at | 0.550 | HIF1-α | Hypoxia-inducible factor 1-α |
| 6 h p.i. | |||
| Ssc.12512.1.A1_at | 1.544 | DDX17 | Probable RNA-dependent helicase p72 (DEAD-box protein 17) |
| Ssc.20344.1.S1_at | 0.667 | WBP2 | WW domain-binding protein 2 |
| Ssc.13400.2.S1_at | 0.666 | C3orf4 | Protein C3orf4 (membrane protein GENX-3745) (HSPC174) |
| Ssc.8570.1.A1_at | 0.665 | DLC1 | Rho-GTPase-activating protein 7 |
| Ssc.13657.1.A1_at | 0.663 | ATF2 | Cyclic-AMP-dependent transcription factor ATF-2 |
| Ssc.16363.1.S1_at | 0.662 | TMOD3 | Ubiquitous tropomodulin |
| Ssc.3420.1.S1_at | 0.660 | C14orf111 | Protein C14orf111 (CGI-35) |
| Ssc.7164.1.A1_at | 0.660 | NP_060530 | Mitochondrial isoleucine tRNA synthetase (Homo sapiens) |
| Ssc.22634.1.S1_at | 0.654 | RB1CC1 | Rb1-inducible coiled coil protein 1 (Homo sapiens) |
| Ssc.1672.2.A1_at | 0.652 | HNRPLL | Heterogeneous nuclear ribonucleoprotein L-like |
| Ssc.16335.1.S2_at | 0.651 | LPL | Lipoprotein lipase precursor |
| Ssc.16422.2.A1_at | 0.647 | PLAA | Phospholipase A-2-activating protein |
| Ssc.30752.2.A1_at | 0.645 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 |
| Ssc.2354.1.S1_at | 0.637 | GPR160 | Probable G protein-coupled receptor 160 |
| Ssc.17091.2.A1_s_at | 0.637 | C3orf52 | TPA-induced transmembrane protein |
| Ssc.23248.1.S1_at | 0.625 | PTPRC | Leukocyte common antigen precursor |
| Ssc.1333.1.A1_at | 0.611 | ZCCHC11 | Zinc finger, CCHC domain containing 11 isoform b |
| Ssc.26084.1.S1_at | 0.607 | ATP2B1 | Plasma membrane calcium-transporting ATPase 1 |
| Ssc.22221.2.S1_at | 0.600 | GKP3 | Glycerol kinase, testis-specific 1 |
| Ssc.13219.1.S1_at | 0.599 | NP_689905 | Core 1 UDP-galactose : N-acetylgalactosamine-α-R β1,3-galactosyltransferase 2; core 1 β3-galactosyltransferase-specific molecular chaperone (Homo sapiens) |
| Ssc.19975.1.S1_at | 0.592 | TEBP | Telomerase-binding protein p23 (Hsp90 co-chaperone) (progesterone receptor complex p23) (Homo sapiens) |
| Ssc.4004.1.A1_at | 0.577 | SYNE2 | Nesprin 2 (nuclear envelope spectrin repeat protein 2) (Syne-2) (synaptic nuclear envelope protein 2) (nucleus and actin connecting element protein) (NUANCE protein) |
| Ssc.10997.1.S1_at | 0.535 | GRP58 | Protein disulfide-isomerase A3 precursor |
| Ssc.4472.1.A1_at | 0.524 | NXT2 | NTF2-related export protein 2 (p15-2 protein) (DC9) (BM-025) |
| Ssc.390.2.S1_at | 0.511 | HIF1-α | Hypoxia-inducible factor 1-α |
| 9 h p.i. | |||
| Ssc.30752.2.A1_at | 3.195 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 |
| Ssc.5085.1.A1_at | 2.794 | TNF-αIP3 | Tumour necrosis factor, alpha-induced protein 3 |
| Ssc.29006.1.S1_at | 2.634 | IFN-β1 | IFN-β precursor |
| Ssc.30532.1.A1_at | 2.546 | XRCC2 | DNA-repair protein XRCC2 (X-ray repair cross-complementing protein 2) |
| Ssc.286.1.S1_s_at | 2.182 | cig5 | Viperin; similar to inflammatory response protein 6 (Homo sapiens) |
| AFFX-Ss_IRP_3_at | 1.919 | IRG6 | Sus scrofa inflammatory response protein 6 |
| Ssc.15761.1.A1_at | 1.881 | TCRA | T-cell receptor α-chain C region (Homo sapiens) |
| Ssc.314.1.S1_at | 1.745 | ADM | ADM precursor [contains adrenomedullin (AM)] |
| Ssc.29054.1.A1_at | 1.639 | GBP1 | IFN-induced guanylate-binding protein 1 |
| Ssc.14474.1.S1_at | 1.538 | LOC396897 | Sus scrofa apomucin |
| Ssc.336.1.S1_at | 1.521 | USP18 | Ubl carboxyl-terminal hydrolase 18 |
| Ssc.11901.1.S1_at | 0.652 | C10orf22 | Chromosome 10 open reading frame 22 |
| Ssc.4462.1.S1_at | 0.630 | SDHC | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial precursor (integral membrane protein CII-3) |
| Ssc.17091.2.A1_s_at | 0.621 | C3orf52 | TPA-induced transmembrane protein |
| Ssc.5353.1.S1_at | 0.620 | ZDHHC3 | Zinc finger DHHC domain containing protein 3 (zinc finger protein 373) (DHHC1 protein) |
| Ssc.15559.1.A1_s_at | 0.552 | NP_060114 | DRE1 protein (Homo sapiens) |
| 12 h p.i. | |||
| Ssc.29006.1.S1_at | 20.150 | IFN-β1 | IFN-β precursor |
| Ssc.30752.2.A1_at | 4.917 | IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 |
| Ssc.30532.1.A1_at | 4.079 | XRCC2 | DNA-repair protein XRCC2 (X-ray repair cross-complementing protein 2) |
| AFFX-Ss_IRP_3_at | 3.827 | IRG6 | Sus scrofa inflammatory response protein 6 |
| Ssc.286.1.S1_s_at | 3.778 | cig5 | Viperin; similar to inflammatory response protein 6 (Homo sapiens) |
| Ssc.5085.1.A1_at | 2.912 | TNF-αIP3 | Tumour necrosis factor-α-induced protein 3 |
| Ssc.15761.1.A1_at | 2.689 | TCR-α | T-cell receptor α-chain C region (Homo sapiens) |
| Ssc.336.1.S1_at | 2.034 | USP18 | Ubl carboxyl-terminal hydrolase 18 |
| Ssc.148.1.S1_at | 1.934 | IL-10 | Interleukin-10 precursor |
| Ssc.12284.1.A1_at | 1.923 | SGK | Serine/threonine-protein kinase Sgk1 |
| Ssc.11048.1.S1_at | 1.918 | PLAC8 | Placenta-specific gene 8 protein |
| Ssc.16288.1.S1_at | 1.859 | IGHM | Ig α-1 chain C region |
| Ssc.29054.3.S1_at | 1.738 | GBP1 | IFN-induced guanylate-binding protein 1 |
| Ssc.314.1.S1_at | 1.718 | ADM | ADM precursor [contains adrenomedullin (AM)] |
| Ssc.18038.1.A1_at | 1.642 | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 |
| Ssc.10754.1.A1_at | 1.621 | PIK3R1 | Phosphatidylinositol 3-kinase regulatory α subunit (PI3-kinase p85-alpha subunit) |
| Ssc.26507.2.S1_at | 1.585 | NP_073596 | Endo-β-N-acetylglucosaminidase (Homo sapiens) |
| Ssc.25855.1.S1_at | 1.531 | XP_846553 | PREDICTED: hypothetical protein |
| Ssc.1701.2.S1_at | 1.529 | Q6PK96 | Cytochrome b, ascorbate-dependent 3 |
| Ssc.100.1.S1_at | 1.513 | TNF-α | Tumour necrosis factor precursor (TNF-α) |
| Ssc.13657.1.A1_at | 0.665 | ATF2 | Cyclic-AMP-dependent transcription factor ATF-2 (activating transcription factor 2) |
| Ssc.4498.1.S1_at | 0.663 | IXL | Intersex-like |
| Ssc.3420.1.S1_at | 0.661 | C14orf111 | Protein C14orf111 (CGI-35) |
| Ssc.8311.1.A1_at | 0.660 | MPI | Mannose-6-phosphate isomerase |
| Ssc.8541.1.A1_at | 0.659 | STAG2 | Cohesin subunit SA-2 (Stromal antigen 2) |
| Ssc.16422.2.A1_at | 0.658 | PLAA | Phospholipase A-2-activating protein |
| Ssc.21796.1.S1_at | 0.657 | SORL1 | Sortilin-related receptor precursor (sorting protein-related receptor containing LDLR class A repeats) |
| Ssc.5404.1.S1_at | 0.657 | MOSPD1 | Motile sperm domain-containing 1 (Homo sapiens) |
| Ssc.27060.1.A1_at | 0.656 | SSSCA1 | Sjogren's syndrome/scleroderma autoantigen 1 (autoantigen p27). |
| Ssc.26553.1.S1_at | 0.655 | AP1-γ1 | Adaptor-related protein complex 1 γ1 subunit (γ-adaptin) (adaptor protein complex AP-1 γ1 subunit) |
| Ssc.3656.1.S1_at | 0.654 | KHLX | Kelch-like protein X (Homo sapiens) |
| Ssc.24239.1.S1_at | 0.653 | C10orf45 | PREDICTED: Pan troglodytes hypothetical protein LOC737061 |
| Ssc.9314.2.S1_at | 0.653 | TP53RK | TP53 regulating kinase |
| Ssc.17314.1.S1_at | 0.653 | C3orf10 | Probable protein BRICK1 |
| Ssc.16057.2.S1_a_at | 0.650 | GANC | Calpain 3 (EC 3.4.22.-) (Calpain L3) |
| Ssc.2756.1.A1_at | 0.650 | MRPL22 | Mitochondrial ribosomal protein L22 (Homo sapiens) |
| Ssc.26084.1.S1_at | 0.649 | ATP2B1 | Plasma membrane calcium-transporting ATPase 1 |
| Ssc.8283.1.A1_at | 0.649 | PTPN11 | Protein-tyrosine phosphatase, non-receptor type 11 |
| Ssc.26735.1.A1_at | 0.649 | Q96BP3 | Peptidylprolyl isomerase domain and WD repeat containing 1 (Bos taurus) |
| Ssc.8430.1.A1_at | 0.649 | Q86W74 | Ankyrin repeat domain 46 (ANKRD46) (Bos taurus) |
| Ssc.1441.1.S1_at | 0.649 | DCTN3 | Dynactin 3 isoform 1; dynactin light chain (Homo sapiens) |
| Ssc.10542.1.S1_at | 0.647 | EXOSC1 | 3′→5′ ExoRNase CSL4 homologue |
| Ssc.772.1.S1_at | 0.643 | CARHSP1 | Calcium-regulated heat-stable protein 1 |
| Ssc.3281.1.S1_at | 0.643 | C11orf10 | UPF0197 protein C11orf10 (HSPC005) |
| Ssc.16495.1.A1_at | 0.643 | DF5L | Gasdermin domain containing protein 1 (Homo sapiens) |
| Ssc.4306.1.A1_at | 0.642 | MESDC1 | Mesoderm development candidate 1 |
| Ssc.26318.1.S1_at | 0.638 | DNCL1 | Dynein light chain 1, cytoplasmic |
| Ssc.24811.1.A1_at | 0.638 | Q6NSI4 | Nuclear receptor-binding protein 2 (NRBP2) (Bos taurus) |
| Ssc.1153.1.A1_at | 0.638 | C9orf28 | C9orf28 protein |
| Ssc.22634.1.S1_at | 0.637 | RB1CC1 | Rb1-inducible coiled-coil protein 1 (Homo sapiens) |
| Ssc.19778.1.S1_at | 0.633 | TNF-αIP8L2 | Tumour necrosis factor-α-induced protein 8-like 2 (Bos taurus) |
| Ssc.8604.1.A1_at | 0.631 | SNX24 | Sorting nexin 24 (SBBI31) |
| Ssc.5353.1.S1_at | 0.627 | ZDHHC3 | Zinc finger DHHC domain-containing protein 3 |
| Ssc.3154.1.S1_at | 0.627 | GRM5 | Metabotropic glutamate receptor 5 precursor (mGluR5) |
| Ssc.6833.1.S1_at | 0.625 | BTG1 | B-cell translocation protein 1 (Homo sapiens) |
| Ssc.1160.1.S1_at | 0.624 | PSMC3 | 26S protease regulatory subunit 6A (TAT-binding protein 1) (TBP-1) (proteasome subunit P50) |
| Ssc.12944.1.A1_at | 0.623 | RPA3 | Replication protein A 14 kDa subunit |
| Ssc.10037.1.A1_at | 0.622 | NLK | Serine/threonine kinase NLK |
| Ssc.22120.1.S1_a_at | 0.617 | RYR2 | PREDICTED: similar to RIKEN cDNA 3110009E18 (Homo sapiens) |
| Ssc.18253.1.S1_at | 0.616 | F8 | Coagulation factor VIII precursor |
| Ssc.24739.1.A1_at | 0.616 | SLC16A7 | Monocarboxylate transporter 2 (MCT 2) |
| Ssc.16936.2.S1_a_at | 0.615 | Q9P0T8 | Similar to hypothetical protein HSPC111 (Bos taurus) |
| Ssc.16691.1.S1_at | 0.609 | H2AF-J | H2A histone family, member J isoform 1 (Homo sapiens) |
| Ssc.30182.1.A1_at | 0.607 | RER1 | RER1 protein (Homo sapiens) |
| Ssc.21559.1.S1_at | 0.606 | ANKRD10 | Ankyrin repeat domain protein 10 |
| Ssc.11369.1.S1_at | 0.605 | NP_077271 | Derlin-1 (Der1-like protein 1) |
| Ssc.1029.1.S1_at | 0.603 | PHF6 | PHD finger protein 6 (PHD-like zinc finger protein) |
| Ssc.13954.1.A1_at | 0.602 | Q5VV17 | PREDICTED: similar to hypothetical protein DKFZp761A052 |
| Ssc.11878.1.S1_at | 0.601 | HMBS | Porphobilinogen deaminase |
| Ssc.16677.1.S1_a_at | 0.599 | C17orf37 | Uncharacterized protein C17orf37 (protein C35) (HBV X-transactivated gene 4 protein) |
| Ssc.24943.1.S1_at | 0.597 | NDUFA11 | NADH-ubiquinone oxidoreductase subunit B14.7 |
| Ssc.3426.1.A1_at | 0.595 | MAPK6 | Mitogen-activated protein kinase 6 |
| Ssc.21783.1.S1_at | 0.595 | MRPL2 | Mitochondrial ribosomal protein L2 (Homo sapiens) |
| Ssc.13370.1.A1_at | 0.595 | Q8NA66 | RIKEN cDNA 1810054D07 gene (1810054D07Rik) (Mus musculus) |
| Ssc.1206.1.A1_at | 0.595 | ADAMTS19 | ADAMTS-19 precursor |
| Ssc.6979.1.A1_at | 0.587 | TPP2 | Tripeptidyl-peptidase II |
| Ssc.13218.1.A1_at | 0.587 | NP_660155 | Testis development protein NYD-SP29 (Homo sapiens) |
| Ssc.19975.1.S1_at | 0.582 | TEBP | Telomerase-binding protein p23 (Hsp90 co-chaperone) (Homo sapiens) |
| Ssc.16392.2.A1_a_at | 0.574 | MKNK2 | MAP kinase-interacting serine/threonine kinase 2 |
| Ssc.6230.2.A1_at | 0.573 | SDCCAG3 | Serologically defined colon cancer antigen 3 (Homo sapiens) |
| Ssc.21987.1.A1_at | 0.57 | IFRD1 | IFN-related developmental regulator 1 |
| Ssc.5163.1.S1_at | 0.569 | GCNT2 | N-Acetyllactosaminide β-1,6-N-acetylglucosaminyl-transferase |
| Ssc.6189.1.A1_at | 0.565 | SLC7A11 | Cystine/glutamate transporter (amino acid transport system xc−) |
| Ssc.1333.1.A1_at | 0.565 | ZCCHC11 | Zinc finger, CCHC domain containing 11 isoform b |
| Ssc.29047.1.S1_at | 0.561 | HIG2 | Hypoxia-inducible protein 2 (Homo sapiens) |
| Ssc.22287.1.S1_at | 0.561 | GABR-α3 | γ-Aminobutyric-acid receptor α-3 subunit precursor [GABA(A) receptor] |
| Ssc.2354.1.S1_at | 0.554 | GPR160 | Probable G protein-coupled receptor 160 |
| Ssc.4004.1.A1_at | 0.552 | SYNE2 | Nesprin 2 (nuclear envelope spectrin repeat protein 2) |
| Ssc.13400.2.S1_at | 0.550 | C3orf4 | Protein C3orf4 (membrane protein GENX-3745) |
| Ssc.26309.1.A1_at | 0.548 | CHES1 | Checkpoint suppressor 1 (Forkhead box protein N3) |
| Ssc.6513.1.S1_at | 0.542 | LRRC28 | Leucine-rich repeat-containing 28 (Homo sapiens) |
| Ssc.3451.1.S1_at | 0.540 | SLC11A2 | Natural resistance-associated macrophage protein 2 (NRAMP2) |
| Ssc.4462.1.S1_at | 0.536 | SDHC | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial precursor (integral membrane protein CII-3) |
| Ssc.14114.1.A1_at | 0.525 | ABCD3 | ATP-binding cassette, subfamily D, member 3 (70 kDa peroxisomal membrane protein) (PMP70) |
| Ssc.16563.1.S1_at | 0.513 | NP_067050 | DC2 protein (Homo sapiens) |
| Ssc.1527.1.A1_at | 0.503 | SLC20A1 | Solute carrier family 20 (phosphate transporter), member 1; Glvr-1; PiT-1; gibbon ape leukemia virus receptor 1 (Homo sapiens) |
| Ssc.16475.1.S1_at | 0.475 | COL4-α3 | Collagen α3(IV) chain precursor (Goodpasture antigen) |
| Ssc.17091.2.A1_s_at | 0.463 | C3orf52 | TPA-induced transmembrane protein |
| Ssc.4472.1.A1_at | 0.459 | NXT2 | NTF2-related export protein 2 (p15-2 protein) (DC9) (BM-025) |
| Ssc.15559.1.A1_s_at | 0.306 | NP_060114 | DRE1 protein (Homo sapiens) |
Fig. 2.
(a) Distribution of signal intensities of the 100 transcripts differentially expressed at 12 h p.i. over the period of infection. Left, control PAMs; right, infected PAMs. Each line represents a transcript. (b) Hierarchical clustering of the different time point conditions, based on the transcripts differentially expressed at 12 h p.i. in control (C) and infected (I) PAMs. Coloration in both figures refers to the condition of infected cells at 12 h p.i. and is directly proportional to the expression, ranging from red (high expression) to green (low expression).
At early time points (1 and 3 h p.i.), the profiles of gene expression in the control and infected conditions were very similar and clustered together, i.e. only two (1 h p.i.) and five (3 h p.i.) transcripts were significantly altered. The expression profiles clearly changed between 3 and 6 h p.i., with greater differences detected at the later time points (9 and 12 h p.i.), when PRRSV has been shown to complete its replication (Halbur, 2001; Rossow et al., 1995).
The 6 h p.i. time point was characterized by a consistent downregulation of gene expression in the infected cells. The 24 downregulated transcripts represented genes with functions related to RNA processing (HNRPLL and NXT2), regulation of biological processes (ATF2, PTPRC, HIF1-α, DLC1 and RB1CC1) and signal transduction (PLAA, PTPRC, HIF1-α, DLC1 and GPR160). Only one transcript, an RNA-dependent helicase (DDX17), was upregulated.
The 9 h p.i. time point was the only one at which most transcripts (11/16) were upregulated. These represented genes (IFIT1, GBP1, USP18 and cig5) that encode accessory proteins related to the immune response, and in particular to the pro-inflammatory cytokine IFN-β, but also genes with a known anti-apoptotic function (ADM and TNF-αIP3).
At 12 h p.i., the downregulated transcripts were also largely predominant over the upregulated ones (80 vs 20, respectively). The latter confirmed the main pattern of anti-apoptotic and antiviral response already observed at 9 h p.i., with the addition of two new transcripts representing TNF-α and IL-10. The overall highest fold change (FC) was observed for IFN-β (FC=20.15 at 12 h p.i.), whilst the most downregulated transcript was NP_060114 (FC=0.306 at 12 h p.i.). NP_060114 corresponds to the human DRE1 protein, a member of the kelch-repeat family, which modulates host immune response to viral infection (Prag & Adams, 2003). KHLX belongs to the same family and also showed a consistent downregulation at 12 h (FC=0.654).
The GO analysis assigned the 100 differentially expressed transcripts at 12 h p.i. to 34 biological processes, five molecular functions and three cellular components (Table 2), with the best ranked biological processes (response to stimulus, response to stress and immune response) effectively representing the general pattern of cell response to infection. This was independently confirmed by assigning the same 100 transcripts to regulatory pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The most represented pathways were all related to the immune response and included mitogen-activated protein kinase (MAPK), JAK–STAT, natural killer cell-mediated cytotoxicity, T-cell receptor signalling, cytokine–cytokine receptor interaction and Toll-like receptor signalling (data not shown).
Table 2.
GO analysis and ranking of the 100 transcripts differentially expressed at 12 h p.i
Ranking and assignment is given for the differentially expressed transcripts at 12 h p.i. to the three GO categories: biological process, molecular function and cellular component. The number of transcripts for each process is shown, with the corresponding e-values.
| GO category | Ranking | Number of transcripts | e-value |
|---|---|---|---|
| Biological process | |||
| Response to stimulus | 1 | 22 | 6.50×10−4 |
| Response to stress | 2 | 14 | 6.10×10−3 |
| Immune response | 3 | 11 | 6.60×10−3 |
| Physiological process | 4 | 68 | 8.30×10−3 |
| Defence response | 5 | 11 | 1.20×10−2 |
| Response to biotic stimulus | 6 | 11 | 1.60×10−2 |
| Anion transport | 7 | 5 | 2.00×10−2 |
| Regulation of apoptosis | 8 | 7 | 2.30×10−2 |
| Regulation of programmed cell death | 9 | 7 | 2.40×10−2 |
| Meiosis | 10 | 3 | 3.00×10−2 |
| M phase of meiotic cell cycle | 11 | 3 | 3.00×10−2 |
| Macromolecule metabolism | 12 | 34 | 3.10×10−2 |
| Meiotic cell cycle | 13 | 3 | 3.10×10−2 |
| Organismal physiological process | 14 | 16 | 3.60×10−2 |
| Anti-apoptosis | 15 | 4 | 4.30×10−2 |
| M phase | 16 | 5 | 4.40×10−2 |
| Inorganic anion transport | 17 | 4 | 5.30×10−2 |
| Response to virus | 18 | 3 | 5.40×10−2 |
| Negative regulation of apoptosis | 19 | 4 | 6.00×10−2 |
| Negative regulation of programmed cell death | 20 | 4 | 6.10×10−2 |
| Apoptosis | 21 | 8 | 6.20×10−2 |
| Programmed cell death | 22 | 8 | 6.30×10−2 |
| Activation of NF-κβ transcription factor | 23 | 2 | 6.40×10−2 |
| Response to wounding | 24 | 6 | 7.00×10−2 |
| Cell death | 25 | 8 | 7.30×10−2 |
| Death | 26 | 8 | 7.50×10−2 |
| Protein metabolism | 27 | 25 | 7.60×10−2 |
| Negative regulation of cell proliferation | 28 | 4 | 7.80×10−2 |
| Positive regulation of transcription factor activity | 29 | 2 | 7.90×10−2 |
| Leukocyte adhesion | 30 | 2 | 7.90×10−2 |
| B-cell proliferation | 31 | 2 | 7.90×10−2 |
| Negative regulation of cellular process | 32 | 9 | 8.30×10−2 |
| Interaction between organisms | 33 | 3 | 8.60×10−2 |
| Ion transport | 34 | 8 | 9.20×10−2 |
| Molecular function | |||
| Antigen binding | 1 | 3 | 1.50×10−2 |
| Receptor binding | 2 | 8 | 6.00×10−2 |
| Phosphatase binding | 3 | 2 | 8.70×10−2 |
| Tumour necrosis factor receptor binding | 4 | 2 | 8.70×10−2 |
| Cytokine activity | 5 | 4 | 8.80×10−2 |
| Cellular component | |||
| Integral to membrane | 1 | 26 | 6.90×10−2 |
| Intrinsic to membrane | 2 | 26 | 7.10×10−2 |
| Extracellular space | 3 | 6 | 8.80×10−2 |
Validation of the microarray data by real-time PCR
The genes tested by real-time PCR (see Supplementary Table S1) were selected to validate the microarray results and to confirm the involvement of key biological pathways. The set included the differentially expressed genes IFN-β, cig5, TNF-α, TNF-αIP3, IL-10, USP18 and GRP58, as well as three additional genes selected from recent literature (IFN-α, IFN-αR1 and sialoadhesin), which were represented on the array but had not shown differential expression (Table 3). Pearson's correlation coefficient (r) showed that both the microarray and real-time PCR data were highly correlated for the genes modulated between 6 and 12 h p.i., with a high consistency between pools and with r values ranging from 0.95 (IFN-β) to 0.88 (USP18 and IL-10) (Table 3). Only GRP58, shown by microarrays to be upregulated at 1 h p.i. but downregulated at 6 h p.i., showed inconsistencies between pools and a very poor r value.
Table 3.
Real-time PCR results of genes differentially expressed following PRRSV infection of PAMs
Real-time PCR results of ten selected genes in two pools of PAMs infected with PRRSV (I) compared with control PAMs (C). The reported values are the means±sd of technical triplicates and were calculated as described in Methods; values that significantly differ between infected and control PAMs are indicated in bold (*, P<0.10; **, P<0.05). The last column gives the Pearson's correlation coefficient (r) between real-time and microarray data.
| Gene symbol | Status | Pool I | Pool II | r | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 h | 3 h | 6 h | 9 h | 12 h | 1 h | 3 h | 6 h | 9 h | 12 h | |||
| IFN-β | I | 0.01±0.005 | 0.04±0.016* | 0.01±0.006 | 0.13±0.032** | 2.03±0.402** | 0.01±0.003 | 0.05±0.011* | 0.03±0.005 | 0.17±0.032** | 2.76±0.556** | 0.95 |
| C | 0.02±0.008 | 0.01±0.005* | 0.004±0.002 | 0.01±0.002** | 0.01±0.006** | 0.01±0.005 | 0.01±0.007* | 0.03±0.013 | 0.02±0.006** | 0.03±0.01** | ||
| TNF-α | I | 4.02±1.614 | 1.37±0.467 | 0.3±0.106 | 0.25±0.072 | 1.16±0.425 | 5.95±2.325 | 1.76±0.679 | 0.59±0.102 | 0.89±0.236 | 2.55±0.728** | 0.94 |
| C | 7.52±2.786 | 0.99±0.28 | 0.32±0.088 | 0.16±0.05 | 2.09±0.598 | 5.89±2.119 | 1.45±0.462 | 0.62±0.221 | 0.41±0.139 | 0.36±0.138** | ||
| TNF-αIP3 | I | 2.1±0.854 | 0.59±0.2 | 0.32±0.039 | 0.75±0.203** | 1.31±0.4* | 4.01±2.172 | 1.19±0.461 | 0.44±0.09 | 0.81±0.16** | 3.17±1.232* | 0.94 |
| C | 1.76±0.484 | 0.33±0.061 | 0.22±0.035 | 0.2±0.026** | 0.42±0.12* | 3.59±1.403 | 0.81±0.255 | 0.47±0.137 | 0.33±0.078** | 0.24±0.071* | ||
| USP18 | I | 0.25±0.085 | 0.41±0.125 | 0.78±0.12 | 0.92±0.166* | 1.61±0.393** | 0.34±0.12 | 0.69±0.227 | 1.06±0.221 | 1.25±0.205* | 2.37±0.868** | 0.88 |
| C | 0.27±0.079 | 0.3±0.036 | 0.74±0.121 | 0.54±0.038* | 0.53±0.106** | 0.29±0.069 | 0.45±0.132 | 0.93±0.195 | 0.52±0.14* | 0.44±0.123** | ||
| cig5 | I | 0.13±0.019 | 0.48±0.065 | 0.97±0.183 | 0.91±0.083* | 1.72±0.102** | 0.15±0.021 | 1.16±0.290 | 1.15±0.192 | 1.33±0.274** | 2.37±0.137** | 0.89 |
| C | 0.14±0.019 | 0.50±0.102 | 0.79±0.151 | 0.58±0.123* | 0.86±0.258** | 0.11±0.031 | 0.88±0.116 | 0.90±0.180 | 0.40±0.054** | 0.45±0.164** | ||
| IL-10 | I | 0.62±0.228 | 0.52±0.199 | 0.19±0.094 | 0.49±0.151 | 1.6±0.415* | 1.08±0.246 | 0.68±0.14 | 0.38±0.074 | 0.67±0.166 | 2.05±0.567** | 0.88 |
| C | 0.57±0.107 | 0.18±0.087 | 0.18±0.047 | 0.23±0.04 | 0.47±0.06* | 1.17±0.294 | 0.43±0.119 | 0.45±0.196 | 0.44±0.091 | 0.7±0.177** | ||
| GRP58 | I | 1.92±0.103** | 0.99±0.175* | 0.66±0.144** | 1.82±0.248 | 2.22±0.307** | 1.22±0.187 | 1.55±0.195* | 1.28±0.088 | 0.66±0.042** | 1.13±0.147 | 0.09 |
| C | 1.00±0.046** | 1.61±0.179* | 1.49±0.268** | 1.19±0.161 | 0.66±0.043** | 1.31±0.166 | 0.92±0.123* | 1.40±0.183 | 1.29±0.154** | 1.16±0.126 | ||
| IFN-α | I | 0.07±0.013 | 0.18±0.028** | 0.03±0.002 | 0.09±0.012 | 0.13±0.020 | 0.05±0.002 | 0.16±0.023** | 0.14±0.018 | 0.09±0.005** | 0.14±0.022 | – |
| C | 0.08±0.015 | 0.03±0.002** | 0.03±0.002 | 0.06±0.013 | 0.08±0.013 | 0.05±0.004 | 0.04±0.004** | 0.11±0.007 | 0.04±0.005** | 0.11±0.004 | ||
| IFN-αR1 | I | 0.93±0.274 | 0.96±0.239 | 0.91±0.165 | 0.85±0.187 | 0.87±0.25 | 1.01±0.139 | 1.06±0.115 | 1.10±0.035 | 1.19±0.10 | 0.95±0.151 | – |
| C | 1.11±0.450 | 1.096±0.265 | 1.12±0.220 | 0.84±0.202 | 1.02±0.192 | 1.06±0.129 | 1.06±0.114 | 0.96±0.133 | 1.20±0,128 | 1.06±0.058 | ||
| Sialoadhesin | I | 0.25±0.016** | 0.17±0.019 | 0.14±0.009 | 0.19±0.026 | 0.22±0.034 | 0.19±0.017 | 0.19±0.016 | 0.21±0.010 | 0.27±0.048* | 0.46±0.071* | – |
| C | 0.17±0.033** | 0.16±0.018 | 0.15±0.005 | 0.23±0.051 | 0.15±0.024 | 0.18±0.008 | 0.21±0.015 | 0.22±0.020 | 0.18±0.025* | 0.28±0.010* | ||
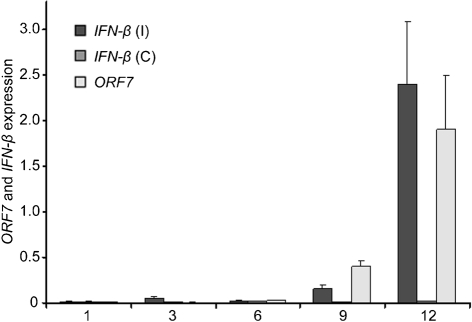
Real-time PCR confirmed that IFN-β was the most upregulated gene, whilst IFN-α was not differentially expressed between control and infected cells at 9 and 12 h p.i. Moreover, real-time PCR analysis of PAMs in an independent challenge experiment, with a different viral strain and lower m.o.i., confirmed that, at 24 h p.i., IFN-β was strongly induced whilst IFN-α was only slightly upregulated (data not shown). The expression of IFN-β increased together with the PRRSV titre, as affirmed by PRRSV ORF7 gene expression (Fig. 3). Interestingly, real-time PCR at 3 h p.i. showed a small but significant peak in IFN-β and IFN-α expression, which was not detected by microarrays. IFN-αR1 was confirmed not to be differentially expressed. Statistically significant differences in values of sialoadhesin expression were found between infected and control samples, but this was inconsistent between pools.
Fig. 3.
Comparison of the time-course expression of host IFN-β and PRRSV ORF7 expression by real-time PCR in control (C) and infected (I) PAMs at five time points p.i. Results are shown as means±sd.
DISCUSSION
The finding that the only gene to be upregulated at 6 h p.i. was a host RNA-dependent helicase (DDX17) indicates that PRRSV does not induce a generalized suppression of host gene transcription. This confirms and reinforces previous observations (Zhang et al., 2000), showing enhanced production of a cellular helicase (RHIV-1) in macrophages in response to PRRSV infection. Other RNA viruses, such as poliovirus and vesicular stomatitis virus, replicating exclusively in the cytoplasm using virus-encoded RNA-dependent RNA polymerases and thus not requiring the host transcriptional apparatus, inhibit nuclear transcription of cellular RNA polymerases (Weidman et al., 2003). This transcription shut-off not only allows the virus to evade cellular responses, but also may favour viral RNA replication by increasing the pool of free ribonucleotides in the cell. The complex expression pattern revealed by the microarrays might suggest that PRRSV blocks specific cellular transcription processes while upregulating others that are potentially beneficial for virus replication. By having a regulatory effect on cellular transcription, viruses may promote their own replication while interfering with the innate and adaptive immune responses that would result in their removal.
The downregulation of four genes encoding mitochondrial proteins (NP_060530, SDHC, MRPL2 and MRPL22) at different time points (6, 9 and/or 12 h p.i.) might add up to the emerging role of mitochondria in antiviral immunity. The mitochondrial antiviral signalling protein MAVS is critical for the IFN-β signalling pathway in response to dsRNA, and is required for both TLR3-mediated and TLR3-independent signalling pathways, such as that triggered by the RNA helicase RIGI (Moore et al., 2008; Xu et al., 2005; Yoneyama et al., 2004). RIGI is the product of DDX58, a member of the DEAD box family of RNA helicases that mediate nucleoside triphosphate-dependent unwinding of dsRNA and are involved in many diverse cellular functions (Lamm et al., 1996). Intriguingly, the only upregulated gene found by microarrays at 6 h p.i. in PAMs (DDX17) belongs to the same family.
The atypical pattern of expression of innate immunity genes indicates that PRRSV has probably developed sophisticated mechanisms to control the antiviral response. Indeed, only a subset (IFIT1, GBP1, USP18 and TNF-αIP3) of genes commonly modulated by pathogens in response to dsRNA and/or stimulated by IFN (Jenner & Young, 2005) were found to be upregulated by PRRSV at 9 and/or 12 h p.i. When the 1.5-fold change threshold was not applied after ANOVA analysis, this subset also included CD44, PML, PRKRA, CCl4, CCl8 and MT2A. Upregulation of USP18 has been observed previously in PAMs following PRRSV infection (Zhang et al., 1999). The same study reported the upregulation of the antiviral gene MX1, but neither MX1 nor MX2 was found to be differentially expressed in the present investigation. Downregulation of NRAMP2 at 12 h p.i. was consistent with the effects observed previously in humans after human immunodeficiency virus infection (reviewed by Jenner & Young, 2005).
Production of IFN-α and IFN-β is a well-known reaction of virus-infected cells; however, only the IFN-β gene was strongly upregulated by PRRSV in PAMs. The induction of IFN-β mRNA, but not IFN-α mRNA, has also been observed in monocyte-derived dendritic cells infected by PRRSV at 12 h p.i. (Loving et al., 2007). Previous studies, both in vitro and in vivo, have also shown that PRRSV is a poor inducer or even a suppressor of IFN-α compared with other respiratory viruses (Albina et al., 1998; Buddaert et al., 1998; Miller et al., 2004; van Reeth et al., 1999). Blocking IFN-α production clearly is beneficial for PRRSV replication, as IFN-α can efficiently block replication when present during infection (Delputte et al., 2007; Loving et al., 2007). IFN-β can also protect macrophages against PRRSV infection (Overend et al., 2007), but it has been suggested that IFN-β alone may be not sufficient to trigger the adaptive immune response (Loving et al., 2007). A recent report has shown that in vitro stimulation of monocytes and macrophages with IFN-α induces expression of sialoadhesin, the main PRRSV receptor in PAMs, and that treatment with IFN-α before inoculation strongly increases PRRSV infection of monocytes (Delputte et al., 2007). In agreement with this, in this study neither the gene encoding sialoadhesin nor that encoding IFN-αR1 (IFN receptor 1) showed consistent differential expression in infected cells.
Despite previous evidence that IFN-β expression by infected cells mediates and potentiates apoptosis (Tanaka et al., 1998), the present study showed a predominance of transcripts leading to prolonged cell survival within 12 h of infection (both upregulation of anti-apoptotic transcripts and downregulation of pro-apoptotic genes). Upregulation was observed for IL-10, ADM and TNF-αIP3. IL-10 has been demonstrated to protect cells against apoptosis (Sieg et al., 1996; Zhou et al., 2001). ADM has been shown (Kubo et al., 1998) to be overproduced by macrophages after inflammation and to modulate cytokine production (specifically TNF-α); several different independent studies support the fact that ADM is an anti-apoptotic peptide on different cell types (Bi et al., 2007; Uzan et al., 2006; Yin et al., 2004). TNF-αIP3 is a cytoplasmic zinc finger protein that inhibits NF-κβ activity and TNF-mediated programmed cell death (Li et al., 2006; Qin et al., 2006). Downregulated genes included those encoding NLK, a stimulator of apoptosis (Yasuda et al., 2003), HIF1-α, which has been suggested to favour apoptosis in the absence of oxygen (Bruick, 2000), and GRM5, known to protect neurons from apoptotic death (Maiese et al., 2000). Taken together, these findings suggest that PRRSV actively induces an anti-apoptotic state in order to complete its virus replication cycle. This is discordant with previous results showing that PRRSV induces infected cells, as well as uninfected bystander cells, to undergo apoptosis (for examples, see Chang et al., 2005; Sirinarumitr et al., 1998), but it should be noted that those data were obtained with in vitro infection treatments much longer than 12 h. On the other hand, the absence of apoptotic induction by PRRSV has been observed in MARC-145 cells (Miller & Fox, 2004) and HeLa cells (Lee et al., 2004). Interestingly, Kim et al. (2002) reported an atypical form of apoptosis that culminates in increased cell membrane permeability and late apoptosis after completion of virus replication.
The upregulation of IL-10 gene expression (FC=1.9) indicates that the IL-10-mediated downregulation of the T-helper cell type 1 (Th1) response may be an important mechanism operated by PRRSV, as well as by other viruses (for reviews, see Fickenscher et al., 2002; Redpath et al., 2001). Upregulation of IL-10 expression was found previously in PRRSV-infected porcine monocytes, macrophages and dendritic cells (Flores-Mendoza et al., 2008; Suradhat et al., 2003) and in vivo in PRRSV-infected pigs (Suradhat & Thanawongnuwech, 2003; Sutherland et al., 2007; Thanawongnuwech & Thacker, 2003; Thanawongnuwech et al., 2004). IL-10 in PRRSV-infected cells seems to be increased concurrent with the onset of viraemia and the development of clinical signs (Díaz et al., 2005). Also, PIK3R1 (upregulated in this study: FC=1.6), is known to positively regulate the production of IL-10 (Saegusa et al., 2007). These findings add to previous studies (Murtaugh et al., 2002; Wang et al., 2007), suggesting that PRRSV causes an imbalanced immune response characterized by an abundance of humoral immunity (Th2-mediated), which is less effective against viral pathogens.
The TNF-α gene was only slightly upregulated at 12 h p.i. (FC=1.5). The role of TNF-α in PRRSV infection is controversial: it has been reported that PRRSV is a potent inducer of TNF-α in PAMs at 18, 36, 54, 72, 90 and 108 h p.i. (Chang et al., 2005) and at 6 and 15 h p.i. (Thanawongnuwech et al., 2004). However, Charerntantanakul et al. (2006) showed that, in porcine monocytes infected by PRRSV, IL-10 gene expression increased, and this response contributed to a reduction in TNF-α production. In fact, crucial anti-inflammatory activities of IL-10 may be due to its inhibitory effects on TNF-α production (Moore et al., 2001). Overall, this suggests that IL-10 may also participate in fine-tuning the production and effects of TNF-α.
Other differentially expressed genes (see Table 1) confirmed that a complex pattern of TNF-α regulation takes place upon PRRSV infection. TNF-αIP3, known to be induced by TNF-α (Dixit et al., 1990; Lee et al., 2000) and suggested to protect against the inflammatory response to influenza virus infection (Onose et al., 2006), was upregulated. STAG2, an enhancer of TNF production (Lara-Pezzi et al., 2004), and the member of the MAPK pathway, ATF2, a transcription activator of both IFN-β and TNF-α in response to virus infection (Biron & Sen, 2001; Tsai et al., 1996), were downregulated. The MAPK pathway was the most highly represented gene network identified in this study, with five differentially expressed genes at 12 h p.i. (ATF2, TNF-α, MAP3K8, MKNK2 and NLK). The MAPK pathway is one of the most important pathways for immune response to infection (Bruder & Kovesdi, 1997; Yang et al., 2007) and has been found to be modulated in PAMs after an antibody-mediated cross-linking treatment of sialoadhesin, the main PRRSV internalization receptor (Genini et al., 2008), although in this case different genes of the pathway were involved.
In conclusion, this work has provided a genome-wide gene expression catalogue of PRRSV pathogenesis and has allowed us to picture how different genes and gene pathways are co-modulated in the physiological context of the PAM. As such, these results provide a large-scale and unbiased basis for further investigations on gene roles and functions. The sequencing of the pig genome currently in progress (www.piggenome.org), besides allowing a complete annotation of all transcripts, will soon give important clues for future genomics studies, for example for the interpretation of genome scan analyses aimed at identifying the genetic components involved in PRRSV resistance/susceptibility in swine populations (Ait-Ali et al., 2007; Lewis et al., 2007).
Supplementary Material
Acknowledgments
The authors are very grateful to Dr Joan K. Lunney for critical reading and revision of this manuscript, to Dr John L. Williams and Dr G. Leone for helpful suggestions, and to C. Vanmaercke and L. Sys for technical assistance in the laboratory. This project was supported by a grant from the Italian Ministry of Research (MIUR project, art.10 D.M. 593/00). S. G. is partially supported by the European Network of Excellence EADGENE (www.eadgene.org). P. L. D. is supported by a grant from the Special Research Fund of Ghent University (grant B/06524). The authors declare no competing financial interests.
Footnotes
The genes and primers used for real-time PCR analysis are available with the online version of this paper.
References
- Ait-Ali, T., Wilson, A., Wescott, D. G., Clapperton, M., Mellencamp, M., Drew, T. W., Bishop, S. C. & Archibald, A. (2007). Innate immune responses to replication of porcine reproductive and respiratory syndrome virus in isolated swine alveolar macrophages. Viral Immunol 20, 105–118. [DOI] [PubMed] [Google Scholar]
- Albina, E., Carrat, C. & Charley, B. (1998). Interferon-α response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J Interferon Cytokine Res 18, 485–490. [DOI] [PubMed] [Google Scholar]
- Allende, R., Laegreid, W. W., Kutish, G. F., Galeota, J. A., Wills, R. W. & Osorio, F. A. (2000). Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J Virol 74, 10834–10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, J., Fichtner, D., Schirrmeier, H., Polster, U., Weiland, E. & Wege, H. (2000). Porcine reproductive and respiratory syndrome virus (PRRSV): kinetics of infection in lymphatic organs and lung. J Vet Med B Infect Dis Vet Public Health 47, 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, G. R., Zhang, H., Zhou, H. J., Bai, L. J., Zhang, H. M., Hai, H. & Fang, X. B. (2007). Effect of adrenomedulin on neuron apoptosis and early growth response gene-1 after focal ischemia/reperfusion in rats. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue 19, 353–357. [PubMed] [Google Scholar]
- Biron, A. B. & Sen, G. C. (2001). Interferons and other cytokines. In Fields Virology, 4th edn, pp. 321–351. Edited by B. N. Fields & D. M. Knipe. Philadelphia: Lippincott Williams & Wilkins.
- Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A. & other authors (2001). Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nat Genet 29, 365–371. [DOI] [PubMed] [Google Scholar]
- Bruder, J. T. & Kovesdi, I. (1997). Adenovirus infection stimulates the Raf/MAPK signaling pathway and induces interleukin-8 expression. J Virol 71, 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick, R. K. (2000). Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A 97, 9082–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddaert, W., van Reeth, K. & Pensaert, M. (1998). In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv Exp Med Biol 440, 461–467. [DOI] [PubMed] [Google Scholar]
- Chang, H. W., Jeng, C. R., Liu, J. J., Lin, T. L., Chang, C. C., Chia, M. Y., Tsai, Y. C. & Pang, V. F. (2005). Reduction of porcine reproductive and respiratory syndrome virus (PRRSV) infection in swine alveolar macrophages by porcine circovirus 2 (PCV2)-induced interferon-α. Vet Microbiol 108, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charerntantanakul, W., Platt, R. & Roth, J. A. (2006). Effects of porcine reproductive and respiratory syndrome virus-infected antigen-presenting cells on T cell activation and antiviral cytokine production. Viral Immunol 19, 646–661. [DOI] [PubMed] [Google Scholar]
- Costers, S., Delputte, P. L. & Nauwynck, H. J. (2006). Porcine reproductive and respiratory syndrome virus-infected alveolar macrophages contain no detectable levels of viral proteins in their plasma membrane and are protected against antibody-dependent, complement-mediated cell lysis. J Gen Virol 87, 2341–2351. [DOI] [PubMed] [Google Scholar]
- Delputte, P. L. & Nauwynck, H. J. (2004). Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol 78, 8094–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delputte, P. L., van Breedam, W., Barbé, F., van Reeth, K. & Nauwynck, H. J. (2007). IFN-α treatment enhances porcine arterivirus infection of monocytes via upregulation of the porcine arterivirus receptor sialoadhesin. J Interferon Cytokine Res 27, 757–766. [DOI] [PubMed] [Google Scholar]
- Dennis, G., Jr, Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C. & Lempicki, R. A. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4, R60. [PubMed] [Google Scholar]
- Díaz, I., Darwich, L., Pappaterra, G., Pujols, J. & Mateu, E. (2005). Immune responses of pigs after experimental infection with a European strain of porcine reproductive and respiratory syndrome virus. J Gen Virol 86, 1943–1951. [DOI] [PubMed] [Google Scholar]
- Dixit, V. M., Green, S., Sarma, V., Holzman, L. B., Wolf, F. W., O'Rourke, K., Ward, P. A., Prochownik, E. V. & Marks, R. M. (1990). Tumor necrosis factor-α induction of novel gene products in human endothelial cells including a macrophage-specific chemotaxin. J Biol Chem 265, 2973–2978. [PubMed] [Google Scholar]
- Duan, X., Nauwynck, H. J. & Pensaert, M. B. (1997a). Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol 142, 2483–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, X., Nauwynck, H. J. & Pensaert, M. B. (1997b). Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Microbiol 56, 9–19. [DOI] [PubMed] [Google Scholar]
- Fickenscher, H., Hor, S., Kupers, H., Knappe, A., Wittmann, S. & Sticht, H. (2002). The interleukin-10 family of cytokines. Trends Immunol 23, 89–96. [DOI] [PubMed] [Google Scholar]
- Flores-Mendoza, L., Silva-Campa, E., Reséndiz, M., Osorio, F. A. & Hernández, J. (2008). Porcine reproductive and respiratory syndrome virus (PRRSV) infects mature porcine dendritic cells and up-regulates IL-10 production. Clin Vaccine Immunol 15, 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genini, S., Malinverni, R., Delputte, P. L., Fiorentini, S., Stella, A., Botti, S., Nauwynck, H. J. & Giuffra, E. (2008). Gene expression profiling of porcine alveolar macrophages after antibody-mediated crosslinking of sialoadhesin (Sn, Siglec-1). J Recept Signal Transduct Res 28, 185–243. [DOI] [PubMed] [Google Scholar]
- Halbur, P. G. (2001). Emerging and recurring diseases in growing pigs. In Animal Health, Biotechnology and Trade, Annual Meeting of the National Institute for Animal Agriculture Proceedings, Colorado Springs, CO, USA, 3 April 2001. http://animalagriculture.com/Proceedings/2001 %20Proc/Halbur.htm.
- Han, E. S., Wu, Y., McCarter, R., Nelson, J. F., Richardson, A. & Hilsenbeck, S. G. (2004). Reproducibility, sources of variability, pooling, and sample size: important considerations for the design of high-density oligonucleotide array experiments. J Gerontol A Biol Sci Med Sci 59, 306–315. [DOI] [PubMed] [Google Scholar]
- Jenner, R. G. & Young, R. A. (2005). Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 3, 281–294. [DOI] [PubMed] [Google Scholar]
- Kim, T. S., Benfield, D. A. & Rowland, R. R. (2002). Porcine reproductive and respiratory syndrome virus-induced cell death exhibits features consistent with a nontypical form of apoptosis. Virus Res 85, 133–140. [DOI] [PubMed] [Google Scholar]
- Kubo, A., Minamino, N., Isumi, Y., Katafuchi, T., Kangawa, K., Dohi, K. & Matsuo, H. (1998). Production of adrenomedullin in macrophage cell line and peritoneal macrophage. J Biol Chem 273, 16730–16738. [DOI] [PubMed] [Google Scholar]
- Lamm, G. M., Nicol, S. M., Fuller-Pace, F. V. & Lamond, A. I. (1996). p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res 24, 3739–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi, E., Pezzi, N., Prieto, I., Barthelemy, I., Carreiro, C., Martínez, A., Maldonado-Rodríguez, A., López-Cabrera, M. & Barbero, J. L. (2004). Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem 279, 6553–6559. [DOI] [PubMed] [Google Scholar]
- Lee, E. G., Boone, D. L., Chai, S., Libby, S. L., Chien, M., Lodolce, J. P. & Ma, A. (2000). Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289, 2350–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Bachand, A., Murtaugh, M. P. & Yoo, D. (2004). Differential host cell gene expression regulated by the porcine reproductive and respiratory syndrome virus GP4 and GP5 glycoproteins. Vet Immunol Immunopathol 102, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, C. R., Ait-Ali, T., Clapperton, M., Archibald, A. L. & Bishop, S. (2007). Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS). Viral Immunol 20, 343–358. [DOI] [PubMed] [Google Scholar]
- Li, H. L., Wang, A. B., Zhang, R., Wei, Y. S., Chen, H. Z., She, Z. G., Huang, Y., Liu, D. P. & Liang, C. C. (2006). A20 inhibits oxidized low-density lipoprotein-induced apoptosis through negative Fas/Fas ligand-dependent activation of caspase-8 and mitochondrial pathways in murine RAW264.7 macrophages. J Cell Physiol 208, 307–318. [DOI] [PubMed] [Google Scholar]
- Lopez, O. J. & Osorio, F. A. (2004). Role of neutralizing antibodies in PRRSV protective immunity. Vet Immunol Immunopathol 102, 155–163. [DOI] [PubMed] [Google Scholar]
- Loving, C. L., Brockmeier, S. L. & Sacco, R. E. (2007). Differential type I interferon activation and susceptibility of dendritic cell populations to porcine arterivirus. Immunology 120, 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese, K., Vincent, A., Lin, S.-H. & Shaw, T. (2000). Group I and group III metabotropic glutamate receptor subtypes provide enhanced neuroprotection. J Neurosci Res 62, 257–272. [DOI] [PubMed] [Google Scholar]
- Mateu, E. & Diaz, I. (2007). The challenge of PRRS immunology. Vet J 177, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, W. A., Galeota, J., Osorio, F. A., Husmann, R. J., Schnitzlein, W. M. & Zuckermann, F. A. (2003). Gradual development of the interferon-γ response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology 309, 18–31. [DOI] [PubMed] [Google Scholar]
- Mengeling, W. L. & Lager, K. M. (2000). A brief review of procedures and potential problems associated with the diagnosis of porcine reproductive and respiratory syndrome. Vet Res 31, 61–69. [DOI] [PubMed] [Google Scholar]
- Miller, L. C. & Fox, J. M. (2004). Apoptosis and porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 102, 131–142. [DOI] [PubMed] [Google Scholar]
- Miller, L. C., Laegreid, W. W., Bono, J. L., Chitko-McKown, C. G. & Fox, J. M. (2004). Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch Virol 149, 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, K. W., de Waal Malefyt, R., Coffman, R. L. & O'Garra, A. (2001). Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19, 683–765. [DOI] [PubMed] [Google Scholar]
- Moore, C. B., Bergstralh, D. T., Duncan, J. A., Lei, Y., Morrison, T. E., Zimmermann, A. G., Accavitti-Loper, M. A., Madden, V. J., Sun, L. & other authors (2008). NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577. [DOI] [PubMed] [Google Scholar]
- Murtaugh, M. P., Xiao, Z. & Zuckermann, F. (2002). Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol 15, 533–547. [DOI] [PubMed] [Google Scholar]
- Neumann, E. J., Kliebenstein, J. B., Johnson, C. D., Mabry, J. W., Bush, E. J., Seitzinger, A. H., Green, A. L. & Zimmerman, J. J. (2005). Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227, 385–392. [DOI] [PubMed] [Google Scholar]
- Nodelijk, G. (2002). Porcine reproductive and respiratory syndrome (PRRS) with special reference to clinical aspects and diagnosis. A review. Vet Q 24, 95–100. [DOI] [PubMed] [Google Scholar]
- Onose, A., Hashimoto, S., Hayashi, S., Maruoka, S., Kumasawa, F., Mizumura, K., Jibiki, I., Matsumoto, K., Gon, Y. & other authors (2006). An inhibitory effect of A20 on NF-κB activation in airway epithelium upon influenza virus infection. Eur J Pharmacol 541, 198–204. [DOI] [PubMed] [Google Scholar]
- Overend, C., Mitchell, R., He, D., Rompato, G., Grubman, M. J. & Garmendia, A. E. (2007). Recombinant swine beta interferon protects swine alveolar macrophages and MARC-145 cells from infection with Porcine reproductive and respiratory syndrome virus. J Gen Virol 88, 925–931. [DOI] [PubMed] [Google Scholar]
- Prag, S. & Adams, J. C. (2003). Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinform 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y. J., Zhang, Z. L., Yu, L. Y., He, J. W., Hou, Y. N., Liu, T. J., Wu, J. C., Wu, S. H. & Guo, L. H. (2006). A20 overexpression under control of mouse osteocalcin promoter in MC3T3-E1 cells inhibited tumor necrosis factor-α-induced apoptosis. Acta Pharmacol Sin 27, 1231–1237. [DOI] [PubMed] [Google Scholar]
- Redpath, S., Ghazal, P. & Gascoigne, N. R. (2001). Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol 9, 86–92. [DOI] [PubMed] [Google Scholar]
- Rossow, K. D., Collins, J. E., Goyal, S. M., Nelson, E. A., Christopher-Hennings, J. & Benfield, D. A. (1995). Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet Pathol 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Rowland, R. R., Lawson, S., Rossow, K. & Benfield, D. A. (2003). Lymphoid tissue tropism of porcine reproductive and respiratory syndrome virus replication during persistent infection of pigs originally exposed to virus in utero. Vet Microbiol 96, 219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royaee, A. R., Husmann, R. J., Dawson, H. D., Calzada-Nova, G., Schnitzlein, W. M., Zuckermann, F. A. & Lunney, J. K. (2004). Deciphering the involvement of innate immune factors in the development of the host response to PRRSV vaccination. Vet Immunol Immunopathol 102, 199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa, K., Yotsumoto, S., Kato, S. & Aramaki, Y. (2007). Phosphatidylinositol 3-kinase-mediated regulation of IL-10 and IL-12 production in macrophages stimulated with CpG oligodeoxynucleotide. Mol Immunol 44, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Shen, G., Xu, C., Hu, R., Jain, M. R., Nair, S., Lin, W., Yang, C. S., Chan, J. Y. & Kong, A. N. (2005). Comparison of (−)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/ 6J/Nrf2 (−/−) mice. Pharm Res 22, 1805–1820. [DOI] [PubMed] [Google Scholar]
- Sieg, S., King, C., Huang, Y. & Kaplan, D. (1996). The role of interleukin-10 in the inhibition of T-cell proliferation and apoptosis mediated by parainfluenza virus type 3. J Virol 70, 4845–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirinarumitr, T., Zhang, Y., Kluge, J. P., Halbur, P. G. & Paul, P. S. (1998). A pneumo-virulent United States isolate of porcine reproductive and respiratory syndrome virus induces apoptosis in bystander cells both in vitro and in vivo. J Gen Virol 79, 2989–2995. [DOI] [PubMed] [Google Scholar]
- Snijder, E. J. & Meulenberg, J. J. M. (1998). The molecular biology of arteriviruses. J Gen Virol 79, 961–979. [DOI] [PubMed] [Google Scholar]
- Suradhat, S. & Thanawongnuwech, R. (2003). Upregulation of interleukin-10 gene expression in the leukocytes of pigs infected with porcine reproductive and respiratory syndrome virus. J Gen Virol 84, 2755–2760. [DOI] [PubMed] [Google Scholar]
- Suradhat, S., Thanawongnuwech, R. & Poovorawan, Y. (2003). Upregulation of IL-10 gene expression in porcine peripheral blood mononuclear cells by porcine reproductive and respiratory syndrome virus. J Gen Virol 84, 453–459. [DOI] [PubMed] [Google Scholar]
- Sutherland, M. A., Niekamp, S. R., Johnson, R. W., van Alstine, W. G. & Salak-Johnson, J. L. (2007). Heat and social rank impact behavior and physiology of PRRS-virus-infected pigs. Physiol Behav 90, 73–81. [DOI] [PubMed] [Google Scholar]
- Tanaka, N., Sato, M., Lamphier, M. S., Nozawa, H., Oda, E., Noguchi, S., Schreiber, R. D., Tsujimoto, Y. & Taniguchi, T. (1998). Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3, 29–37. [DOI] [PubMed] [Google Scholar]
- Teifke, J. P., Dauber, M., Fichtner, D., Lenk, M., Polster, U., Weiland, E. & Beyer, J. (2001). Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J Comp Pathol 124, 238–245. [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech, R. & Thacker, E. L. (2003). Interleukin-10, interleukin-12, and interferon-γ levels in the respiratory tract following mycoplasma hyopneumoniae and PRRSV infection in pigs. Viral Immunol 16, 357–367. [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech, R., Thacker, B., Halbur, P. & Thacker, E. L. (2004). Increased production of proinflammatory cytokines following infection with porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Clin Diagn Lab Immunol 11, 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, E. Y., Yie, J., Thanos, D. & Goldfeld, A. E. (1996). Cell-type-specific regulation of the human tumor necrosis factor α gene in B cells and T cells by NFATp and ATF-2/JUN. Mol Cell Biol 16, 5232–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S., Cassady, J. P., Freking, B. A., Nonneman, D. J., Rohrer, G. A. & Piedrahita, J. A. (2006). Annotation of the Affymetrix porcine genome microarray. Anim Genet 37, 423–424. [DOI] [PubMed] [Google Scholar]
- Uzan, B., Ea, H. K., Launay, J. M., Garel, J. M., Champy, R., Cressent, M. & Lioté, F. (2006). A critical role for adrenomedullin-calcitonin receptor-like receptor in regulating rheumatoid fibroblast-like synoviocyte apoptosis. J Immunol 176, 5548–5558. [DOI] [PubMed] [Google Scholar]
- van Reeth, K., Labarque, G., Nauwynck, H. & Pensaert, M. (1999). Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voicu, I. L., Silim, A., Morin, M. & Elazhary, M. A. (1994). Interaction of porcine reproductive and respiratory syndrome virus with swine monocytes. Vet Rec 134, 422–423. [DOI] [PubMed] [Google Scholar]
- Wang, X., Eaton, M., Mayer, M., Li, H., He, D., Nelson, E. & Christopher-Hennings, J. (2007). Porcine reproductive and respiratory syndrome virus productively infects monocyte-derived dendritic cells and compromises their antigen-presenting ability. Arch Virol 152, 289–303. [DOI] [PubMed] [Google Scholar]
- Weidman, M. K., Sharma, R., Raychaudhuri, S., Kundu, P., Tsai, W. & Dasgupta, A. (2003). The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res 95, 75–85. [DOI] [PubMed] [Google Scholar]
- Wensvoort, G., Terpstra, C., Pol, J. M. A., ter Laak, E. A., Bloemraad, M., de Kluyver, E. P., Kragten, C., van Buiten, L., den Besten, A. & other authors (1991). Mystery swine disease in The Netherlands: the isolation of the Lelystad virus. Vet Q 13, 121–130. [DOI] [PubMed] [Google Scholar]
- Wills, R. W., Doster, A. R., Galeota, J. A., Sur, J. H. & Osorio, F. A. (2003). Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J Clin Microbiol 41, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z., Batista, L., Dee, S., Halbur, P. & Murtaugh, M. P. (2004). The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. J Virol 78, 5923–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z. & Shu, H. B. (2005). VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell 19, 727–740. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Mosser, D. M. & Zhang, X. (2007). Activation of the MAPK, ERK, following Leishmania amazonensis infection of macrophages. J Immunol 178, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, J., Tsuchiya, A., Yamada, T., Sakamoto, M., Sekiya, T. & Hirohashi, S. (2003). Nemo-like kinase induces apoptosis in DLD-1 human colon cancer cells. Biochem Biophys Res Commun 308, 227–233. [DOI] [PubMed] [Google Scholar]
- Yin, H., Chao, L. & Chao, J. (2004). Adrenomedullin protects against myocardial apoptosis after ischemia/reperfusion through activation of Akt-GSK signaling. Hypertension 43, 109–116. [DOI] [PubMed] [Google Scholar]
- Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S. & Fujita, T. (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Shin, J., Molitor, T. W., Schook, L. B. & Rutherford, M. S. (1999). Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology 262, 152–162. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Wang, C., Schook, L. B., Hawken, R. J. & Rutherford, M. S. (2000). An RNA helicase, RHIV-1, induced by porcine reproductive and respiratory syndrome virus (PRRSV) is mapped on porcine chromosome 10q13. Microb Pathog 28, 267–278. [DOI] [PubMed] [Google Scholar]
- Zhou, J. H., Broussard, S. R., Strle, K., Freund, G. G., Johnson, R. W., Dantzer, R. & Kelley, K. W. (2001). IL-10 inhibits apoptosis of promyeloid cells by activating insulin receptor substrate-2 and phosphatidylinositol 3′-kinase. J Immunol 167, 4436–4442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.