Abstract
Determination of side-chain conformations is an important step in protein structure prediction and protein design. Many such methods have been presented, although only a small number are in widespread use. SCWRL is one such method, and the SCWRL3 program (2003) has remained popular due to its speed, accuracy, and ease-of-use for the purpose of homology modeling. However, higher accuracy at comparable speed is desirable. This has been achieved through: 1) a new backbone-dependent rotamer library based on kernel density estimates; 2) averaging over samples of conformations about the positions in the rotamer library; 3) a fast anisotropic hydrogen bonding function; 4) a short-range, soft van der Waals atom-atom interaction potential; 5) fast collision detection using k-discrete oriented polytopes; 6) a tree decomposition algorithm to solve the combinatorial problem; and 7) optimization of all parameters by determining the interaction graph within the crystal environment using symmetry operators of the crystallographic space group. Accuracies as a function of electron density of the side chains demonstrate that side chains with higher electron density are easier to predict than those with low electron density and presumed conformational disorder. For a testing set of 379 proteins, 86% of χ1 angles and 75% of χ1+2 are predicted correctly within 40° of the X-ray positions. Among side chains with higher electron density (25th–100th percentile), these numbers rise to 89% and 80%. The new program maintains its simple command-line interface, designed for homology modeling, and is now available as a dynamic-linked library for incorporation into other software programs.
Keywords: homology modeling, side-chain prediction, protein structure, rotamer library, graph decomposition, SCWRL
Introduction
The side-chain conformation prediction problem is an integral component of protein structure determination, protein structure prediction, and protein design. In single-site mutants and in closely related proteins, the backbone often changes little and structure prediction can be accomplished by accurate side-chain prediction1. In docking of ligands and other proteins, taking into account changes in side-chain conformation is often critical to accurate structure predictions of complexes2–4. Even in methods that take account of changes in backbone conformation, one step in the process is recalculation of side-chain conformation or “repacking.”5 Because many backbone conformations may be sampled in model refinements, side-chain prediction must also be very fast. In protein design, as changes in the sequence are proposed by Monte Carlo steps or other algorithms, conformations of side chains need to be predicted accurately in order to determine whether the change is favorable or not6–8.
Most side-chain prediction methods are based on a sample space that depends on a rotamer library, which is a statistical clustering of observed side-chain conformations in known structures9. Such rotamer libraries can be backbone-independent, lumping all side chains together regardless of the local backbone conformation10, or backbone-dependent, such that frequencies and dihedral angles vary with the backbone dihedral angles φ and ψ11,12. An alternative to using statistical rotamer libraries is to use conformer libraries, which are samples of side chains from known structures, usually in the form of Cartesian coordinates, thus accounting for bond length, bond angle, and dihedral angle variability13–16. Once a search space in the form of rotamers (and samples around rotamers in some cases) or conformers is defined, a scoring function is required to evaluate the suitability of the sampled conformations. These may include the negative ogarithm of the observed rotamer library frequencies17–20, van der Waals or hard sphere steric interactions of side chains with other side chains or the backbone, and sometimes electrostatic, hydrogen bonding, and solvation terms20–24. Many search algorithms have been applied, including cyclic optimization of single residues or pairs of residues11,16, Monte Carlo5,18,25, dead-end elimination26,27, self-consistent mean field optimization28, integer programming29, and graph decomposition17,30,31. These methods vary in how fast they can solve the combinatorial problem, and whether they guarantee a global minimum of the given energy function or instead search for a low energy without such a guarantee. In general, such a guarantee is not necessary, given the approximate nature of the energy functions, and it is the overall prediction accuracy and speed that are more important features of a prediction method. In recent years, it has become clear that some flexibility around rotameric positions15,16,32 and more sophisticated energy functions20,33 are needed for improved side-chain packing and prediction.
SCWRL3 is one of the most widely used programs of its type with 2986 licenses in 72 countries as of April 30, 2009. It uses a backbone-dependent rotamer library12, a simple energy function based on the library rotamer frequencies and a purely repulsive steric energy term, and a graph decomposition to solve the combinatorial packing problem30. It has a number of features that have made it widely used. The first of these is speed, which has enabled the program to be used on a number of web servers that predict protein structure from sequence-structure alignments34 and may perform many hundreds of predictions per day. The second is accuracy. At the time of its publication, it was one of the most accurate side-chain prediction methods. However, a number of other methods have appeared claiming higher accuracy15,18,20,35, although often at much longer CPU times. The third feature of SCWRL3 is usability. The program takes input PDB coordinates for the backbone, optionally a new sequence, and outputs coordinates for the structure with predicted side chains using the same residue numbering and chain identifiers as the input structure. This feature is simple but in fact many if not most side-chain prediction programs renumber the residues of the input structure and strip the chain identifiers, making them difficult to use in homology modeling. One unfortunate feature of SCWRL3 is that the graph decomposition method used may not always result in a combinatorial optimization that can be solved quickly. In such cases, the program may go on for many hours instead of finishing in a few seconds, since it lacks any heuristic method for simplifying the problem and finishing quickly.
In developing a new generation of SCWRL, called SCWRL4, we had several goals. First, we wanted to increase the accuracy over SCWRL3 such that SCWRL4’s accuracy would be comparable or better than programs developed in the last several years. Second, we wanted to maintain the speed advantage that SCWRL has over most similar programs. Third, we wanted to maintain the usability of the program for homology modeling and other purposes. As part of this, we wanted to make sure that the program always solves the structure prediction problem in a reasonable time, even if the graph is not sufficiently decomposable. This is accomplished with an approximation, that while not guaranteeing a global minimum of the energy function given the rotamer search space, does complete the calculation quickly in all cases tested.
In this paper, we describe the development of the SCWRL4 program for prediction of protein side-chain conformations. We used a number of different approaches to accomplish the goals described above. We have improved the SCWRL energy function using a new backbone-dependent rotamer library (Shapovalov and Dunbrack, in preparation) which uses kernel density estimates and kernel regressions to provide rotamer frequencies, dihedral angles, and variances that vary smoothly as a function of the backbone dihedral angles φ and ψ. SCWRL4 also uses a short-range, soft van der Waals interaction potential between atoms rather than a linear repulsive-only function used in SCWRL3, as well as an anisotropic hydrogen bond function similar to that used in Rosetta36 (but using a different functional form that is faster to evaluate). To account for variation of dihedral angles around the mean values given in the rotamer library, we used the approach of Mendes et al.32, which samples χ angles around the library values and averages the energy of interaction between rotamers of different side chains over these samples, resulting in a free-energy-like scoring function. In order to determine the interaction graph, as used in SCWRL3, we implemented a fast method for detecting collisions (i.e., atom-atom interactions less than some distance) using k-discrete oriented polytopes (“kDOPs”). kDOPs are three-dimensional shapes with faces perpendicular to common fixed axes, such that kDOPs around two groups of atoms can be rapidly tested for overlap37.
In SCWRL3, we used a graph decomposition method that broke down the interaction graph of residues into biconnected components, which overlap by single residues called articulation points. In most cases, this solves the graph quickly. However, with a longer-range energy function and sampling about the rotameric dihedral angles, this is no longer true. We therefore implemented our own version of a tree decomposition of the graphs, as suggested by Jinbo Xu for the side-chain prediction problem31. This is almost always successful but in a small number of cases may still not result in an easily solvable combinatorial problem. We therefore added a heuristic projection of the pairwise energies onto self-energies within some threshold. This approximation of the full prediction problem always results in a solution, even if it is not guaranteed to find the global minimum. Finally, the new program has been developed as a library, so that its functions can be called easily by other programs such as loop modeling and protein design.
Methods
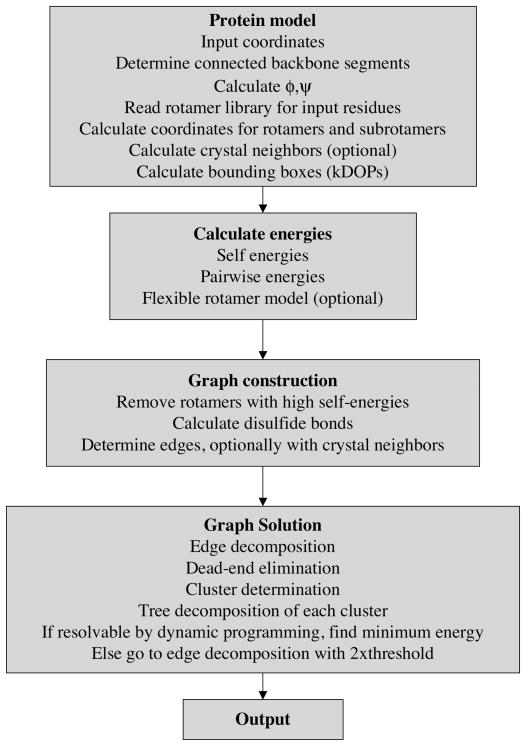
In Figure 1 we show a flowchart of the basic steps in SCWRL4 to solve the side-chain prediction problem. These will be discussed further below. The major steps are 1) inputting the data and constructing the side-chain coordinates; 2) calculating energies; 3) graph computation, with symmetry operators if any; 4) combinatorial optimization via edge decomposition, dead-end elimination, and tree decomposition; 5) outputting the results. SCWRL4 runs on a command line with a number of required and optional flags. A number of other options and parameters are specified in a required initialization file with extension.ini, which uses a standard name=value format (see http://en.wikipedia.org/wiki/INI_file).
Figure 1.
Steps in SCWRL4 side-chain conformation prediction
lnput and construction of coordinates
An individual residue position is defined by specifying four backbone atoms (N,Cα,C,O) in a PDB-format input file. These individual residue sites can comprise one or more polypeptide chains, from which the backbone dihedral angles φ and ψ are calculated for each residue. For purposes of looking up residues in the rotamer library, the N-terminal residue φ is set to −60°. Similarly for C-terminal residues, ψ is set to 60°. These values are those for which there is weak dependence of the rotamer probabilities on the missing dihedral angle38. The Ci−1-Ni atom distances are checked to determine whether there are missing internal residues in a chain.
For each residue, rotamers are read from a new version of the backbone-dependent rotamer library (Shapovalov and Dunbrack, in preparation). This rotamer library is based on a much larger data set, and is derived using kernel density estimates and kernel regressions. The rotamer library includes rotamer frequencies and mean dihedral angles and their standard deviations over a discrete (φ,ψ)-grid. This library offers much greater detail for non-rotameric degrees of freedom, in particular χ2 for Asn, Asp, His, Phe, Trp, and Tyr and χ3 for Glu and Gln. Optionally SCWRL4 can determine frequencies and dihedral angle parameters by bilinear interpolation from the four neighboring φ,ψ grid points in the library. For each χ1 rotamer of Ser and Thr, SCWRL4 generates three rotamers for the hydroxyl hydrogen with χ2 dihedral set to −60°, +60° and 180° and the variance set to 10° times the corresponding parameter given in the configuration file. For each χ1, χ2 rotamer of Tyr, two rotamers are generated for the hydroxyl hydrogen with χ3 dihedral set to 0° and 180°, which are the values observed in neutron diffraction studies39. For His, extra rotamers are created for the singly protonated states (proton on ND1 or NE2). Rotamers that represent positively charged His can be enabled in the program using a command-line option.
Side-chain coordinates are built for all rotamers and for subrotamers about these rotamers used by the Flexible Rotamer Model (FRM, see below). Subrotamers as used here are conformations with dihedral angles ± one standard deviation (or a fixed proportion thereof) away from rotamer values given in the rotamer library. For subrotamers, only one dihedral at a time differs from the library value, since we found that allowing multiple deviations did not noticeably improve the accuracy but did slow the calculation (data not shown). Side chains are represented in a tree-like structure, so that atoms common to more than one subrotamer (e.g., same CG position for different χ2 conformers) are calculated and stored only once40. Coordinates are built using a fast incremental torsion to Cartesian conversion method41.
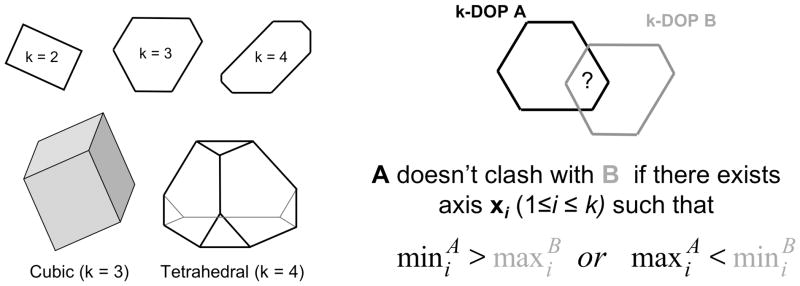
Because SCWRL4 uses a large number of rotamers and subrotamers, we implemented a fast collision detection algorithm based on k-dimensional Discrete Oriented Polytopes or kDOPs37. The kDOP algorithm is based on two key ideas. The first idea is to enclose each geometric object into a convex polytope of a special kind and use these as bounding boxes for clash checks. A particular class of kDOPs is defined by a set of k pairwise non-collinear unit vectors, and consists of all convex polytopes with 2k facets such that any facet is perpendicular to one of these vectors. Examples are shown in Figure 2. For instance, if k=3, these could be the x, y, and z-axes, so that all bounding boxes are rectangular parallelepipeds whose faces are perpendicular to the Cartesian axes. The second key idea is to organize all bounding boxes related to a particular group of geometric objects as a hierarchy. These hierarchies can then be used for efficient search of all possible clashes between individual bounding boxes.
Figure 2. k-Dimensional Oriented Polytopes (kDOPs).
Left: examples of kDOPs in the plane (k=2,3,4) and in three dimensions (k=3,4). Right: Overlap test for kDOP A (black) and kDOP B (gray). The objects enclosed within the kDOPs may clash if one of the conditions shown is satisfied.
The advantage of using a single set of vectors for defining the bounding boxes is that two bounding boxes of the same kDOP class can be efficiently checked for clashes. As illustrated in Figure 2, this can be accomplished by testing for overlaps of the real intervals that represent their projections across the corresponding basic axes. Let and be the sets of these intervals for two kDOPs “A” and “B” respectively. “A” does not clash with “B” if there exists I ∈ {1..k} such that min (ai)>max (bi) or if there exists j ∈ {1..k} such that min (bi)> max (ai). If neither of these conditions are met, then the underlying objects enclosed inside of these two boxes may clash, but do not necessarily do so. The last is to be checked by pairwise distance calculations of the objects enclosed in the bounding boxes.
Building a kDOP around a geometric object consists of finding projections of the object onto the basic axes. Both the van der Waals function and the hydrogen bond potentials described in the next section have a certain boundary distance beyond which the potential is zero. These distances are used to represent each atom as a sphere with a certain radius. To build a bounding box around a whole side chain, each atom is enclosed into a kDOP and then the elementary shells are merged. The rotamers and subrotamers of a side chain can be enclosed into a single kDOP, such that all residue-residue interactions can be checked very quickly. In SCWRL4, we use four basic axes and construct bounding boxes around individual atoms, backbone atoms of each residue, parts of each side chain, individual rotamers (i.e. entire set of its subrotamers) and every residue (all rotamers). The basic axes form a tetrahedral geometry:
Using these four axes results in somewhat faster calculations, by about 15%, than using three axes along the x,y, and z axes, despite some overhead involved in calculating the projections.
Calculating of self energies and pairwise energies via modified flexible rotamer model
SCWRL4 uses both a rigid rotamer model (RRM), as in SCWRL3, and a flexible rotamer model (FRM)32. In the rigid rotamer model, the total energy of the system is expressed as:
where the vector r specifies a single rotamer for each of N residues in the system. In this case, the self energy of each rotamer is:
where the first term expresses the rotamer energy relative to the most populated rotamer, rmax, given the backbone dihedrals φ and ψ of residue i and the frame term expresses interaction of the side chain with the backbone and any ligand or other fixed atoms present. We allow the value of the constant in front of the log term to be residue-type dependent.
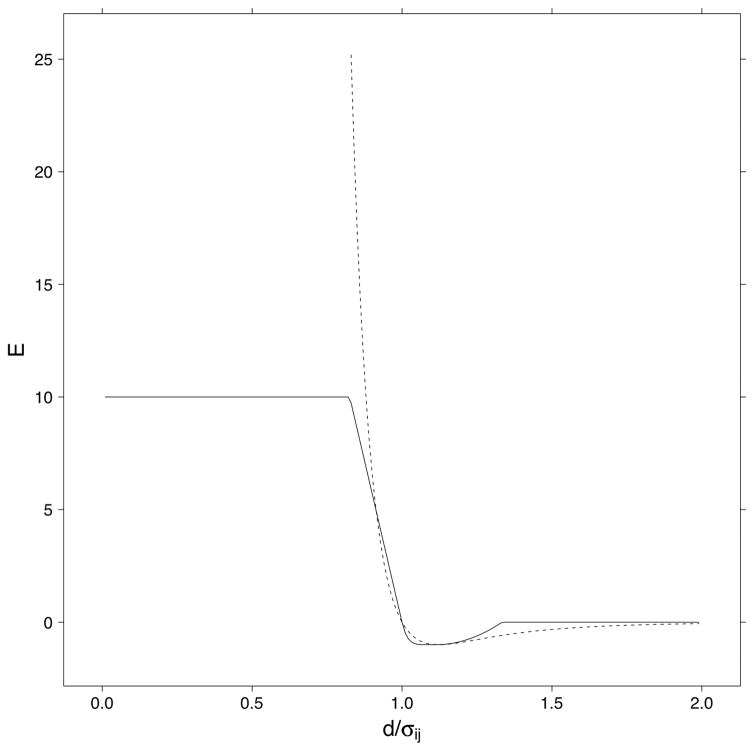
In contrast to SCWRL3, in SCWRL4 the frame and pairwise rotamer energies consist of repulsive and attractive van der Waals terms as well as a hydrogen bonding term. The repulsive van der Waals term is the same as the piecewise linear term used in SCWRL3, but is combined with a short-range attractive potential as follows. If σij is the sum of the hard-sphere radii of atoms i and j and Eij is , where the Ei values are the Emin values from the CHARMM param19 potential42, and d is the distance between the two atoms, then
This potential is shown in Figure 3 along with the standard Lennard-Jones potential with the same Eij and Rij. The hard-sphere radii were manually optimized for the training set accuracies. The minimum energy occurs at which is close to the standard Lennard-Jones parameterization in which the minimum occurs at rmin = 21/6σij = 1.12σij. The parameters are given in Supplemental Information.
Figure 3. SCWRL4 van der Waals potential.
The van der Waals potential used in SCWRL4 is shown (solid line) with a standard Lennard-Jones 6–12 potential (dotted line) with Eij = 1.
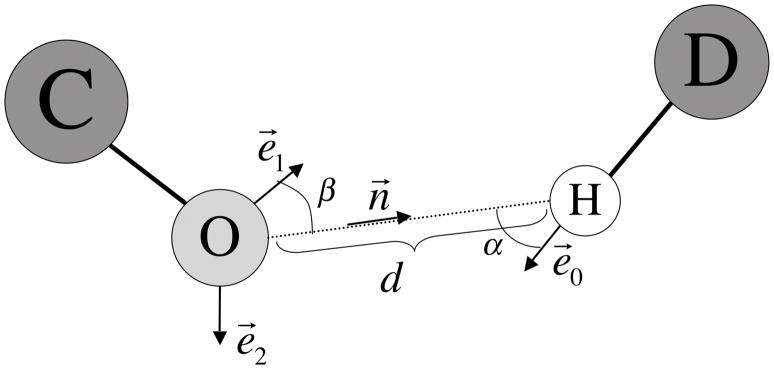
The hydrogen bonding term in SCWRL4 is similar to the one used in Rosetta36, although it is parameterized in a different way, as shown in Figure 4. We define d in this case as the distance between a polar hydrogen (HN- or HO-) and a hydrogen bond acceptor (oxygen), n⃗ as a unit vector from O acceptor to H, e⃗0 as a unit vector along the covalent bond from the hydrogen bond donor heavy atom D to H, and two unit vectors and e⃗1 and e⃗2 from the hydrogen bond acceptor O toward the middle of the oxygen lone-pair electron clouds. For carbonyl oxygens these two vectors are 120° apart from the double bond and coplanar with the carbonyl carbon substituents. For hydroxyl oxygens, these two vectors are 109.5° from each other and from the other two oxygen substituents (H and C), forming a tetrahedral arrangement. The hydrogen bond function is evaluated first for e⃗1, and if no hydrogen bond is found, then for e⃗2. For e⃗1, the weight w for the hydrogen bond is defined:
where is the angle between the D-H bond and the H…O vector and is the angle between the O-lone pair and the O…H vector. If the multiplicand under the square root is negative then the score is set to zero. The calculation of this score enables an efficient implementation and together with the distance d and vector n⃗ can be done within 30 arithmetic operations, one division, and two square root evaluations.
Figure 4. Hydrogen bond potential.
Interaction of hydrogen bond acceptor O and hydrogen bond donor, D. Unit vector n⃗ is the vector from atom O to atom H. Unit vectors e⃗1 and e⃗2 are placed from atom O along each lone pair of electrons. Unit vector e⃗0 connects the hydrogen bond donor D to the hydrogen atom. is the angle between the D-H bond and the H…O vector and is the angle between the O-lone pair and the O…H vector.
After the weight w has been computed it is used to derive the final energy of oxygen-hydrogen interaction by balancing the default van der Waals energy and pure hydrogen-bond attraction terms:
where qH and qO are the charges from the CHARMM param19 potential. The formulas above include five atom-independent coefficients:d0, σd, αmax, βmax, B. The values of these coefficients were optimized on the training set proteins and are given in the Supplemental Data.
Using single rotamers sometimes results in poor packing predictions, due to fluctuations in the dihedral angles and imprecise representations of the backbone in homology modeling. We investigated the use of subrotamers, which we define as conformations that differ in one or more dihedral angles by one standard deviation (or some constant times this value) from the mean values given in the rotamer library:
If we allowed variations in all dihedral angles in this manner, treating the subrotamers as additional rotamers resulted in intractable calculations using the graph decomposition algorithm used in SCWRL3. Even with the tree decomposition of Xu31, implemented in SCWRL4 (see below), the calculations often remained intractable. So we implemented the flexible rotamer model of Mendes et al.32, in which the subrotamers are integrated to produce an approximate free energy using the Kirkwood superposition approximation43:
We treat the first term as the “self free energy” and the second term as the “pairwise free energy”. Aself (ri) and Apair, (rj, rj)are defined as:
The terms Eframe (ri, si) and Aframe (ri)contain only the van der Waals and hydrogen bond energies. In our implementation, each residue type has a separately optimized temperature, and for the pairwise free energy, Tij = (Ti + Tj)/2.
Graph construction
As with SCWRL3, some rotamers with high self-energy are removed from the calculation, since they are very unlikely to be part of the predicted structure. These rotamers are marked as inactive. In SCWRL3, rotamers with self-energy above a certain residue-independent bound were inactivated. However it sometimes happens that all rotamers have self-energy above this bound. In this case all rotamers were reactivated. In SCWRL4 we replaced this heuristic by making the bound relative to the lowest energy rotamer for each residue. This approach guarantees that at least one rotamer will remain active. After some study the value of this threshold was set to 30. The exact value of the threshold can be customized through the configuration file.
Before the graph is constructed, disulfide bonds are resolved. SCWRL4 uses the same criterion as SCWRL3 to identify if two cysteine side chains can form a disulfide bond, but introduces a new procedure for resolving ambiguities. An ambiguity occurs when more than one rotamer of a particular cysteine residue can form a disulfide bond or when one rotamer can form disulfide bonds with more than one other cysteine side chain. To select a particular collision-free combination of disulfide bonds, SCWRL4 finds the minimum total energy out of all possible combinations of feasible disulfide bonds. To do this we use an objective function in the form:
where the summations run over all possible disulfide bonds, C is a large positive constant and η is a binary function that evaluates to one if a particular disulfide bond is switched on and to zero otherwise. The functional above is of the same form as the one used to compute the total energy of rotamer assignment. Therefore we can minimize it for function η via the same optimization procedure. Doing this yields a list of the optimal disulfide bonds that do not have collisions. If for a particular cysteine residue one of the rotamers is part of an optimal disulfide bond then all other rotamers are inactivated for that residue. Energies of interaction of cysteines in disulfides are added to the self-energies of rotamers of other side chains within interacting distance.
SCWRL uses an interaction graph to represent the side-chain placement problem17,30. In this graph, vertices represent residues while edges between vertices indicate that at least one rotamer of one residue has a non-zero interaction with rotamers from another residue connected by the edge. For a single protein or protein complex, the graph is constructed by checking for overlap of the kDOP around whole residues. If at least one rotamer or subrotamer of one residue can interact with non-zero energy with a rotamer or subrotamer of another residue, then an edge is added to the graph between the vertices in the graph representing these residues.
SCWRL4 is able to model side chains in symmetric complexes using symmetry operators. These rotation-translation operators can be generated from the CRYST1 record in the input PDB file or specified explicitly by the user in a separate input file. For crystals, if the input PDB file contains the asymmetric unit, all residues in asymmetric units that may contact the input coordinates are constructed, as described previously44. Bounding boxes are constructed around the residues, rotamers, subrotamers, and atoms of the symmetry copies. Interactions between atoms in the input structure and its side chains and atoms in the symmetry copies and their side chains are determined. If side chains in the input structure interact with the backbone or ligands of the symmetry copies, then the static frame energies of these residues are modified accordingly. If the side chains of the input structure interact with the side chains of the symmetry copies, then an edge is created between the corresponding residues, if it does not already exist. If it does, then the pairwise energies are modified to account for the additional interactions between symmetry-related proteins. Thus a residue on one side of a protein may have an edge with a residue on the other side of a protein because of symmetry.
Graph solution via tree decomposition
Before the major optimization via dynamic programming is launched the interaction graph undergoes some preprocessing consisting of edge decomposition and dead-end elimination. Typically this eliminates a significant number of rotamers as well as some edges and nodes. Because edges were formed based on overlapping of bounding boxes some of them may contain only zeros as pairwise rotamer-rotamer energies. If this is the case or if the actual energies of interactions are very close to zero then the edge is removed.
Edge decomposition removes edges that can be approximated as the sum over single-residue energies. If this representation is feasible within a certain threshold then the corresponding self-energies are modified and the edge is removed. With larger thresholds, more edges may be removed. In this preprocessing stage, the threshold is set to a very small value, ε =0.02 kcal/mol.
The pairwise energies of two residues, (Epair ri, rj), in the rigid rotamer model or free energies, (Apair ri, rj), in the flexible rotamer model, may be represented by a matrix of real numbers for rotamers k=1..m and l=1..n. Edge decomposition consists of finding two sets of real numbers and which minimize the average deviation:
By setting the partial derivatives of δ with respect to ak and bl to zero, we find that these two sets should satisfy the following equations:
The initial task is not well defined as its solution is not unique. Thus adding some value to all ak and subtracting the same value from all bl does not change the sum ak + bl Therefore we can set ā to an arbitrary value. For example we can set:
Substituting this value into the second equation we find the corresponding value for b̄:
Using these values, we can determine ak and bl and evaluate the maximal absolute deviation:
SCWRL4 checks if this deviation is less than a certain threshold and if so then we remove the corresponding edge and modify the self-energies of the kth rotamer of residue i and the lth rotamer of residue j:
As stated earlier, the initial value of the threshold is 0.02, which enables the algorithm to eliminate almost all redundant “near-zero” edges. We remove from the graph those nodes that now have zero edges; its assigned rotamer is that of lowest Eself.
The next step is to perform dead-end elimination (DEE) that identifies and removes rotamers that cannot be the part of the global solution. These rotamers are identified via Goldstein’s criterion that was used in SCWRL327. If for a certain residue only one rotamer is left after DEE then that rotamer is part of the solution. If this residue has adjacent edges then all pairwise energies with the remaining rotamer are incorporated into self-energies of the corresponding rotamers from adjacent residues and these edges are removed. This makes the residue isolated, which means that it can be removed from the graph; the self-energy of its single rotamer is added to the total value of the minimal energy. The edge decomposition and DEE steps are repeated until nothing further is removed.
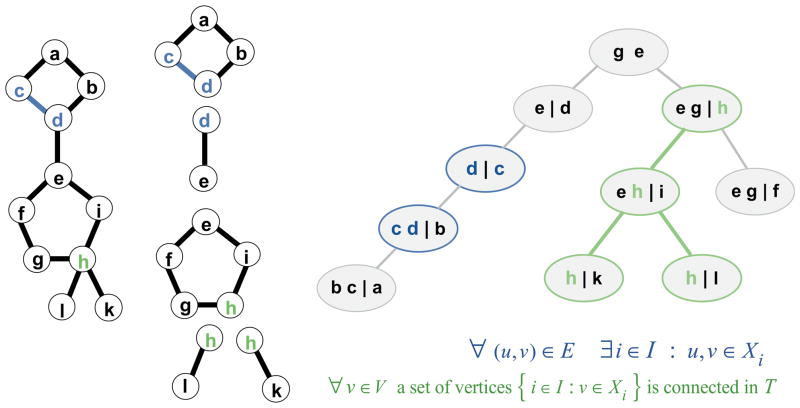
As in SCWRL3, the resulting graph may contain separated graphs or clusters with no edges between them; each of these clusters is then subject to graph decomposition. In SCWRL3, the graph decomposition was based on the determination of biconnected components, which are subgraphs that cannot be broken into parts by the removal of a single node. The graph is then a set of biconnected components connected by single nodes called articulation points. Tree decomposition can be viewed as a generalization of graph decomposition based on biconnected components31. To see this, we show in Figure 5 the same graph as described in the SCWRL3 paper, its decomposition into biconnected components, and its tree decomposition. The nodes of the graph on the left are gathered into “bags” which are nodes of the tree shown on the right. Every node of the graph is represented in one or more of the bags (condition 1 of the definition given below). Every edge of the graph on the left is also represented in one or more of the bags, so that the two nodes of an edge are together in at least one bag (condition 2). Finally, for any vertex of the graph, all those bags on the tree that contain the vertex form a connected subtree (condition 3). More formally:
Figure 5. Tree decomposition as generalization of biconnected component decomposition.
At left, the graph used in the SCWRL3 paper is shown along with its biconnected component decomposition. At right, a tree decomposition of the same graph is shown. Residues in blue and green illustrate conditions 2 and 3 of a tree decomposition being satisfied. The relevant conditions are shown below the tree decomposition. At each node of the tree, those residues that are members of set L are shown to the left of the vertical bar, and those of set R are shown to the right. Set L consists of those residues shared with the parent of each node, and R the remaining residues of the node.
Definition
A tree-decomposition of a graph G=(V, E) is a pair (T, Z), where T=(W, F) is a tree (i.e., a graph with no cycles) with vertex set W and edge set F and Z = {Zw : Zw ⊆ V}w∈W is a family of subsets of the set V associated one-to-one with the vertices of T that satisfies the conditions:
∀(u, v) ∈ E ∃ w ∈ W : u, v ∈ Zw
∀ v ∈ V a set of vertices{w ∈ W : v ∈ Zw}is connected in T
Due to the one-to-one correspondence between sets Z and W, we will denote the vertices of tree T as “bags”. Figure 5 shows that condition 1 is satisfied by this tree decomposition, since all the residues are present in one or more bags. Residues c,d illustrate that condition 2 is satisfied since the edge c-d is contained in at least one bag (in this case, two). Residue h illustrates condition 3, since all the bags that contain h are connected in a single subtree.
Typically several different tree-decompositions can be built for a given graph. The width of a particular tree-decomposition is the size of the largest bag minus one. For a given graph a tree-decomposition with the minimal possible width is the optimal one and its width is called the treewidth of the graph. This characteristic indicates how well a graph is tree-decomposable. For example if a graph has no cycles (and thus is a tree) then its treewidth equals one, while for a simple cycle the treewidth equals two.
In SCWRL3, the graph solution begins with any biconnected component with a single articulation point by finding the minimum energy of the biconnected component residues for each rotamer of the articulation point. This energy is then added to the self-energy of the articulation point rotamer, and the rotamers of the biconnected component that achieve this minimum energy are assigned to the articulation point rotamer. The biconnected component can then be removed, and the process continues for all biconnected components with one articulation point in the remaining graph. The combinatorial problem is thus reduced to the order of the largest biconnected component (i.e., the one with the largest number of rotamer combinations).
In a tree decomposition, instead of using single nodes to separate the graph, the graph can be separated by removing one, two, or more nodes. To see this, in the tree decomposition in Figure 5, each bag w is broken up into two sets of residues, Lw and Rw, where the residues in Lw are those residues in the bag that are shared between the bag and its immediate parent bag. For each bag in the figure, these are listed to the left of a vertical bar. The remaining residues in the bag, the set Rw, are those not in the parent and are placed to the right of the vertical bar. Each set Lw is a separation set of the graph G45; that is removing the residues in Lw breaks the original residue graph into two or more separate unconnected graphs. For instance, removing residues b and c breaks the original graph into two graphs, one consisting of residue a and the other the rest of the graph below residues b and c.
Solving for the minimum energy of the graph proceeds as it does in SCWRL3. Starting at a leaf (a bag with no children), y, e.g. the one containing “b c | a”, find the lowest energy of the residue(s) in Ry (in this case residue a) for each combination of the rotamers in Ly (in this case, residues b and c), saving the corresponding assignment of rotamers of Ry. Then add these energies to that rotamer combination in the parent bag, which by the definition of tree decomposition contains b and c. The procedure continues up the tree to the parent node of y (let us call it node z). Again, the minimum energy of all the rotamer combinations of those residues in Rz is calculated for each rotamer combination in set Lz. These energies need to include the energies for b,c calculated for the child node y. This procedure continues until only the root bag is left. We provide a more formal description of this procedure below.
The complexity of the solution is associated with the width of the tree, since all the rotamer combinations of the residues in each bag need to be enumerated. It is in general difficult to compute a treewidth and to find the optimal tree-decomposition, and it has been proved to be NP-hard for an arbitrary graph46. For building a tree-decomposition we have developed a heuristic algorithm. Our algorithm is similar to the one suggested by Xu31 who referred to it as a “minimal degree heuristic.”
In the first step, the family of sets Z is built. The input graph is gradually disassembled using a loop of the following steps:
Select any vertex with the minimal number of adjacent edges.
Form a bag of the tree from this vertex and all its neighbors. The selected vertex we will denote as the primary vertex of the corresponding bag.
Add edges into the graph being processed so that the neighbors of the selected vertex become a clique (a subgraph where all nodes have edges to each other).
Remove the selected vertex and all adjacent edges from the graph.
Repeat from the first step until there are no more vertices left in the graph.
Thus we obtain a set of “bags” which represent the vertices of the tree-decomposition.
Bags are numbered in the order of their construction. It is important to notice here that within any iteration the intersection of the bag w with the vertices of the remaining graph (Sw) consists solely of the neighbors (Nw) of the initial vertex of the bag concerned, Zw ∩ = Sw = Nw. The one-to-one correspondence between vertices and the bags verifies that the first condition in the definition of tree decomposition is automatically satisfied. Also it is clear that the edges are removed solely during the bag construction and that when any edge is removed both adjacent vertices are included into a bag. This guarantees that the second condition in the definition of a tree-decomposition is satisfied.
The second step is to connect the “bags” to obtain a tree that meets the definition of tree decomposition. This is done by sequentially fastening these bags to the tree in the reverse order in which they were constructed. Thus the bag that was created last becomes the root of the tree. The next bag becomes connected to it and thus becomes its immediate child. For the next bag, there are two choices for where to attach it. However, the appropriate node of the tree-decomposition must meet the following condition:
According to this condition a set of vertices in the appropriate node must contain all vertices from the bag to be added that are already present in the tree.
The tree decomposition just obtained undergoes some additional minor processing. This consists of two normalization rules, which are applied until they cannot be applied further: 1) If all vertices associated with some bag belong to the vertex set of its immediate parent then this node is removed and all its immediate children are reconnected to the parent node. 2) If some bag contains all vertices associated with its parent node then the parent bag is substituted by this bag which thus moves up one edge towards the root.
The minimum energy rotamer configuration is calculated as follows. Starting with a leaf node consisting of sets L and R, the left and right portions of each bag as defined above, we define 〈L〉 as the set of all rotamer combinations of the residues in the set L, and similarly define 〈R〉 for set R. A single member of 〈L〉 we denote as l, which is a vector of rotamer assignments, one rotamer li for each residue i in the set L; similarly define r for 〈R〉. For a leaf node, we calculate energies for each vector l :
where
and
For fixed rotamers in L, only the pairwise interactions with rotamers in R are included, while both self and pairwise interactions among the rotamers in R must be added. For the flexible rotamer model, the values of Aself and Apair are used instead Eself and Epair. For an inner node of the tree decomposition, we need to add in the energies assigned to rotamer combinations in the node via its children:
where
where the sum is over the children c of the inner node, and lc is a vector of the rotamers of the residues in the set Lc, such that these rotamer are in the set {li : i ∈ L; rj : j ∈ R}. By definition of the tree decomposition, all the residues in Lc are in {L∪R}. In order to calculate the energies of the internal nodes, the nodes must be traversed in leaf to root order. Since the root has no parent, it has no set L (equivalently, its set L is empty). Its energies are given by
In the last part of the algorithm the nodes of the tree-decomposition are traversed in the root-to-leaves order to assemble the global assignment of rotamers for the cluster being processed. For any node except the root we have a local partial solution that lets us obtain an optimal assignment of rotamers of R for any rotamer assignment of rotamers over L. But by construction L belongs to the parent bag, which means that we can easily retrieve the optimal assignment over all residues in some bag if we know the optimal assignment at the parent node. Thus we have a recursive procedure that gradually extends an assignment for the entire cluster starting from the root node:
For each child c of the root, the optimal assignment of rotamers to the residues in Rc may be made, given the assignment of rotamers of the root, and the minimum energy added to the total:
where the assignments in lc are already known from rroot. The rotamers are assigned and the energies updated for each child of each c, and so on, following from the root to all leaves in a depth-first search.
The actual search for the local solution is done via exhaustive direct enumeration, which is quite affordable if the product of rotamer numbers within the corresponding bag is not very large. The number of possible rotamer assignments over a particular bag we will refer to as local complexity of the node. The sum of the local complexities of all nodes gives the overall computational complexity of the optimization. Typically tree-decompositions of smaller width yield lower complexity. In order to limit the time required by SCWRL4 for a single rotamer assignment we introduced an upper bound for the overall complexity of 108. If the actual complexity exceeds this bound then the optimization is treated as not tractable and SCWRL4 returns to the graph construction step (edge decomposition and DEE) after doubling the value of the edge decomposition threshold. This process continues until a solution is found.
Outputting the results
The optimization resolves both the minimal total energy of the entire model and the corresponding assignment of rotamers. The SCWRL4 executable saves the resolved optimal conformation of the whole protein model into PDB file. The corresponding value of the total energy is printed into the standard output, which can be redirected to a file for further analysis. If the task was set up and solved via the API of the SCWRL4 library then the corresponding workspace with or without modifications can be used in subsequent calculations.
Training and test sets
We constructed a training set of proteins for optimizing the parameters and procedures, and a separate testing set for reporting the accuracy of SCWRL4. Because we wanted to use electron density calculations to estimate the reliability of side-chain coordinates, we started with the list of PDB entries with electron densities available from the Uppsala Electron Density Server47, generally those with deposited structure factors. We removed entries with ligands other than water, so that side chains could be predicted without requiring charges, hydrogen positions, or van der Waals radii of ligands. This set was culled using the PISCES server48,49 at maximum mutual sequence identity 30%, ≤1.8 Å resolution, and maximum R-factor of 20%. Because we planned to optimize the energy function by predicting side chains in the crystal form, we checked whether the CRYST1 records and scale matrices produced viable crystals. We removed some entries that produced extensive clashes of protein atoms when crystal neighbors of the asymmetric unit were constructed (e.g. PDB entry 1RWR). The resulting list of proteins was broken up into the training and testing set, with the training set consisting of monomeric asymmetric units to speed the optimization procedures described below.
For all complete side chains in the resulting protein lists, we calculated the geometric mean of the electron density, as described previously50. In this prior work, low values of mean electron density were correlated with non-rotameric side chains and conformationally disordered side chains. For each residue type, mean electron densities for the training and testing sets were sorted and turned into percentiles, with 0% for the lowest electron density side chain and 100% for the highest.
For both sets, we used the program SIOCS (Heisen and Sheldrick, unpublished) available with SHELX51 to resolve the ambiguity in the flip state of Asn and His χ2 and Gln χ3. SIOCS uses hydrogen bonding and crystal contacts to indicate whether these side chains are correctly placed in crystal structures, or if it is likely that the terminal dihedrals should be flipped by 180°.
In crystal mode, calculations were performed on the asymmetric unit with inclusion of interactions with crystal neighbors. However, accuracy was only assessed on one protein of the asymmetric unit, as provided by PISCES.
Optimization of the energy parameters
The accuracy of SCWRL4 depends on the choice of rotamer library, the non-bonded energy functions and their parameters, the parameters used in the flexible rotamer model (FRM), and several other procedural choices. We first needed to decide on an objective function to be optimized. There are a number of possible choices, including RMSD, percent χ1 correct within some threshold (typically 40° in previous literature), percent χ1+2 correct, and percent of side chains correct (side chains with all χ angles within some threshold). We decided to use the average absolute accuracy. For a side-chain type such as Lys, this is an average of percent χ1, percent χ1+2, percent χ1+2+3, and percent χ1+2+3+4 correct:
where N12 for instance is the number of lysine side chains with both χ1 and χ2 correct within 40°. This value gives added weight to the more reliably determined degrees of freedom closer to the backbone. To obtain an accuracy across all side-chain types, we weight PC for each amino acid type by its frequency:
We have a large number of parameters that can be optimized. First, the χ-angle deviations δi for the sub-rotamers can be set individually for each dihedral degree of freedom. SCWRL4 uses a constant times the standard deviation provided in the rotamer library, where the constant is specific for each degree of freedom for each side-chain type:
We also optimize the “temperature” used in the FRM procedure separately for each amino acid type. For calculation of pairwise rotamer-rotamer energies the temperature is taken as the arithmetic average of the temperatures of the corresponding amino-acid types. The last parameter is the coefficient in front of the rotamer library term which balances the influence of the static frame and the rotamer library in the self-energy of a rotamer. Thus for every amino-acid type we obtain l+2 parameters, where l is the number of χ angles required to specify the conformation of the side-chain of a certain amino-acid type. These parameters form a 78-dimensional space that can be searched to improve the quality of prediction. In addition, we also have the hydrogen bond parameters and the atomic radii that can be optimized to improve the prediction accuracy.
We used a special technique to perform the optimization in the 78-dimensional space of the parameters concerned. The main idea is similar to classical block-coordinate descent methods52, where on each iteration an optimum along one or several axes is resolved. In our case, the search space of any iteration consists of the parameters of a single amino-acid type, while the objective function is maximized within some vicinity of the current values of these parameters. Every iteration updates only the parameters associated with the corresponding amino-acid type while the parameters of the other amino-acid types remain unchanged. In this method, any iteration updates the parameters in a way that increases the value of the objective functions, otherwise keeping the parameters unchanged. Within each iteration, the approximate solution of the underlying optimization task is obtained using a special technique as described in the Supplementary Material. Changing the amino-acid types from one iteration to another will impose a sequence of points in the original 78-dimensional space along which the value of the objective function will gradually increase (or at least will remain unchanged). Iterations are grouped into rounds – a randomly shuffled sequence of 18 iterations, which contains all residue types except ALA and GLY. Our experiments showed that a few rounds are typically enough to find some (not necessarily global) maximum of the objective function.
The key advantage of the protocol is that it lets us utilize specific properties of the internal structure of the rotamer assignment procedure. When the flexible rotamer model is enabled, the most CPU-consuming part of the whole prediction is the calculation of the interaction graph and especially the calculation of the pairwise energies. However for some pair of amino acids, if neither of the rotamers has been changed then the current energy of the pairwise interaction between them is valid and does not need to be recalculated. Conversely, changing the parameters of some amino acid type affects only the rotamers of that type and their interactions with rotamers of any other type. During a single iteration only side chains of one amino-acid type are modified and so most of the interaction graph can be preserved while only a minor part has to be recomputed. Moreover if only the weight of the rotamer library’s energy term is changed, then neither the static frame energies nor the energies of pairwise interactions have to be recomputed. The parameters were optimized such that all side chains were in their crystal environment, interacting with crystal neighbors of the asymmetric unit. This is particularly important for polar side chains with contacts between asymmetric units.
Accuracy vs. accessible surface area and percentile electron density
The smoothed curves in Figure 8 and 9 were calculated using kernel density estimates53 by calculating probability density estimates of correctly and incorrectly predicted side chains as a function of relative accessible surface area (RSA) and percentile of electron density:
where Ai is the RSA of residue i or its electron density percentile, and K is a Gaussian kernel with bandwidth h:
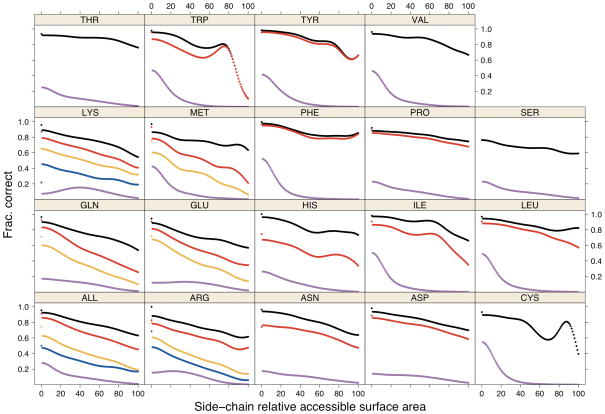
Figure 8. Accuracy vs. relative surface area.
Accuracy of SCWRL4 predictions is shown as a function of side-chain relative accessible surface area, calculated with kernel density estimates (see Methods): χ1 (black), χ1+2 (red), χ1+2+3 (orange), χ1+2+3+4 (blue) within 40°. The data points for 0% RSA were calculated separately from the kernel density estimates. The magenta curves are the probability density estimates of all side chains of each type in the crystal.
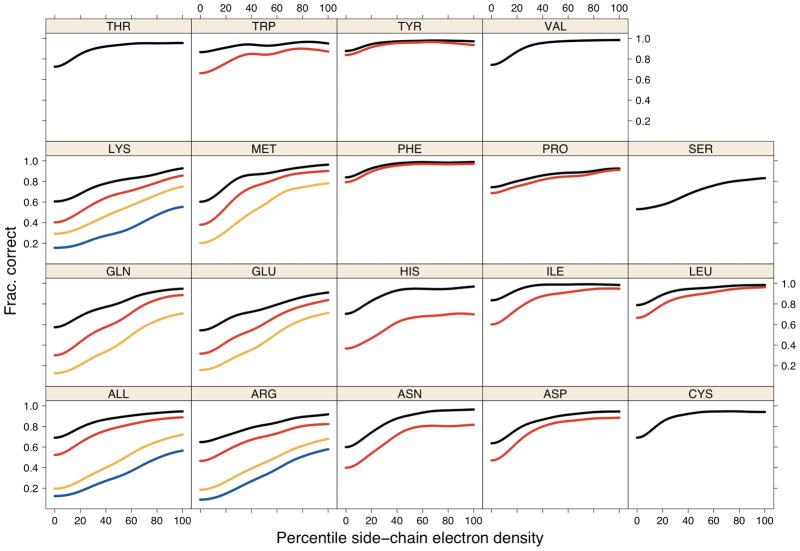
Figure 9. Accuracy vs. percentile of electron density.
Accuracy of SCWRL4 predictions is shown as a function of electron density percentile calculated for each residue type, calculated with kernel density estimates (see Methods). Curves for χ1 (black), χ1+2 (red), χ1+2+3 (orange), χ1+2+3+4 (blue) within 40° are shown.
The prediction rate at A is calculated using Bayes’ rule:
where p(A|corr) and p(A|incorr) are calculated using the expression for f(A) using the correctly predicted and incorrectly predicted side chains respectively. p(corr) and p(incorr) are just the frequencies of correctly and incorrectly predicted side chains overall. Kernel density estimates exhibit unfavorable behavior at boundaries, and so the data were reflected across A=0 to account for this. No correction was made at A=100.
Library
SCWRL4 has been redesigned as a library, so that its optimization engine can be used in other scenarios such as protein design. It utilizes a delayed computation model. At first the model data (referred to as the workspace) is defined by specifying the location of amino-acid residues with appropriate rotamers via calls to functions of the library. After that the calling program uses the SCWRL4 engine in the library to derive the optimal assignment of rotamers. This will request that SCWRL4 calculate all the required energies and perform combinatorial optimization after which for each residue one of its rotamers will be marked as optimal. This information can then be used in other applications. The SCWRL4 library keeps the model alive for further usage even after the optimal assignment has been found. This means that after the optimal rotamers have been resolved, some modifications can be introduced into the model and the optimization requested again. In this case SCWRL4 will recalculate only those energies that need to be modified due to changes in the model, while for energies between persistent objects it will use cached values.
Availability
SCWRL4 is available at http://dunbrack.fccc.edu/scwrl4.
Results
Training and testing sets
We used separate training and testing sets of 100 and 379 proteins respectively to optimize and test various parameters and algorithmic choices for development of SCWRL4. Details of these sets are given in Supplementary Data. The resolution cutoff for both sets was 1.8 Å, and the maximum mutual sequence identity was 30%. All calculations below are performed either on the asymmetric unit or using crystal symmetry, although in each case the accuracy results are compiled on sets consisting of only one chain of each sequence.
Accuracy of SCWRL4
The overall accuracy of SCWRL4 is presented in Table I for those side chains with electron density above the 25th percentile. The accuracy is reported in three different ways. First, for each side-chain type, the conditional accuracy for each dihedral degree of freedom, χi, is reported. This is the percent of χi that is correct, given that the χ angles closer to the backbone, χi−1,…, χ1 are also correct. So for instance, for Met, for those residues with both χ1 and χ2 correct, 77.1% have χ3 correct. The column “ALL” counts only those residue types with that degree of freedom. Second, for each side-chain the absolute accuracy at each degree of freedom is the percentage of all residues of that type such that χi,…, χ1 are all correct. So for instance, for Met, 60.9% of all residues are predicted correctly for χ1,χ2, and χ3. Finally, the average RMSDs are given for each side chain type. For a set of residues of the same type, there are two ways to calculate the RMSD. First, one can calculate the RMSD for each residue, and then average these values. Second, one can do the sum of square distances over all of the atoms of all of the residues, take the mean, and then the square root. The values are not the same. We use the former definition.
Table I.
Accuracy of SCWRL4
| ALL | ARG | ASN | ASP | CYS | GLN | GLU | HIS | ILE | LEU | LYS | MET | PHE | PRO | SER | THR | TRP | TYR | VAL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | 45216 | 2803 | 2238 | 3161 | 805 | 1934 | 3579 | 1202 | 3043 | 5096 | 2996 | 1107 | 2115 | 2489 | 3229 | 2935 | 758 | 1828 | 3898 | |
| Conditional | Avg. | 88.1 | 77.6 | 86.6 | 90.5 | 92.7 | 77.7 | 78.4 | 79.8 | 95.4 | 95.4 | 78.1 | 84.9 | 97.3 | 92.1 | 75.8 | 94.0 | 91.1 | 96.6 | 97.1 |
| Chi_1 | 89.3 | 81.8 | 90.1 | 88.8 | 92.7 | 84.6 | 78.3 | 91.1 | 98.6 | 95.4 | 81.9 | 89.0 | 96.9 | 88.2 | 75.8 | 94.0 | 93.0 | 95.6 | 97.1 | |
| Chi_2 | 89.1 | 86.2 | 83.2 | 92.2 | 79.9 | 81.5 | 68.4 | 92.2 | 95.4 | 85.0 | 88.7 | 97.8 | 96.1 | 89.2 | 97.5 | |||||
| Chi_3 | 73.5 | 66.4 | 68.6 | 75.5 | 79.6 | 77.1 | ||||||||||||||
| Chi_4 | 70.7 | 76.1 | 65.7 | |||||||||||||||||
| Absolute | Avg. | 82.4 | 58.7 | 82.5 | 85.3 | 92.7 | 66.2 | 63.4 | 76.7 | 94.7 | 93.2 | 60.8 | 76.3 | 95.8 | 86.5 | 75.8 | 94.0 | 88.0 | 94.4 | 97.1 |
| Chi_1 | 89.3 | 81.8 | 90.1 | 88.8 | 92.7 | 84.6 | 78.3 | 91.1 | 98.6 | 95.4 | 81.9 | 89.0 | 96.9 | 88.2 | 75.8 | 94.0 | 93.0 | 95.6 | 97.1 | |
| Chi_2 | 79.7 | 70.5 | 74.9 | 81.8 | 67.6 | 63.8 | 62.3 | 90.9 | 91.0 | 69.6 | 79.0 | 94.8 | 84.7 | 83.0 | 93.2 | |||||
| Chi_3 | 50.5 | 46.8 | 46.3 | 48.2 | 55.4 | 60.9 | ||||||||||||||
| Chi_4 | 36.0 | 35.5 | 35.1 | |||||||||||||||||
| RMSD | Avg. | 0.82 | 2.15 | 0.79 | 0.68 | 0.41 | 1.43 | 1.34 | 1.14 | 0.33 | 0.48 | 1.58 | 1.09 | 0.65 | 0.24 | 0.70 | 0.31 | 1.27 | 0.81 | 0.22 |
| Sigma | 1.05 | 1.52 | 0.89 | 0.88 | 0.66 | 1.16 | 1.10 | 1.16 | 0.42 | 0.64 | 1.20 | 0.96 | 0.80 | 0.25 | 0.91 | 0.53 | 1.55 | 1.04 | 0.39 | |
Percent accuracy is given for side chains with electron density from 25th to 100th percentiles. Calculations were performed on the asymmetric units of the 379 PDB testing set.
The numbers most frequently cited for side-chain prediction accuracy are the χ1 and χ1+2 rates over all side-chain types, where χ1+2 is the absolute accuracy at the χ2 degree of freedom in Table I. These values are 89.3% and 79.7% respectively for the 25–100th percentiles of electron density. For all side chains, these values fall to 86.1% and 74.8%. We exclude the bottom 25% because of the inherent uncertainty in these conformations (see below). For 10 of 18 residue types, the χ1 accuracies exceed 90%, and these are predominantly the aliphatic and aromatic residue types. Ser is the most difficult to predict, with an accuracy rate of 75.8%.
In Table II, we show the improvement in prediction accuracy of SCWRL4 over SCWRL3 for each residue type and for the conditional and absolute accuracy measures. The overall improvement in χ1 accuracy is 3.5% (SCWRL4 accuracy - SCWRL3 accuracy on the same test set of 379 proteins). The largest improvements are in Trp, Arg, Gln, Glu, Met, Asp, Asn, and Ser, all of which exceed 6% improvement in average absolute accuracy.
Table II.
Improvement of SCWRL4 over SCWRL3
| ALL | ARG | ASN | ASP | CYS | GLN | GLU | HIS | ILE | LEU | LYS | MET | PHE | PRO | SER | THR | TRP | TYR | VAL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | 45216 | 2803 | 2238 | 3161 | 805 | 1934 | 3579 | 1202 | 3043 | 5096 | 2996 | 1107 | 2115 | 2489 | 3229 | 2935 | 758 | 1828 | 3898 | |
| Conditional | Avg. | 3.9 | 9.0 | 4.7 | 3.9 | 1.9 | 10.0 | 6.4 | 4.8 | 2.5 | 2.0 | 0.8 | 4.4 | 2.5 | 1.6 | 6.2 | 1.9 | 7.7 | 2.6 | 2.0 |
| Chi_1 | 3.5 | 5.3 | 4.9 | 5.1 | 1.9 | 5.3 | 3.5 | 2.1 | 2.1 | 2.4 | 3.1 | 4.3 | 2.1 | 3.3 | 6.2 | 1.9 | 6.5 | 2.7 | 2.0 | |
| Chi_2 | 3.0 | 1.4 | 4.6 | 2.8 | 6.2 | 5.7 | 7.5 | 2.9 | 1.6 | 0.4 | 4.1 | 2.9 | −0.1 | 8.9 | 2.5 | |||||
| Chi_3 | 8.7 | 10.4 | 18.3 | 10.1 | 0.8 | 4.7 | ||||||||||||||
| Chi_4 | 8.7 | 19.0 | −1.0 | |||||||||||||||||
| Absolute | Avg. | 4.8 | 9.0 | 6.4 | 6.1 | 1.9 | 10.5 | 7.2 | 5.1 | 3.5 | 3.1 | 2.6 | 6.9 | 3.5 | 3.2 | 6.2 | 1.9 | 10.0 | 3.8 | 2.0 |
| Chi_1 | 3.5 | 5.3 | 4.9 | 5.1 | 1.9 | 5.3 | 3.5 | 2.1 | 2.1 | 2.4 | 3.1 | 4.3 | 2.1 | 3.3 | 6.2 | 1.9 | 6.5 | 2.7 | 2.0 | |
| Chi_2 | 5.7 | 5.6 | 8.0 | 7.0 | 9.2 | 7.1 | 8.1 | 4.8 | 3.8 | 2.9 | 7.3 | 4.9 | 3.1 | 13.5 | 5.0 | |||||
| Chi_3 | 9.7 | 10.5 | 17.0 | 11.1 | 2.9 | 9.0 | ||||||||||||||
| Chi_4 | 7.9 | 14.8 | 1.4 | |||||||||||||||||
Percent accuracy improvement of SCWRL4 over SCWRL3 is given for side chains with electron density from 25th to 100th percentiles. Calculations were performed on the asymmetric units of the 379 PDB testing set.
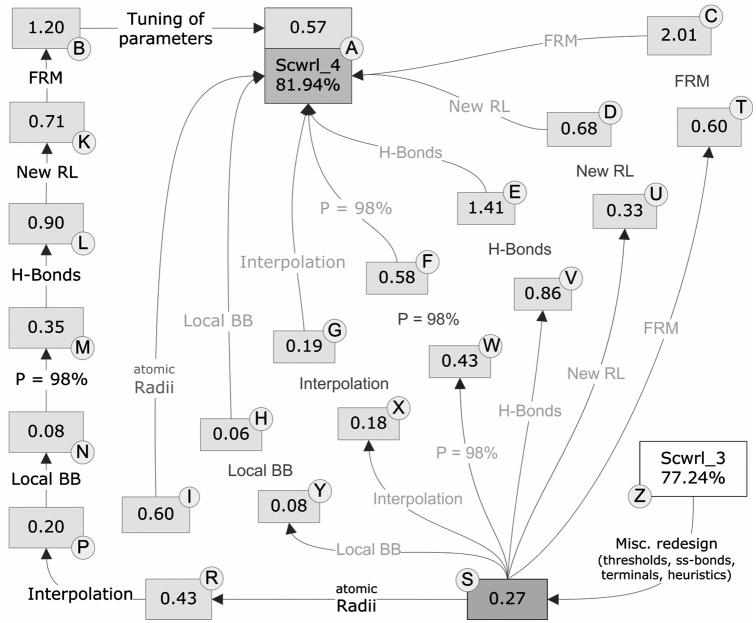
The improvement in accuracy in SCWRL4 over SCWRL3 was achieved through a number of different changes in the sampling space, the energy function, and the algorithm. Each of these feature changes was chosen and/or optimized on the basis of improvement in the training set of 100 proteins. In Figure 6, we show the effects of each change added to SCWRL3 (boxes R through T) or each change removed from the final SCWRL4 protocol (boxes A through I). The figure demonstrates that the effect of each feature is context-dependent; that is, the effect is different when added to SCWRL3, which contains none of the new features vs. when it is removed from the final SCWRL4, which contains all of the new features. The directed graph leading from SCWRL3 to SCWRL4 along the outside of the figure shows the improvements as each feature is added consecutively. The most important changes include the flexible rotamer model, the new rotamer library, the addition of a hydrogen bonding function, changes in the atomic radii used, and using a larger percentage of the rotamer library (the top 98% of rotamer density, instead of 90% as used in SCWRL3). The rigid rotamer model (RRM, box C) is 2.01% less accurate in average absolute accuracy than the full flexible rotamer model (FRM, box A). The decrease in χ1 and χ1+2 accuracies are 1.4% and 2.8% respectively.
Figure 6. Effect of SCWRL4 features on differences in SCWRL3 and SCWRL4 accuracy.
The accuracy shown is average absolute accuracy of the training set, which covers all side-chain dihedral angles (see text). “Atomic radii” = use of optimized radii; “Interpolation” = interpolation of rotamer library probabilities and dihedral angles; “Local BB” = adding interaction between side chain and atoms N,HN of residue i−1 and C,O of residue i+1, previously neglected in SCWRL3; “P=98%” = reading in top 98% of probability from rotamers sorted in descending order of frequency (90% in SCWRL3); “H-bonds” = new hydrogen bond potential; “New RL” = new rotamer library; “FRM” = Flexible rotamer model; “Tuning of parameters” = tuning of FRM parameters and rotamer library weights.
Prediction of side-chain conformation in crystals
We enabled consideration of crystal symmetry in side-chain conformation prediction in SCWRL4. This is accomplished by determining the interaction graph in the context of neighboring chains to the asymmetric unit within the crystal. Thus, a residue in the graph may have a neighbor on the other side of the protein, if that residue makes contact with that residue in a crystal neighbor. Crystals were built and neighbors determined as described in previous work44. It is of interest to determine the effect on prediction accuracy when crystal symmetry is taken into account. It should be noted that this is a bona fide prediction within the crystal, since the side chains in the neighboring asymmetric units have the same conformations as the asymmetric unit whose structure is being predicted.
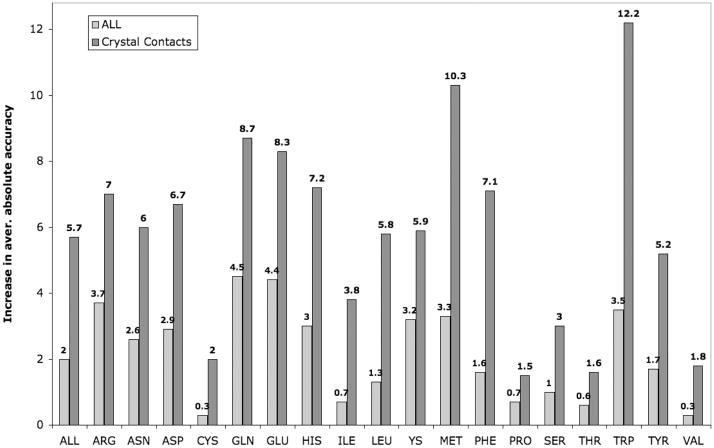
In Figure 7, we show the improvement in accuracy for all side chains and for those in crystal contacts when the crystal symmetry feature is enabled. The accuracy values shown are average absolute accuracy, which are averages of the χ1, χ2, χ3, and χ4 absolute accuracies shown in Table I. Improvement occurs for all side-chain types. Among all side chains, not just those in crystal contacts, the effect is strongest for those most likely to be on the surface, in particular the longer side chains, Arg, Lys, Glu, and Gln. However, when other side chain types are in crystal interfaces, their accuracy is also strongly affected by the presence of the crystal neighbors. This is especially true for Trp and Met. The χ1 and χ1+2 accuracy in the crystal for side chains with electron densities in the 25–100% percentiles are 90.9% and 82.6% respectively. For all side chains, the values are 87.4% and 77.1%
Figure 7. Improvement in accuracy due to inclusion of crystal neighbors.
The accuracy figures shown reflect the differences in average absolute accuracy for the testing set as described in the text.
Prediction of side-chain conformation vs. accessible surface area and electron density
Exposed side chains have fewer steric constraints and are more difficult to predict accurately. We have calculated the accuracy of predictions (within 40°) as a function of the relative surface accessibility (RSA) of side chains calculated with the program naccess54. Both the predictions and the surface area calculations were performed in the crystal environment, so that side chains in protein interfaces in the crystal are considered buried. The results are shown in Figure 8. The accuracy vs. surface area was calculated using kernel density estimates as described in the Methods section. In Figure 8, the probability density of each residue type as a function of accessible surface area is shown in magenta (multiplied by 20). These curves show that in the crystal, most residue types are predominantly buried (RSA<40%) with maxima at 0% RSA. The only exceptions are Lys, Arg, and Glu with density modes at 30–40% exposure.
The frequency of accurate predictions is shown for each side-chain type for all side chains with RSA>0%: χ1 (black), χ1+2 (red), χ1+2+3 (orange), χ1+2+3+4 (blue). As expected, less accessible side chains are predicted more accurately than accessible side chains. Note that at high RSA, the estimates can be quite noisy due to very small counts, especially for Cys and Trp. Separate points are plotted for those side chains with 0% RSA (using the same color scheme), calculated separately from the kernel density estimates shown in the curves, For completely buried side chains in the crystal, 96.0% of χ1 and 91.5% of χ1+2 dihedrals are correctly predicted. All side-chain types are predicted with greater than 95% accuracy for χ1 except Cys (94.4%), Pro (91.9%), and Ser (79.1%).
We have previously shown that rotameric side chains have higher electron density than non-rotameric side chains, and that non-rotameric side chains are likely to be disordered in a manner similar to side chains that are annotated in PDB files as existing in more than one χ1 rotamer in the crystal50. Since SCWRL4 predicts only one conformation per side chain, it seems likely that side chains with lower electron density should be harder to predict, since they may be placed in only a portion of the observable electron density. In Figure 9, we show prediction accuracy as a function of the percentile of electron density of the entire side chain, using the same color scheme as that in Figure 8. The curves are also calculated with kernel density estimates. Low percentiles correspond to low electron density, disordered side chains, and high percentiles correspond to high electron density, well-ordered side chains. Some low-density side chains may be incorrectly placed in the density.
For all side-chain types and all degrees of freedom, the accuracy rises with increasing electron density. For some degrees of freedom, this is only true at low electron density, and the accuracy plateaus above the 30th percentile. But for the longer side chains, the increase in accuracy extends from 0 to 100%. For all side chains, the χ1 accuracy increases from 69.0% to 94.6% at 0th and 100th percentiles of electron density. For χ1+2, the equivalent numbers are 52.4% and 82.3%. At the highest electron densities, the χ1+2+3 and χ1+2+3+4 accuracies are 71.9% and 56.5%.
CPU time
In Table III, we show a comparison of the CPU time for the testing set of 379 proteins for SCWRL3 and SCWRL4, for both the rigid rotamer model (RRM) and the flexible rotamer model (FRM) and for the asymmetric units and crystals. The mean, median, and maximum CPU times over the testing set reveal that the different calculations have different properties. SCWRL3 is very fast on most proteins, but on a small number of structures takes exceedingly long times. On two ASUs, SCWRL3 failed to finish and on one protein took 1409 seconds. SCWRL4 with the RRM model takes slightly longer than SCWRL3 with median times of 1.51 and 1.27 seconds respectively. This is due to slightly longer times required for the more complicated energy function and denser and larger graphs that result. However, the maximum time for the SCWRL4 RRM is 72 seconds and the mean time only 4 seconds.
Table III.
CPU Time Comparison of SCWRL3 and SCWRL4
| Program | Model | Target | Mean (sec) | Median (sec) | Max (sec) |
|---|---|---|---|---|---|
| SCWRL3 | RRM | ASU | 8.03** | 1.27 | 1409** |
| SCWRL4 | RRM | ASU | 4.17 | 1.51 | 72 |
| Crystal | 11.31 | 4.56 | 158 | ||
| FRM | ASU | 12.15 | 7.94 | 99 | |
| Crystal | 20.66 | 14.55 | 98 |
Test set of 379 proteins. RRM=rigid rotamer model. FRM=flexible rotamer model.
ASU=asymmetric unit. Crystal=including crystal symmetry.
Calculations were performed on a machine running Windows XP with an AMD Opteron
For SCWRL3, two PDB entries did not finish after several hours and are excluded from mean and maximum.
For the FRM model, SCWRL4 takes a median of 7.9 seconds, a mean of 12.2 seconds, and a maximum of 98 seconds. Thus the median time is about 6.3 times slower for the SCWRL4 FRM calculation compared to SCWRL3, and the mean time is 1.5 times slower. SCWRL4 is also able to calculate the conformations of proteins in the crystal environment, taking account of crystal symmetry. Calculation with crystal symmetry takes 1.7–1.8 times that of the ASU for the FRM model of SCWRL4. The effect on the RRM model is somewhat larger.
The calculations were performed on a machine with an AMD Athlon 64 X2 Dual Core Processor 4400+ at 2.21GHz, with 3.25 GB of RAM, and running by 32-bit Microsoft Windows XP Professional Service Pack 3.
Discussion
While the process of sequence-structure alignment is well represented by many available web servers and downloadable programs, relatively few programs exist for producing three-dimensional coordinates from target-template alignments55. We have previously developed the MolIDE program that takes an input target sequence and produces alignments of this target sequence to available homologous proteins of known structure56,57. In a graphical environment, it is then possible to use the SCWRL3 program to produce a model of the target sequence retaining the input sequence numbering. We intend for SCWRL4 to perform the same function within the existing MolIDE, but with higher accuracy than SCWRL3. It may also be used in existing web servers that perform searches for remote homologues such as FFAS0334. Using the rigid rotamer model, SCWRL4 is about the same speed as SCWRL3 in most cases but is able to complete all test cases in a reasonable time, while SCWRL3 sometimes does not converge. With the flexible rotamer model, the median value of SCWRL4 is slower by a factor of about 6, but with convergence in all cases tested. SCWRL4 has a similar ease of use, and therefore will function in similar environments as SCWRL3.
There are a number of potential sources of disagreement between predicted side-chain conformations based on native backbones and experimental structures. In this paper, we have explored several of these. The first and most obvious is the scoring function that must realistically represent the physical forces that position side chains in proteins. We have improved the rotamer library that SCWRL depends on, especially in those degrees of freedom that are not strictly rotameric. These include the amide and carboxylate moieties of Asn, Asp, Glu, and Gln, and the aromatic rings of Phe, Tyr, His, and Trp (Shapovalov and Dunbrack, in preparation). Second, we have also explored issues of sampling by including conformations near the rotameric positions using the flexible rotamer model approach suggested Mendes et al.32 Each of these issues can be explored further, for instance by including solvation energy terms as well as continuous dihedral angle minimization, as performed in Rosetta.19 For the latter, the new rotamer libraries may afford an opportunity by providing continuous energy functions as a function of the side-chain χ dihedral angles, independent of rotamer definitions.
We have explored two other aspects of side-chain prediction that affect the overall accuracy. The first of these comprise the interactions of side chains within the crystal. For the first time, we have developed a side-chain prediction program that can account for arbitrary symmetry, that is, predicting the conformation of all side chains within the crystal. We find increases in accuracy of almost all side-chain types, especially for those most likely to be in crystal contacts. Improvement in the crystal is interesting for two reasons. First, if side-chain prediction is used for molecular replacement or structure refinement, then the ability to consider the crystal symmetry will be very useful. Second, this is some indication that the prediction of side-chain conformation in protein-protein interfaces with SCWRL4 is likely to be significantly better than for side chains on the surface but not in protein interfaces.
The second important issue is the apparent disorder of many side chains in crystals that we have studied previously50. We have shown that prediction accuracy monotonically improves with increasing electron density, such that side chains that are clearly positioned in one conformation in the crystal are the easiest to predict. Similarly, side chains that are buried within the crystal (either within single proteins or within asymmetric-unit or crystal interfaces) are much better predicted than those that are exposed to the solvent.
In this paper we have explored the properties of SCWRL4 and we hope that users will find it beneficial for predicting the structures of proteins.
Supplementary Material
Acknowledgments
We thank Adrian A. Canutescu and Nikolay A. Kudryashov for their advice during the development of SCWRL4. This work was funded by NIH grants R01 HG02302, R01 GM84453, and P20 GM76222.
References
- 1.Veenstra DL, Kollman PA. Modeling protein stability: a theoretical analysis of the stability of T4 lysozyme mutants. Protein Eng. 1997;10(7):789–807. doi: 10.1093/protein/10.7.789. [DOI] [PubMed] [Google Scholar]
- 2.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 3.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65(3):538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 4.Leach AR. Ligand docking to proteins with discrete side-chain flexibility. J Mol Biol. 1994;235(1):345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 5.Rohl CA, Strauss CE, Chivian D, Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55(3):656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 6.Jones DT. De novo protein design using pairwise potentials and a genetic algorithm. Prot Eng. 1994;3:567–574. doi: 10.1002/pro.5560030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahiyat BI, Mayo SL. Protein design automation. Prot Science. 1996;5:895–903. doi: 10.1002/pro.5560050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302(5649):1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 9.Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12(4):431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 10.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40(3):389–408. [PubMed] [Google Scholar]
- 11.Dunbrack RL, Jr, Karplus M. Backbone-dependent rotamer library for proteins. Application to side- chain prediction. J Mol Biol. 1993;230(2):543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 12.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6(8):1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Maeyer M, Desmet J, Lasters I. All in one: a highly detailed rotamer library improves both accuracy and speed in the modelling of sidechains by dead-end elimination. Fold Des. 1997;2(1):53–66. doi: 10.1016/s1359-0278(97)00006-0. [DOI] [PubMed] [Google Scholar]
- 14.Shetty RP, De Bakker PI, depristo MA, Blundell TL. Advantages of fine-grained side chain conformer libraries. Protein Eng. 2003;16(12):963–969. doi: 10.1093/protein/gzg143. [DOI] [PubMed] [Google Scholar]
- 15.Peterson RW, Dutton PL, Wand AJ. Improved side-chain prediction accuracy using an ab initio potential energy function and a very large rotamer library. Protein Sci. 2004;13(3):735–751. doi: 10.1110/ps.03250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Z, Honig B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol. 2001;311(2):421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 17.Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267(5):1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 18.Liang S, Grishin NV. Side-chain modeling with an optimized scoring function. Protein Sci. 2002;11(2):322–331. doi: 10.1110/ps.24902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Dousis AD, Ma J. OPUS-Rota: a fast and accurate method for side-chain modeling. Protein Sci. 2008;17(9):1576–1585. doi: 10.1110/ps.035022.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson RM, Gabb HA, Sternberg MJ. Rapid refinement of protein interfaces incorporating solvation: application to the docking problem. J Mol Biol. 1998;276(1):265–285. doi: 10.1006/jmbi.1997.1519. [DOI] [PubMed] [Google Scholar]
- 22.Mendes J, Baptista AM, Carrondo MA, Soares CM. Implicit solvation in the self-consistent mean field theory method: side-chain modeling and prediction of folding free energies of protein mutants. J Comp Aided Mol Design. 2001;15:721–740. doi: 10.1023/a:1012279810260. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320(3):597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 24.Camacho CJ. Modeling side-chains using molecular dynamics improve recognition of binding region in CAPRI targets. Proteins. 2005;60(2):245–251. doi: 10.1002/prot.20565. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Evaluation of protein models by atomic solvation preference. J Mol Biol. 1992;225(1):93–105. doi: 10.1016/0022-2836(92)91028-n. [DOI] [PubMed] [Google Scholar]
- 26.Desmet J, De Maeyer M, Hazes B, Lasters I. The dead-end elimination theorem and its use in protein sidechain positioning. Nature. 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein RF. Efficient rotamer elimination applied to protein side-chains and related spin glasses. Biophys J. 1994;66(5):1335–1340. doi: 10.1016/S0006-3495(94)80923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehl P, Delarue M. Application of a self-consistent mean field theory to predict protein side-chains conformation and estimate their conformational entropy. J Mol Biol. 1994;239(2):249–275. doi: 10.1006/jmbi.1994.1366. [DOI] [PubMed] [Google Scholar]
- 29.Kingsford CL, Chazelle B, Singh M. Solving and analyzing side-chain positioning problems using linear and integer programming. Bioinformatics. 2005;21(7):1028–1036. doi: 10.1093/bioinformatics/bti144. [DOI] [PubMed] [Google Scholar]
- 30.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12(9):2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J. Rapid protein side-chain packing via tree decomposition. 9th Annual International Conference on Research in Computational Molecular Biology (RECOMB); 2005. pp. 423–439. [Google Scholar]
- 32.Mendes J, Baptista AM, Carrondo MA, Soares CM. Improved modeling of side-chains in proteins with rotamer-based methods: a flexible rotamer model. Proteins. 1999;37(4):530–543. doi: 10.1002/(sici)1097-0134(19991201)37:4<530::aid-prot4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson MP, Kaminski GA, Friesner RA, Rapp CS. Force Field Validation Using Protein Side Chain Prediction. Journal of Physical Chemistry B. 2002;106:11673–11680. [Google Scholar]
- 34.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33(Web Server issue):W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Jiang L, Gao Y, Liang S, Chen H, Han Y, Lai L. Beyond the rotamer library: genetic algorithm combined with the disturbing mutation process for upbuilding protein side-chains. Proteins. 2003;50(1):49–62. doi: 10.1002/prot.10253. [DOI] [PubMed] [Google Scholar]
- 36.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J Mol Biol. 2003;326(4):1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 37.Klosowski JT, Held M, Mitchell JB, Sowizral H, Zikan K. Efficient collision detection using bounding volume hierarchies of k-dops. IEEE Trans Visualization Comp Graphics. 1998;4:21–36. [Google Scholar]
- 38.Dunbrack RL, Jr, Karplus M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nature Struct Biol. 1994;1:334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 39.Kossiakoff AA, Shpungin J, Sintchak MD. Hydroxyl hydrogen conformations in trypsin determined by the neutron diffraction solvent difference map method: relative importance of steric and electrostatic factors in defining hydrogen-bonding geometries. Proc Natl Acad Sci U S A. 1990;87(12):4468–4472. doi: 10.1073/pnas.87.12.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leaver-Fay A, Kuhlman B, Snoeyink J. Algorithms in Bioinformatics. Volume 3692, Lecture Notes in Computer Science. Berlin: Springer; 2005. Rotamer-pair energy calculations using a trie structure; pp. 389–400. [Google Scholar]
- 41.Parsons J, Holmes JB, Rojas JM, Tsai J, Strauss CE. Practical conversion from torsion space to Cartesian space for in silico protein synthesis. J Comput Chem. 2005;26(10):1063–1068. doi: 10.1002/jcc.20237. [DOI] [PubMed] [Google Scholar]
- 42.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 43.Kirkwood JG. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 44.Xu Q, Canutescu AA, Wang G, Shapovalov M, Obradovic Z, Dunbrack RL., Jr Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol. 2008;381(2):487–507. doi: 10.1016/j.jmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller GL, Teng S, Thurston W, Vavasis SA. Separators for sphere-packings and nearest-neighbor graphs. J Assoc Comp Machinery. 1997;44:1–29. [Google Scholar]
- 46.Arnborg S, Corneil DG, Proskurowski A. Complexity of finding embedding in a k-tree. SIAM J Algebraic and Discrete Methods. 1987;8:277–284. [Google Scholar]
- 47.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 48.Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19(12):1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Dunbrack RL., Jr PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33(Web Server issue):W94–98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapovalov MV, Dunbrack RL., Jr Statistical and conformational analysis of the electron density of protein side chains. Proteins. 2007;66(2):279–303. doi: 10.1002/prot.21150. [DOI] [PubMed] [Google Scholar]
- 51.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Briggs WL, Henson VE, mccormick SF. A Multigrid Tutorial. Philadelphia: SIAM; 2000. [Google Scholar]
- 53.Silverman BW. Density Estimation for Statistics and Data Analysis. New York: Chapman & Hall; 1986. p. 175. [Google Scholar]
- 54.Hubbard SJ, Thornton JM. NACCESS. London: Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 55.Wang G, Jin Y, Dunbrack RL., Jr Assessment of fold recognition predictions in CASP6. Proteins. 2005 doi: 10.1002/prot.20721. [DOI] [PubMed] [Google Scholar]
- 56.Canutescu AA, Dunbrack RL., Jr Mollde: a homology modeling framework you can click with. Bioinformatics. 2005;21(12):2914–2916. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, Canutescu AA, Dunbrack RLJ. SCWRL and molide: Programs for protein side-chain prediction and homology modeling. Nature Protocols. 2008 doi: 10.1038/nprot.2008.184. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.