Abstract
The receptor kinase EFR of Arabidopsis thaliana detects the microbe-associated molecular pattern elf18, a peptide that represents the N terminus of bacterial elongation factor Tu. Here, we tested subdomains of EFR for their importance in receptor function. Transient expression of tagged versions of EFR and EFR lacking its cytoplasmic domain in leaves of Nicotiana benthamiana resulted in functional binding sites for elf18. No binding of ligand was found with the ectodomain lacking the transmembrane domain or with EFR lacking the first 5 of its 21 leucine-rich repeats (LRRs). EFR is structurally related to the receptor kinase flagellin-sensing 2 (FLS2) that detects bacterial flagellin. Chimeric receptors with subdomains of FLS2 substituting for corresponding parts of EFR were tested for functionality in ligand binding and receptor activation assays. Substituting the transmembrane domain and the cytoplasmic domain resulted in a fully functional receptor for elf18. Replacing also the outer juxtamembrane domain with that of FLS2 led to a receptor with full affinity for elf18 but with a lower efficiency in response activation. Extending the substitution to encompass also the last two of the LRRs abolished binding and receptor activation. Substitution of the N terminus by the first six LRRs from FLS2 reduced binding affinity and strongly affected receptor activation. In summary, chimeric receptors allow mapping of subdomains relevant for ligand binding and receptor activation. The results also show that modular assembly of chimeras from different receptors can be used to form functional receptors.
Keywords: Innate Immunity, Membrane Proteins, Reactive Oxygen Species (ROS), Receptor Serine Threonine Kinase, Receptors, Chimeric Receptors, Leucine-rich Repeats
Introduction
Receptor-like kinases (RLKs)2 form the biggest family of surface receptors in higher plants. Based on genomic sequence information, more than 600 genes in Arabidopsis thaliana and more than 1,000 genes in rice are predicted to encode RLKs (1, 2). RLKs have a common molecular structure consisting of a C-terminal cytoplasmic Ser/Thr protein kinase domain connected by a single-pass transmembrane motif to different types of N-terminal ectodomains facing extracytoplasmic compartments. Members of the RLK family play fundamental roles for cellular response programs regulating cell growth, morphogenesis, fertilization, abscission, plant defense, and interaction with symbionts (3–7). Nevertheless, the attribute “receptor-like” holds for most of the RLKs that still remain orphan with respect to their biological functions and their potential regulatory ligands. Concerning functional aspects of ligand interaction and transmembrane activation, the receptor kinase BRI1, functioning as the receptor for the growth hormone brassinolide, has been most thoroughly studied (8–10). Thereby, the ligand interaction site could be localized to a small subdomain within the ectodomain of BRI1 (11), and activation was found to involve a ligand-dependent complex formation with a second receptor kinase termed BAK1 for BRI1-associated receptor kinase 1 (9, 10).
Several RLKs have been assigned roles in recognition of pathogen attack. Examples for RLKs that function as pattern recognition receptors for identified ligands include XA21 from rice (12), FLS2 (13), EFR (14), and CERK1 (15) detecting microbe-associated molecular patterns (MAMPs), and AtPEPR1 (16) detecting wound-related, endogenous danger signals. Upon activation with their respective ligands, FLS2, EFR, and AtPEPR1 seem to form heteromeric complexes with the co-receptor BAK1. Thus, intriguingly, BAK1 appears to function in the transmembrane signaling of plant RLKs with very different signal output programs (17–20).
The receptor kinase EFR from A. thaliana responds to bacterial elongation factor Tu (EF-Tu) with the induction of defense and increased resistance (14, 21). Mutant plants lacking this perception system show increased susceptibility to infection by Agrobacterium tumefaciens. The epitope that acts as MAMP was identified as the acetylated N terminus of EF-Tu. Synthetic peptides like elf18 and elf26 which represent the N terminus of EF-Tu with at least 18 amino acids are fully active as MAMPs and stimulate responses at subnanomolar concentrations. A radiolabeled derivative of elf26 was used to demonstrate specific, high affinity binding sites in tissues expressing EFR. In affinity cross-linking experiments this radioligand specifically labeled the EFR protein, providing evidence that EFR is the bona fide receptor and interacts directly with the elf ligand (14, 21).
EFR is a RLK with 21 leucine-rich repeats (LRRs) arranged in tandem in its ectodomain. Here, we studied the function of this ectodomain as the presumptive high affinity binding site for the ligand peptide elf18. Initial attempts to map the interaction site on EFR using a deletional approach indicated that functional binding depends on an intact ectodomain, including even the transmembrane domain of EFR. Loss of functionality could also result from general structural changes that affect the formation or exposure of the binding site in an indirect way. Thus, we tried to complement and substitute parts of the EFR receptor with parts of the structurally and functionally related pattern recognition receptor FLS2. Although some of these replacements, such as swapping the kinase domains, indeed resulted in fully functional EF-Tu receptors, others showed partial functionality as specific binding sites for the elf ligand. Apart from providing an important advancement for the characterization of the interaction sites of EFR with its ligand, this study should also serve as a basis for employing the modular properties of RLKs to study the specificity and molecular functioning of this eminent class of plant receptors.
EXPERIMENTAL PROCEDURES
Cloning and Chimera Construction
All PCRs were performed with the High Fidelity PCR Enzyme Mix from MBI Fermentas. For truncated EFR constructs, templates were amplified with the EFR forward primer (atgaagctgtccttttcacttg) and designed reverse primers for E-TM (ccaacacagagaagccacaattat) and E-oJM (gacaactttctttctaactgacag). PCR products were then directionally cloned to pENT/TEV/D-Topo vector (Gateway vector; Invitrogen) and further recombined to pK7FWG2.0 (Plant Systems Biology, VIB, University of Gent), an in planta expression vector with a CaMV35S promoter and a C-terminal green fluorescent protein (GFP) fusion tag.
For the construction of the internal deletion in EFRΔ5 and the chimeric forms of EFR/FLS2, a PCR-based technique was used with fusions of two single PCR products. The two parts were synthesized separately using primers with 21 nucleotide overhangs corresponding to the respective fusion partners used in the subsequent step of the construction process. Sequences of the 42-nucleotide primers were gtgatctcaccctccattggtaatTTGACTCAAATGGTGTTTTTCCAA for EFRΔ5, cctctgtcagttagaaagaaaGTCATCCTGATTATTCTTGGA for E-oJM/F, agggtgccaacaacaggagtgtttAAAAACATCAACGCCTCTGATCTA for E-21/F, atggaatttctctttatgcaaggaAACAATCTCTCGGGTCATATACCA for E-19/F, and acggatttagacctgagtggtAATAGTTTTTCAGGTGGTTTTCCT for F-6/E. After gel purification the fragments of the first PCR were then put together in an additional PCR using the appropriate forward and reverse primers (FLS2_fw, atgaagttactctcaaagacctt; FLS2_rev, aacttctcgatcctcgttacg; or EFR_fw, atgaagctgtccttttcacttg; EFR_rev, catagtatgcatgtccgtatttaacatc). Chimeras were cloned to pK7FWG2.0 in-frame with a C-terminal GFP fusion tag via Gateway as described above.
Transient Expression in N. benthamiana
A. tumefaciens (strain GV3101) harboring the gene constructs to be expressed were grown for 48 h in LB medium, collected by centrifugation, and transferred to induction medium (22) with 150 μm acetosyringone and 10 mm MgCl2 at an A600 of 0.1. After further incubation at room temperature for 2–3 h, bacteria were pressure-infiltrated into leaves of 4–5-week-old Nicotiana benthamiana plants grown in the greenhouse (16-h day at 22 °C/8-h night at 18 °C). The next day (24–36 h after infiltration), leaves were cut in pieces of ∼3 × 3 mm and floated on water in Petri dishes overnight at room temperature. Leaf pieces (∼44–48 h after infiltration) were then used to study ethylene biosynthesis and oxidative burst as follows. Ethylene biosynthesis was assayed by placing leaf samples (four pieces with ∼25-mg fresh weight) in 6-ml tubes with 500 μl of water or water containing the appropriate concentration of elf18. Tubes were sealed with rubber caps, and ethylene accumulating in the headspace within 3 h of incubation was determined by gas chromatography.
For oxidative burst, leaf pieces (one piece/well) were placed in wells of 96-well plates containing 100 μl of water, ∼10 ng/ml peroxidase (horseradish peroxidase; Applichem), 20 μm luminol, and elf18 at the concentration to be tested. Light emission was measured as relative light units in a 96-well luminometer (Mithras LB 940; Berthold Technologies).
Binding Assays
Binding assays with N. benthamiana plant extracts were performed as described (14). The binding buffer contained 25 mm MES (pH 6.0), 2 mm MgCl2, 5 mm KI, 2 mm KCl and 1 mm dithiothreitol. Radiolabeled peptide elf26-125I (2,000 Ci/mmol specific activity) was used at a concentration of 0.3 nm in standard assays. After incubation at 4 °C for 20 min, unbound ligands were removed by washing on paper filters with 10 ml of binding buffer.
Functionality Assay in A. thaliana Protoplasts
Transient expression in leaf mesophyll protoplasts of efr × fls2 mutant plants (23) was performed as described (24). Aliquots of 80,000 protoplasts were co-transformed with 5 μg of plasmid DNA encoding firefly luciferase under the FRK1 promoter (25) and 20 μg of plasmid DNA encoding EFR or the chimeric receptors. The protoplasts were resuspended in 800 μl of W5 solution with 200 μm luciferin (d-luciferin, firefly, PJK GmbH, Kleinblitterrdorf, Germany) and distributed in aliquots of 100 μl with 10,000 protoplasts/well in a 96-well plate. After a 16-h incubation the cells were treated with elf18. Luminescence of protoplast samples was quantified in vivo using by a 96-well luminometer (Mithras LB 940).
Reproducibility
The figures show representative results obtained in one of several independent repetitions. Within a single set of experiments, little variation in responsiveness to elf18 was observed for oxidative burst in N. benthamiana leaves and for induction of luciferase in A. thaliana protoplasts. In independent transformation experiments performed with different batches of plant material, the maximal responses varied up to 3-fold whereas the relative sensitivity to elf18 remained constant.
RESULTS
N- and C-terminal Truncations of EFR
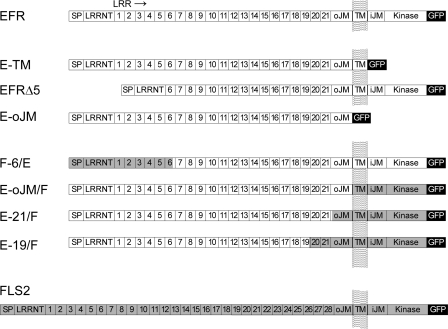
As a first step in analyzing EFR, we wanted to test whether the LRR ectodomain is responsible for specific ligand binding. To this end, we generated gene constructs encoding N- and C-terminally truncated variants of EFR (Fig. 1). All constructs were under control of the CaMV35S promoter and encoded proteins fused to a GFP tag at their C-terminal ends. To test for specific binding to the LRR ectodomain we constructed a gene encoding the ectodomain with the transmembrane domain (E-TM) and one that encoded the ectodomain alone (E-oJM). To build an N-terminally truncated EFR, the first 5 of the 21 LRRs were removed, leaving the N terminus, conserved in many LRR-RLKs, and the signal peptide for protein export intact (EFRΔ5).
FIGURE 1.
Schematic representation of receptor constructs used in this work. EFR (open bars, At5g2480) and FLS2 (gray bars, AT5G46330) encode receptor kinases with a signal peptide (SP), an N terminus conserved in LRR receptors (LRRNT; LRRNT_2 pfam08263), a LRR-domain with 21 repeats in EFR or 28 repeats in FLS2, respectively, an outer juxtamembrane domain (oJM), a transmembrane domain (TM), an inner juxtamembrane domain (iJM) and a serine/threonine protein kinase domain (Kinase, S_TKc). All constructs were C-terminally fused in-frame to a GFP tag.
N. benthamiana lacks an endogenous perception system for EF-Tu-derived peptides like elf18 and is highly suited as experimental system for transient, heterologous expression of fully functional EFR (14). Thus, we used N. benthamiana leaves 2–3 days after infiltration with A. tumefaciens carrying the constructs to be tested. These leaves were assayed for expression of the GFP tag by fluorescence microscopy and Western blotting, for accumulation of specific binding sites for radiolabeled elf peptide, and for functionality as receptor by monitoring elf18-dependent induction of ethylene biosynthesis and oxidative burst.
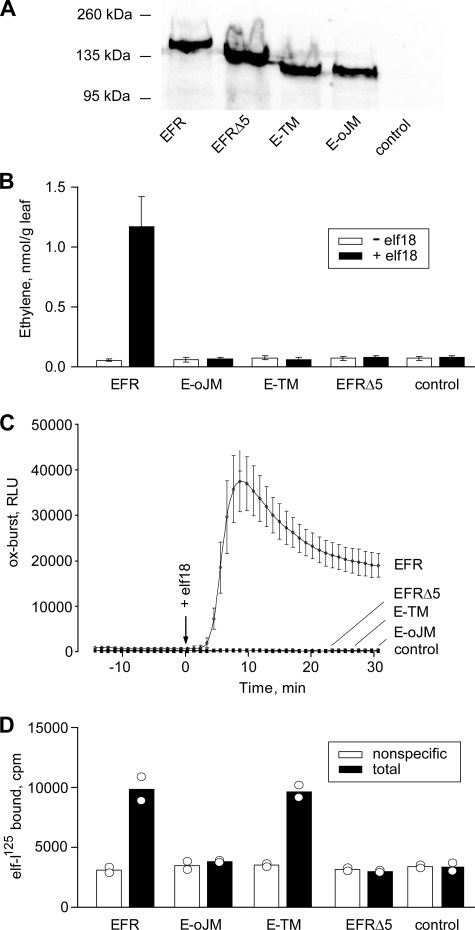
All constructs led to accumulation of similar levels of tagged proteins (Fig. 2A and supplemental Fig. 1). After separation by SDS-PAGE and analysis by Western blotting, these proteins migrated with the sizes expected for the tagged and glycosylated forms of the corresponding proteins (Fig. 2A).
FIGURE 2.
Functionality of intact and truncated forms of EFR after transient expression in leaves of N. benthamiana. A, Western blot with extracts from leaves expressing the different forms of EFR stained with anti-GFP antibodies. B, ethylene biosynthesis in transformed leaf pieces treated for 3 h in the absence or presence of 100 nm elf18. Values show mean ± S.D. (error bars) of n = 3 replicates. C, oxidative burst in leaf pieces of transformed plants treated with 100 nm elf18. Data show mean ± S.D. in Luminol-dependent light emission (RLU, relative light units) of n = 4 replicates. D, binding of elf-125I in the presence (nonspecific binding) and absence (total binding) of 1 μm unlabeled elf18. Measurements were done in duplicate with bars indicating the mean of the two values.
Apart from full-length EFR used as a positive control, none of the truncated EFRs conferred responsiveness to elf18 in transgenic leaf tissue of N. benthamiana when tested for induction of oxidative burst or ethylene production (Fig. 2, B and C). Thus, full functionality of EFR as a receptor depends on the first five LRRs as well as on the cytoplasmic domain of the protein. In a second step, the proteins were tested for their function as specific binding sites for elf-derived ligands. In tests with leaf extracts expressing the different constructs only full-length EFR and E-TM, the ectodomain with transmembrane domain, showed specific binding of radiolabeled ligand peptide elf-125I, whereas EFR lacking the first five LRRs (EFRΔ5) and the EFR-ectodomain (E-oJM) alone did not bind the elf ligand (Fig. 2D). E-oJM lacks a transmembrane domain and should result in an exported and soluble protein that might get washed away in the binding assays with crude extracts. Thus, we tested binding activity of soluble and detergent-solubilized proteins after adsorption of proteins via their glycosylation moieties to ConA beads (14). However, in contrast to EFR used as a positive control, E-oJM did not show binding of the radioligand (supplemental Fig. 2).
The elf18 peptide is a water-soluble molecule that is rather small in size compared with the EFR ectodomain with its 21 LRRs. Thus, the finding that functional binding seems to depend on both the N-terminal 5 LRRs and the transmembrane domain at the C-terminal end of the ectodomain was somewhat unexpected. However, rather than interacting directly with the ligand, these parts of EFR might have an indirect effect on the formation or exposure of the receptor binding sites. To test this, we tried to substitute the missing parts in these EFR constructs with “equivalent ” parts of the structurally related receptor FLS2.
Constructions of Chimeric Receptors between EFR and FLS2
FLS2, the receptor for bacterial flagellin FLS2 (13), is one of the closest relatives of EFR encoded in the A. thaliana genome. EFR and FLS2 both belong to the same subfamily XII of the LRR-RLKs (26). Structurally, the biggest difference between the two proteins is the number of the LRRs, 21 in EFR and 28 in FLS2, respectively. An alignment shows 31% identity of EFR with FLS2 over the whole length of the protein (supplemental Fig. 3). Identity in the LRR domains similarly amounts to 29%. This is mainly due to conservation of the structural residues according to the consensus IPXXLGXLXXLXXLXLXXNXL(S/T)GX of these 24-aa repeats with ∼44% identity between EFR and FLS2. In contrast, the amino acids predicted to be solvent exposed (X in the consensus) show identity of only 14%. This indicates little conservation at the surface of the ectodomains which can be assumed to form the interaction sites for the structurally unrelated ligands elf18 and flg22, respectively.
The kinase domains of the two receptors also exhibit ∼32% sequence identity (supplemental Fig. 3B). Although high homology exists for residues conserved in kinases of all RLKs (supplemental Fig. 3B) there is little sequence homology outside these regions. This is surprising because it is the relatedness of the kinase sequences that was used to place EFR and FLS2 into the same subfamily XII of the LRR receptor like kinases (family 1.12.4 in PlantsP Kinase classification) (2). EFR and FLS2 trigger the same signal outputs leading to a congruent set of responses (14). For the approach involving sequence substitutions between EFR and FLS2 we thus also included swaps of the kinase domains to test for functional complementation.
Functionality of Chimeric Receptors
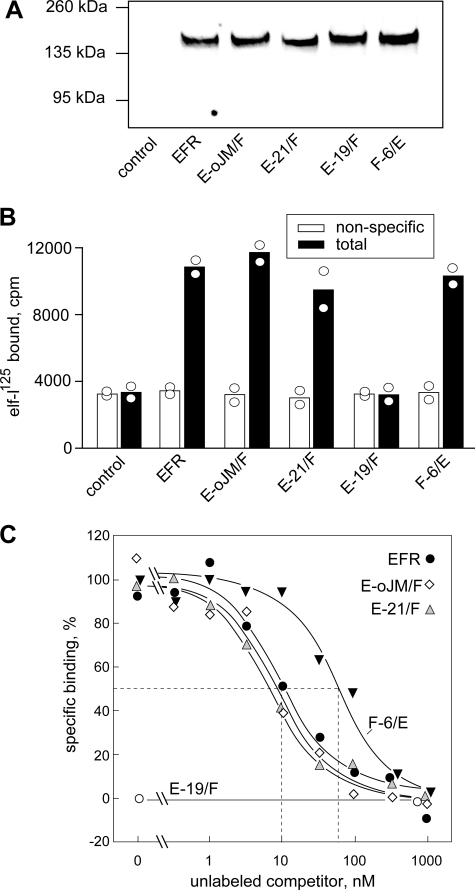
In one of the chimeric proteins, termed F-6/E, the N-terminal part with the first 6 LRRs of FLS2 was swapped to EFR, taking care not to interrupt the repeat structure in the LRR of the protein. Similarly, three chimeric receptors were constructed with N-terminal parts of EFR and swaps to FLS2 in LRR 19 (E-19/F), after the last of the 21 LRRs (E-21/F) or after the outer juxtamembrane domain (E-oJM/F) (Fig. 1 and supplemental Fig. 3). The gene constructs were transiently expressed in N. benthamiana leaves and were found to accumulate to similar levels when checked on Western blots (Fig. 3A). Significant levels of specific, high affinity binding sites for elf-peptides were detectable in extracts with F-6/E, E-21/F, E-oJM/F, and EFR (Fig. 3A). In contrast, no specific binding sites were detectable in extracts with E-19/F (Fig. 3A).
FIGURE 3.
Binding of elf-125I to EFR and chimeric receptors. A, Western blot with extracts from leaves expressing EFR or the different chimeric receptors stained with anti-GFP antibodies. B, binding of elf-125I in the absence (total binding, filled bars) or presence of 1 μm elf18 (nonspecific binding, open bars). Data shown represent two replicates with bars indicating the mean. C, competition of specific binding with different concentrations of unlabeled elf18.
The binding affinities of the different constructs were further analyzed in competition experiments with increasing amounts of unlabeled elf18 (Fig. 3B). In the case of EFR, half-maximal competition of binding (IC50) was reached at a concentration of ∼10 nm elf18. This value corresponds to the IC50 of ∼10 nm reported for EFR binding sites in A. thaliana cells (14). No significant difference was observed for the IC50 values with E-21/F and E-oJM/F indicating that these chimeric receptors have full affinity for the elf18 ligand. The chimera F-6/E, although clearly binding the radioligand, showed a somewhat reduced affinity with an IC50 of ∼70 nm (Fig. 3B).
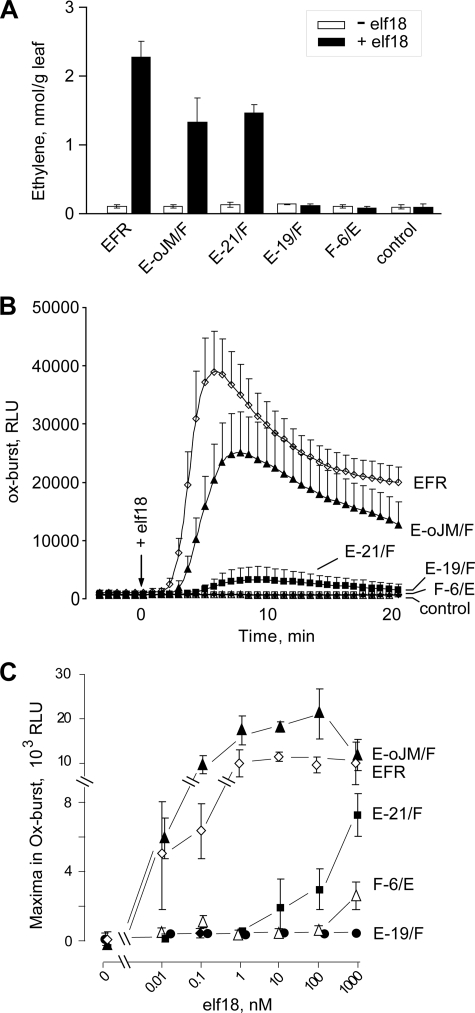
The chimeras E-oJM/F and E-21/F showed functionality as EF-Tu receptors when assayed for activation of ethylene production and oxidative burst in N. benthamiana leaves (Fig. 4). Tissues expressing E-oJM/F were as responsive to elf18 as tissues expressing EFR. Similar to the high sensitivity of A. thaliana cells in alkalinization tests (21), leaf tissues of N. benthamiana expressing EFR or E-oJM/F show a very high sensitivity and respond to elf18 at concentrations as low as 10 pm with induction of an oxidative burst (Fig. 4C).
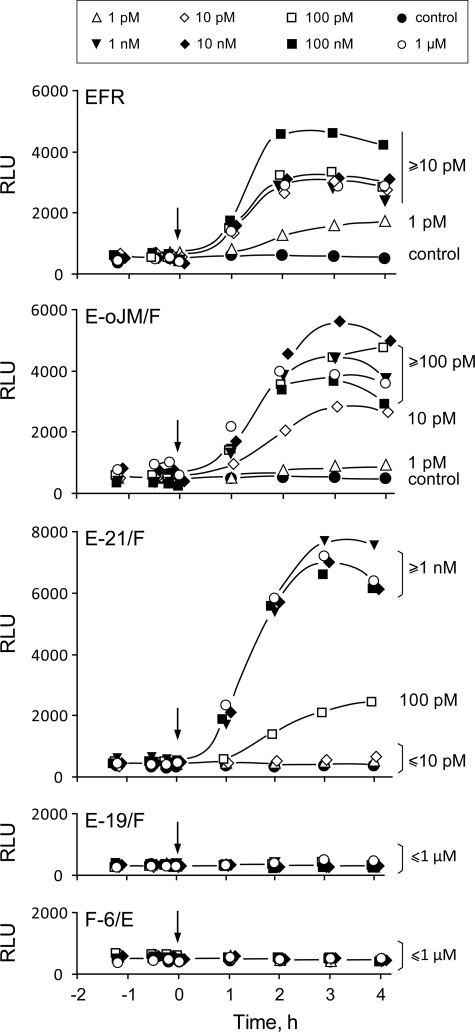
FIGURE 4.
Functionality of chimeric receptors after expression in N. benthamiana leaves. A, ethylene biosynthesis in leaf tissues expressing EFR or chimeric forms of EFR and FLS2 after treatment with 100 nm elf18. B, time courses of oxidative burst after treatment with 100 nm elf18. C, maxima in oxidative bursts after treatment with different concentrations of elf18. Values represent mean ± S.D. (error bars) of n = 4 replicates.
Compared with EFR and E-oJM/F, leaves with E-21/F responded to elf peptides with clearly reduced efficiency, and significant responses occurred only at concentrations of ≥10 nm (Fig. 4). Leaf tissue expressing F-6/E showed no oxidative burst at concentrations up to 100 nm, whereas a small but significant oxidative burst could be reproducibly detected after treatment with 1000 nm elf18 (Fig. 4C). In contrast, no responses to 1,000 nm elf18 were observed in leaves expressing E-19/F (Fig. 4C) or control leaves expressing no EFR constructs.
Functioning of Chimeric receptors in A. thaliana Cells
The functionality of the chimeric receptors was further tested in mesophyll protoplasts of A. thaliana from mutant plants lacking functional EFR. Protoplasts were co-transformed with luciferase under the FRK1 promoter (25) and the different EFR constructs to be tested. Protoplasts expressing EFR, E-oJM/F, and E-21/F showed clear induction of luciferase when exposed to elf18 whereas luciferase in cells expressing either F-6/E or E-19/F did not increase above the background level of untreated control cells. As above with transformed N. benthamiana leaves, the maximal amplitude of the response varied somewhat, probably reflecting the slight variation in number of cells transfected. However, the cells transformed with EFR or E-oJM/F reproducibly showed a very high sensitivity with responses down to 1 or 10 pm elf18, respectively (Fig. 5). Cells expressing E-21/F were somewhat less sensitive and induction of luciferase was detectable only at concentrations ≥100 pm elf18 (Fig. 5).
FIGURE 5.
Functionality of chimeric receptors after transient expression in mesophyll protoplasts from efr × fls2 mutants of A.thaliana. Luciferase activity in protoplasts treated with different concentrations of elf18 is shown.
DISCUSSION
EFR Ectodomain as Interaction Site for the elf Ligands
Mapping of the ligand interaction site by deletional analysis was done successfully for the well characterized plant receptor kinase BRI1 (8). Expressing subdomains of BRI1 in E. coli allowed attribution of the binding site for the hormone ligand brassinolide to a small island domain and the subsequent 22nd of the 25 LRRs of BRI1 (11). In initial experiments we also attempted to narrow the site of interaction for the rather small elf18 peptide to a subdomain within the rather big EFR ectodomain. Removing the cytoplasmic part with the kinase domain did not affect binding. However, further deletions at the N terminus as in EFRΔ5 or further truncation from its C-terminal side resulted in proteins lacking affinity for elf peptides. Recent reports have shown that accumulation of functional EFR strongly depends on quality control steps and glycosylation processes in the secretory pathway of the plant (27, 28). The constructs used in our experiments all contained an N-terminal signal sequence, and they led to accumulation of C-terminally tagged proteins with the size of appropriately glycosylated EFR derivatives (Fig. 2A). Elf peptides are water-soluble, and direct interaction with the transmembrane domain of EFR seems unlikely. Thus, the lack of binding activity observed for the soluble ectodomain (E-oJM) was rather unexpected. However, specific binding was found to be restored in EFR variants containing the transmembrane domain of FLS2 as in the chimeric receptors E-21/F and E-oJM/F. Thus, the transmembrane domain might rather be important for correct folding or the exposure of the interaction site for the elf ligand. Anchoring to the plasma membrane might be important to keep the predicted solenoid structure of the LRR domain in an open, ligand-accessible state. Whether the transmembrane domain is important for a potential oligomerization with further EFR moieties or other proteins acting as a co-receptor remains an open question.
EFR-FLS2 Chimeras
To study the elf binding properties and EFR functionality, we substituted parts of the receptor by corresponding parts of the closely related LRR receptor kinase FLS2. Chimeric constructions with domain swaps between different receptors have previously been used to investigate specificity of ligand interaction and signal output of Toll-like receptors in humans and animals (29, 30). In plants, a much-noticed first example was reported for a chimera of the pattern recognition receptor XA21 from rice and the brassinolide receptor BRI1 from A. thaliana (31). Thereby, the kinase domain of XA21 was swapped to BRI1 and resulted in a receptor that triggered defense rather than growth in response to brassinolide. Although indicating that plant RLKs can indeed be combined in modular form, this pivotal study has not been corroborated with other functional chimeras so far.
A chimera-based technique was recently also used to investigate the S-locus receptor kinases that detect corresponding S-locus cysteine rich ligands and confer self-incompatibility in crucifer plants (32, 33). S-locus receptor kinases also belong to a family of RLKs with ectodomains other than LRRs. Combining parts of ectodomains from two closely related but distinct S-locus receptor kinases in chimeric constructs allowed the identification of two noncontiguous parts of the ectodomain as important for specifying self-incompatibility.
Chimeric approaches were also applied to study tomato resistance proteins that confer resistance to specific races of the leaf mold fungus Cladosporium fulvum. These resistance proteins form a small family of plasma membrane-anchored receptor-like proteins with LRR ectodomains but lacking cytoplasmic kinase domains. Domain swaps between these resistance proteins were used to identify subdomains carrying the specificity for the corresponding effectors from the C. fulvum pathogens (34–36). However, as a limitation of this analysis, for none of these resistance proteins could a direct interaction with the corresponding effector proteins of the pathogen be demonstrated so far (37).
In our work we used the two LRR receptor kinases EFR and FLS2 to create chimeric receptors. These proteins share only ∼31% overall amino acid sequence identity. EFR and FLS2 detect the two distinct bacterial MAMPs elf18 and flg22 as ligands, respectively, and sequence divergence can be expected for their ligand interaction domains. After ligand-induced activation, EFR and FLS2 induce a nearly congruent set of responses (14), and one would expect the kinases of the two receptors to be functionally equivalent. Indeed, swapping the kinase domain of FLS2 to EFR did result in a fully functional receptor (E-oJM/F). However, the kinases of these two receptors are rather divergent in sequence, and the conservation is restricted to residues conserved in kinases of plant RLKs in general (supplemental Fig. 3B). At first glance, the cytoplasmic domains of FLS2, EFR, and PEPR1, which stimulate a common danger or defense program in the cells, share no obvious features in their sequence that would distinguish them from the kinases of BRI1, HAESA, EMS, CLV1, and PSKR involved in other intracellular output programs. Swapping the kinase domain of FLS2 to EFR results in the fully functional receptor E-oJM/F. Indeed, transformed N. benthamiana leaf cells and A. thaliana protoplasts showed responses down to 1–10 pm elf18, indicating excellent functionality of the EFR ectodomain with the EFR kinase or the FLS2 kinase in both expression systems. Competitive binding studies would suggest half-maximal saturation of receptor sites to occur only at around 10 nm elf18 (Fig. 3C). Maximal response output is reached at considerably lower concentrations, indicating that occupancy of a very small percentage of the receptor sites is sufficient to trigger full responses. Such behavior could be explained by the concept of “spare” receptors whereby a pool of functionally equivalent receptor sites signals through a common, limiting element further downstream in the signaling pathway. Strong expression of the receptor, as with the 35S-promoter in this study, might thus contribute to the exquisite sensitivity of the perception system observed in the transformed cells. High sensitivity indicative of spare receptors has been observed before in nontransformed cells responding to other MAMPs like flg22 and chitin fragments (38, 39).
Swapping subdomains within the LRR domain yielded receptors with different degrees of functionality and ligand binding activity. Comparison of E-21/F and E-19/F indicates that the last two LRRs of EFR cannot be replaced without complete loss of binding activity and functionality. The E-19/F construct was designed to maintain the regularity of the LRR domain, and the corresponding gene product accumulated and localized without apparent problems (Fig. 3A and supplemental Fig. 1). These data cannot completely exclude a conformational disturbance introduced at the junction site of the two ectodomains. Nevertheless, the LRRs 19–21 are primary candidates for further analysis on their roles for the interaction with the elf ligand.
Interestingly, the chimera F-6/E with the first six LRRs swapped from FLS2 to EFR does bind elf but with a ∼7–10-fold lower affinity. The functionality of F-6/E is more drastically reduced with only residual activity as inducer of oxidative burst in N. benthamiana leaves and no activity as inducer of FRK1-luciferase in A. thaliana protoplasts. E-21/F exhibits full affinity for the elf ligand. Compared with EFR and E-oJM/F, however, E-21/F is clearly less efficient and requires higher concentrations of elf18 for the induction of oxidative burst in N. benthamiana and FRK1-luciferase in A. thaliana protoplasts. Thus, it seems that the oJM domains of FLS2 and EFR are functionally not equivalent. EFR and FLS2, although detecting different ligands, share all functional aspects of receptor activation and signal output. Activation of both receptors occurs according to the address-message concept, presumably involving ligand-induced, conformational changes that allow heteromerization with co-receptors such as BAK1 (18, 21, 39, 40). Thus, for substitutions with parts of FLS2 that fully retain elf binding, one would rather expect functionality of receptor activation to be retained as well. However, an incompatibility of the outer juxtamembrane domain from FLS2 with the EFR ectodomain could arise if the activation process requires intramolecular interaction of the oJM with a second part in the EFR ectodomain. The search for this interaction site will require construction and testing of further chimeric constructs.
Our data demonstrate that chimeras of EFR and FLS2 can be constructed and used for studying receptor function. The results show that different, noncontiguous parts of the ectodomain from EFR are required to form a functional binding site for the elf ligand. For a more precise localization of the sites involved in this interaction swapping of smaller subdomains are required. The A. thaliana genome encodes a small, four-membered family of RLKs that are very closely related to EFR. Genetic evidence suggests that these RLKs do not function as EF-Tu receptors. Provided that these EFR-likes indeed have no affinity for elf-ligands they are good candidates for further chimeric analysis of the EFR binding site. Targeted mutagenesis of single positions in the LRR, as extensively used to study interaction of flg22 with FLS2 (41), might be an alternative.
Having established procedures to produce functional chimeras with very closely related receptors, it will be challenging to expand this approach to less related receptors. In future, hybrid receptors might also help to characterize orphan receptors and to identify the determinants required for interaction with co-receptors, receptor activation, and induction of different intracellular signaling pathways. In short, experimental approaches with chimeric receptors as described in this report should provide important tools to investigate and to understand the molecular functioning of the important family of plant receptor kinases.
Supplementary Material
Acknowledgments
We thank I. Bock (Zentrum für Molekularbiologie der Pflanzen (ZMBP), Tübingen) for technical assistance and I. Küfner (ZMBP, Tübingen) for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft as part of initiatives SFB 446, SFB 766, and ERA-PG RLKRLP.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- RLK
- receptor-like kinase
- FLS2
- flagellin-sensing 2
- MAMP
- microbe-associated molecular pattern
- EF-Tu
- elongation factor Tu
- LRR
- leucine-rich repeat
- GFP
- green fluorescent protein
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Shiu S. H., Bleecker A. B. (2001) Sci. STKE 2001, re22. [DOI] [PubMed] [Google Scholar]
- 2.Shiu S. H., Karlowski W. M., Pan R., Tzeng Y. H., Mayer K. F., Li W. H. (2004) Plant Cell 16, 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morillo S. A., Tax F. E. (2006) Curr. Opin. Plant Biol. 9, 460–469 [DOI] [PubMed] [Google Scholar]
- 4.De Smet I., Voss U., Jürgens G., Beeckman T. (2009) Nat. Cell Biol. 11, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 5.Tör M., Lotze M. T., Holton N. (2009) J. Exp. Bot. 60, 3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butenko M. A., Vie A. K., Brembu T., Aalen R. B., Bones A. M. (2009) Trends Plant Sci. 14, 255–263 [DOI] [PubMed] [Google Scholar]
- 7.Boller T., Felix G. (2009) Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 8.Belkhadir Y., Chory J. (2006) Science 314, 1410–1411 [DOI] [PubMed] [Google Scholar]
- 9.Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., Walker J. C. (2002) Cell 110, 213–222 [DOI] [PubMed] [Google Scholar]
- 10.Nam K. H., Li J. (2002) Cell 110, 203–212 [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005) Nature 433, 167–171 [DOI] [PubMed] [Google Scholar]
- 12.Lee S. W., Han S. W., Sririyanum M., Park C. J., Seo Y. S., Ronald P. C. (2009) Science 326, 850–853 [DOI] [PubMed] [Google Scholar]
- 13.Gómez-Gómez L., Boller T. (2000) Mol. Cell 5, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 14.Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., Felix G. (2006) Cell 125, 749–760 [DOI] [PubMed] [Google Scholar]
- 15.Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffaker A., Pearce G., Ryan C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006) Plant Cell 18, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010) J. Biol. Chem 285, 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M., He K., Li J., Schroeder J. I., Peck S. C., Rathjen J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinchilla D., Shan L., He P., de Vries S., Kemmerling B. (2009) Trends Plant Sci. 14, 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004) Plant Cell 16, 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimchuk Z., Marois E., Kjemtrup S., Leister R. T., Katagiri F., Dangl J. L. (2000) Cell 101, 353–363 [DOI] [PubMed] [Google Scholar]
- 23.Nekrasov V., Li J., Batoux M., Roux M., Chu Z. H., Lacombe S., Rougon A., Bittel P., Kiss-Papp M., Chinchilla D., van Esse H. P., Jorda L., Schwessinger B., Nicaise V., Thomma B. P., Molina A., Jones J. D., Zipfel C. (2009) EMBO J. 28, 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo S. D., Cho Y. H., Sheen J. (2007) Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 25.Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gómez-Gómez L., Boller T., Ausubel F. M., Sheen J. (2002) Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- 26.Shiu S. H., Bleecker A. B. (2003) Plant Physiol. 132, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010) J. Biol. Chem. 285, 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiya T., DeFranco A. L. (2004) J. Biol. Chem. 279, 19008–19017 [DOI] [PubMed] [Google Scholar]
- 30.Ishii A., Matsuo A., Sawa H., Tsujita T., Shida K., Matsumoto M., Seya T. (2007) J. Immunol. 178, 397–406 [DOI] [PubMed] [Google Scholar]
- 31.He Z., Wang Z. Y., Li J., Zhu Q., Lamb C., Ronald P., Chory J. (2000) Science 288, 2360–2363 [DOI] [PubMed] [Google Scholar]
- 32.Boggs N. A., Dwyer K. G., Nasrallah M. E., Nasrallah J. B. (2009) Curr. Biol. 19, 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boggs N. A., Dwyer K. G., Shah P., McCulloch A. A., Bechsgaard J., Schierup M. H., Nasrallah M. E., Nasrallah J. B. (2009) Genetics 182, 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Hoorn R. A., Kruijt M., Roth R., Brandwagt B. F., Joosten M. H., De Wit P. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Hoorn R. A., Roth R., De Wit P. J. (2001) Plant Cell 13, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wulff B. B., Thomas C. M., Smoker M., Grant M., Jones J. D. (2001) Plant Cell 13, 255–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wulff B. B., Chakrabarti A., Jones D. A. (2009) Mol. Plant Microbe Interact. 22, 1191–1202 [DOI] [PubMed] [Google Scholar]
- 38.Baureithel K., Felix G., Boller T. (1994) J. Biol. Chem. 269, 17931–17938 [PubMed] [Google Scholar]
- 39.Meindl T., Boller T., Felix G. (2000) Plant Cell 12, 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., Felix G., Boller T. (2007) Nature 448, 497–500 [DOI] [PubMed] [Google Scholar]
- 41.Dunning F. M., Sun W., Jansen K. L., Helft L., Bent A. F. (2007) Plant Cell 19, 3297–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.