Abstract
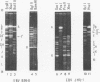
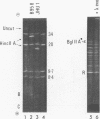
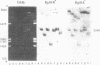
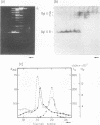
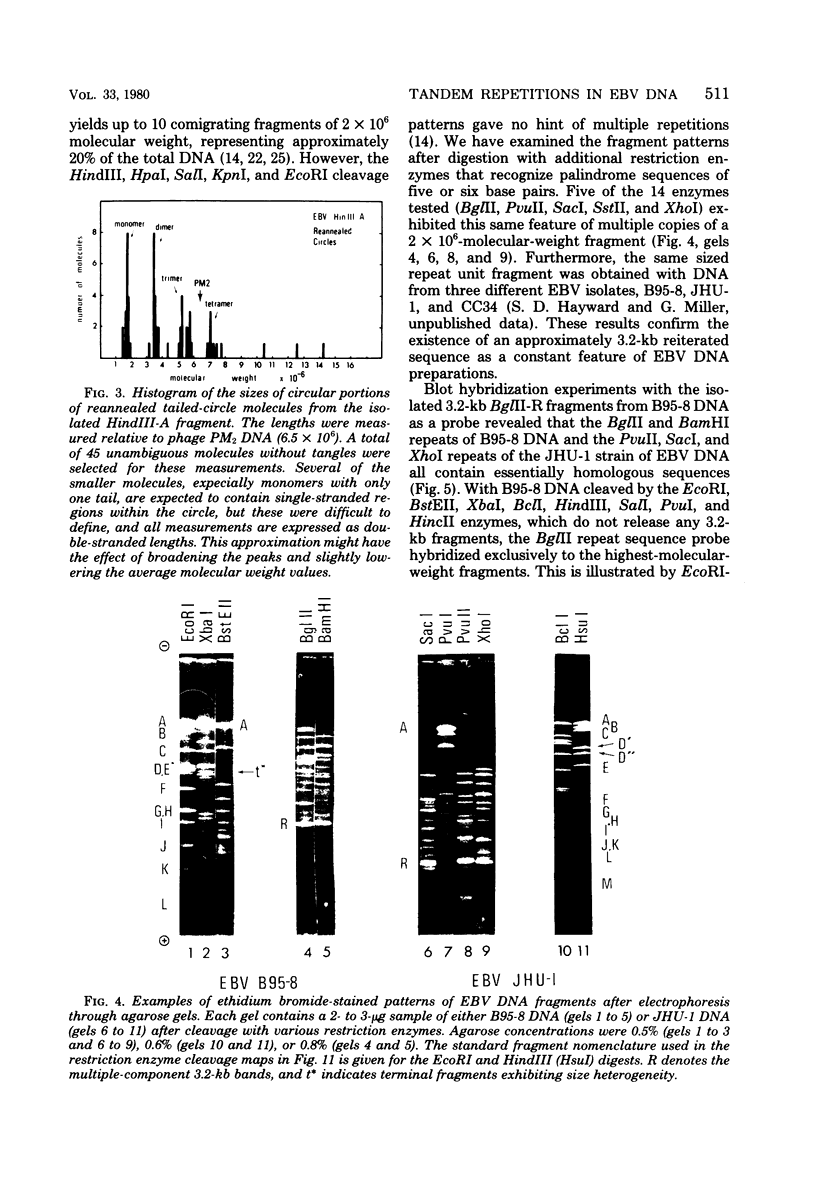
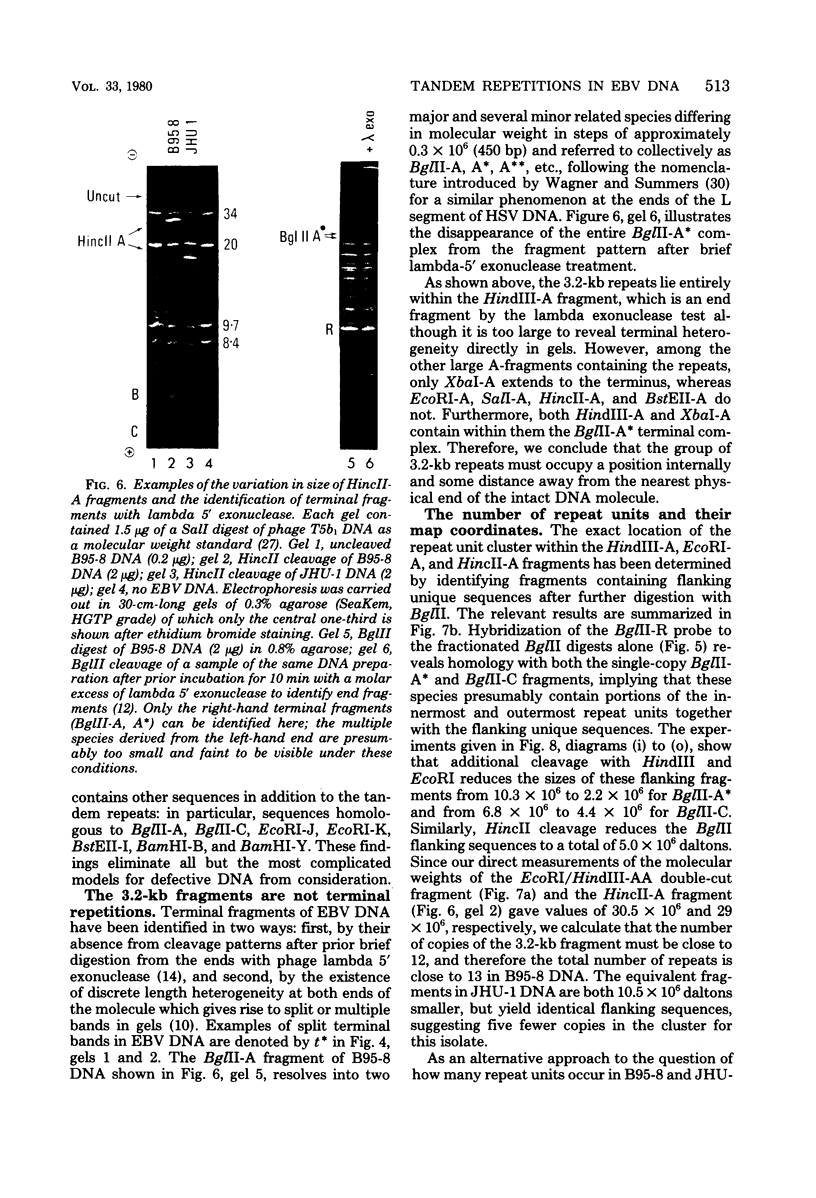
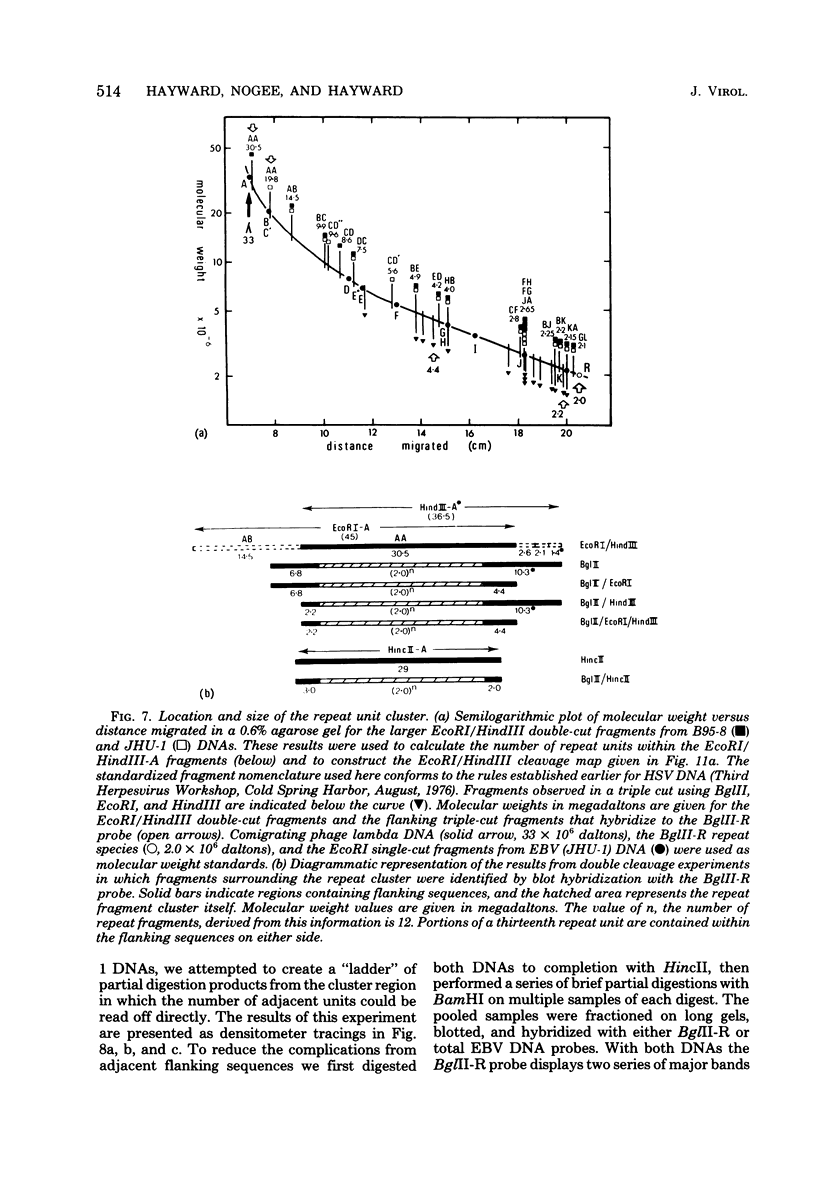
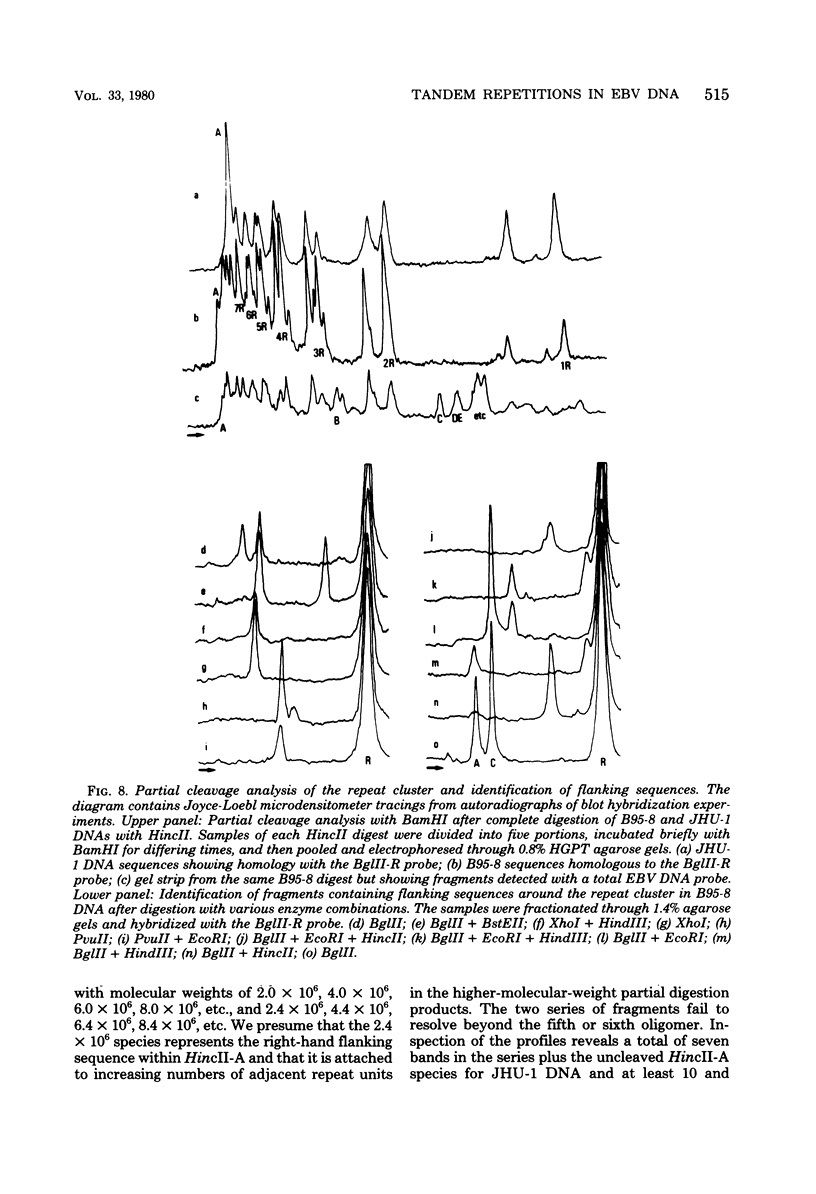
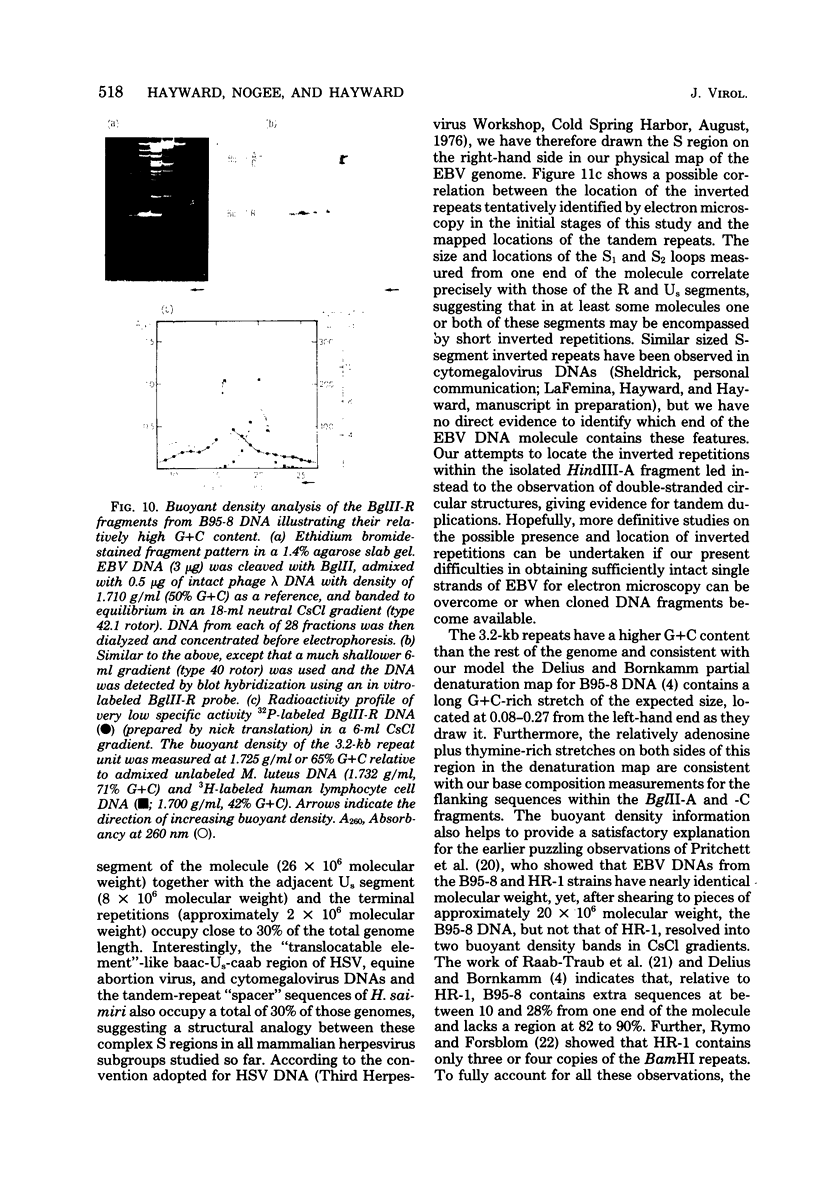
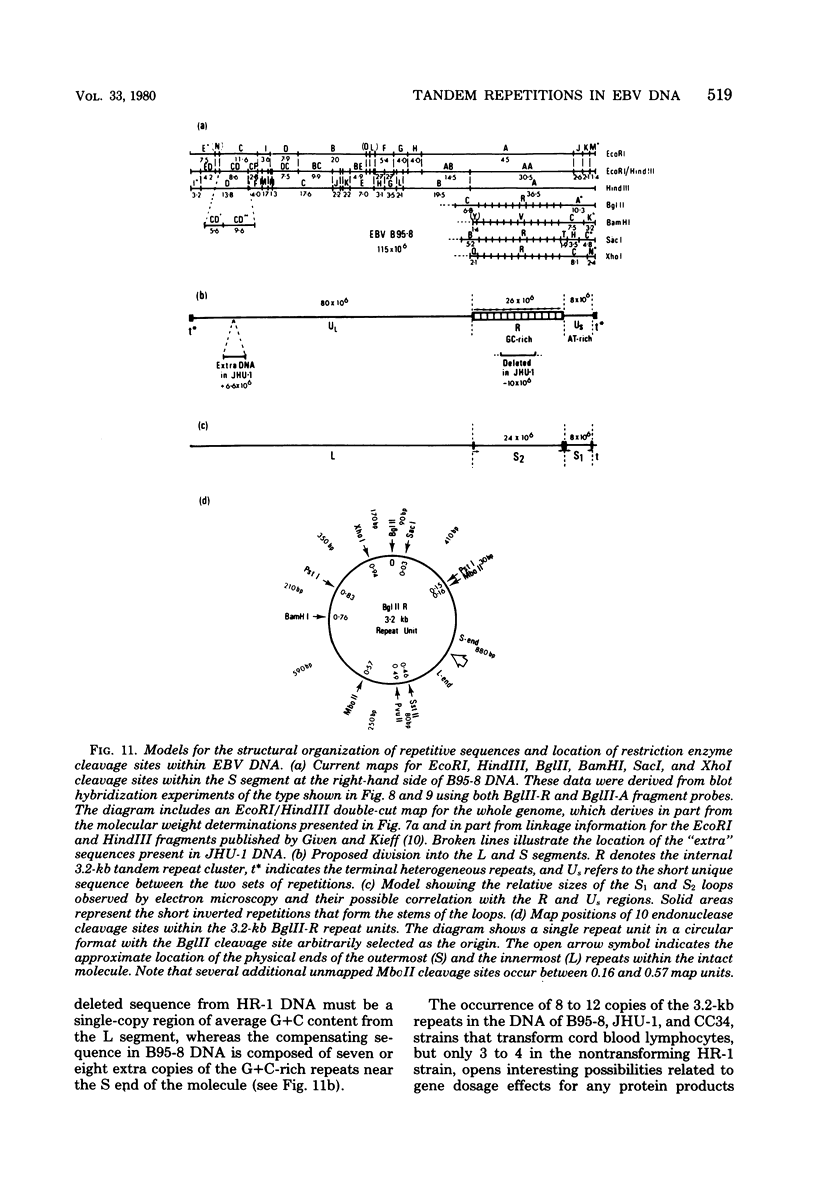
Virions of human Epstein-Barr virus released from the B95-8 line of marmoset lymphoblasts have linear double-stranded DNA molecules of 115 x 10(6) molecular weight (180 +/- 10 kilobase pairs). Approximately 20% of this DNA yields multiple fragments of 3,200 base pairs when cleaved with any one of the BglII, BamHI, PvuII, SacI, SstII, or XhoI restriction enzymes. The results of cleavage site mapping with these and other enzymes, together with blot hybridization experiments using the 3.2-kilobase pair BglII-R fragment as a probe, indicate that these fragments originate from an internal region between 0.710 and 0.915 map units containing a cluster of at least 12 apparently identical repetitions of a sequence with relatively high guanine plus cytosine content. The repeat units are arranged in adjacent tandem array with all copies having the same orientations, and they form a series of oligomers of tailed double-stranded circles when fragments containing portions of the cluster are denatured and reannealed. Physical maps of cleavage sites within the 3.2-kilobase pair repeat units and in the flanking sequences surrounding the repeat cluster have been constructed. We conclude that the Epstein-Barr virus DNA molecule, like those of other mammalian herpesviruses, may be regarded as being divisible into a large L segment and a smaller S segment. However, the detailed arrangement of repetitive sequences within the Epstein-Barr virus S segment differs significantly from that in all other herpesvirus genomes described so far.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A., Bjursell G., Kaschka-Dierich C., Lindahl T. Circular Epstein-Barr virus genomes of reduced size in a human lymphoid cell line of infectious mononucleosis origin. J Virol. 1977 May;22(2):373–380. doi: 10.1128/jvi.22.2.373-380.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Bornkamm G. W. Heterogeneity of Epstein-Barr virus. III. Comparison of a transforming and a nontransforming virus by partial denaturation mapping of their DNAs. J Virol. 1978 Jul;27(1):81–89. doi: 10.1128/jvi.27.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dolyniuk M., Pritchett R., Kieff E. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J Virol. 1976 Mar;17(3):935–949. doi: 10.1128/jvi.17.3.935-949.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Ludwig H. Repetitive sequences in complete and defective genomes of Herpesvirus saimiri. J Virol. 1975 Feb;15(2):398–406. doi: 10.1128/jvi.15.2.398-406.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen J. L., Walig C., Wertheim P., van der Noordaa J. Human cytomegalovirus DNA. I. Molecular weight and infectivity. J Virol. 1978 Jun;26(3):813–816. doi: 10.1128/jvi.26.3.813-816.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given D., Kieff E. DNA of Epstein-Barr virus. IV. Linkage map of restriction enzyme fragments of the B95-8 and W91 strains of Epstein-Barr Virus. J Virol. 1978 Nov;28(2):524–542. doi: 10.1128/jvi.28.2.524-542.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. D., Kieff E. D. Epstein-Barr virus-specific RNA. I. Analysis of viral RNA in cellular extracts and in the polyribosomal fraction of permissive and nonpermissive lymphoblastoid cell lines. J Virol. 1976 May;18(2):518–525. doi: 10.1128/jvi.18.2.518-525.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. D., Kieff E. DNA of Epstein-Barr virus. II. Comparison of the molecular weights of restriction endonuclease fragments of the DNA of Epstein-Barr virus strains and identification of end fragments of the B95-8 strain. J Virol. 1977 Aug;23(2):421–429. doi: 10.1128/jvi.23.2.421-429.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Pagano J. S. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simples I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J Virol. 1974 Mar;13(3):642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick B. A., Huang E. S. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J Virol. 1977 Oct;24(1):261–276. doi: 10.1128/jvi.24.1.261-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Luka J., Lindahl T., Klein G. Purification of the Epstein-Barr virus-determined nuclear antigen from Epstein-Barr virus-transformed human lymphoid cell lines. J Virol. 1978 Sep;27(3):604–611. doi: 10.1128/jvi.27.3.604-611.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A. 1973 Jan;70(1):190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett R. F., Hayward S. D., Kieff E. D. DNA of Epstein-Barr virus. I. Comparative studies of the DNA of Epstein-Barr virus from HR-1 and B95-8 cells: size, structure, and relatedness. J Virol. 1975 Mar;15(3):556–559. doi: 10.1128/jvi.15.3.556-559.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N., Pritchett R., Kieff E. DNA of Epstein-Barr virus. III. Identification of restriction enzyme fragments that contain DNA sequences which differ among strains of Epstein-Barr virus. J Virol. 1978 Aug;27(2):388–398. doi: 10.1128/jvi.27.2.388-398.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo L., Forsblom S. Cleavage of Epstein-Barr virus DNA by restriction endonucleases EcoRI, HindIII and BamI. Nucleic Acids Res. 1978 Apr;5(4):1387–1402. doi: 10.1093/nar/5.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Holthausen H., zur Hausen H. Partial purification of the Epstein-Barr virus and some properties of its DNA. Virology. 1970 Mar;40(3):776–779. doi: 10.1016/0042-6822(70)90229-1. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sugden B. Comparison of Epstein-Barr viral DNAs in Burkitt lymphoma biopsy cells and in cells clonally transformed in vitro. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4651–4655. doi: 10.1073/pnas.74.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Summers W. C. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J Virol. 1978 Aug;27(2):374–387. doi: 10.1128/jvi.27.2.374-387.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gabain A., Hayward G. S., Bujard H. Physical mapping of the HindIII, EcoRI, Sal and Sma restriction endonuclease cleavage fragments from bacteriophage T5 DNA. Mol Gen Genet. 1976 Feb 2;143(3):279–290. doi: 10.1007/BF00269404. [DOI] [PubMed] [Google Scholar]
- zur Hausen H., O'Neill F. J., Freese U. K., Hecker E. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature. 1978 Mar 23;272(5651):373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]