Abstract
Rapid Myc protein turnover is critical for maintaining basal levels of Myc activity in normal cells and a prompt response to changing growth signals. We characterize a new Myc-interacting factor, TRPC4AP (transient receptor potential cation channel, subfamily C, member 4-associated protein)/TRUSS (tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein), which is the receptor for a DDB1 (damage-specific DNA-binding protein 1)–CUL4 (Cullin 4) E3 ligase complex for selective Myc degradation through the proteasome. TRPC4AP/TRUSS binds specifically to the Myc C terminus and promotes its ubiquitination and destruction through the recognition of evolutionarily conserved domains in the Myc N terminus. TRPC4AP/TRUSS suppresses Myc-mediated transactivation and transformation in a dose-dependent manner. Finally, we found that TRPC4AP/TRUSS expression is strongly down-regulated in most cancer cell lines, leading to Myc protein stabilization. These studies identify a novel pathway targeting Myc degradation that is suppressed in cancer cells.
Keywords: Myc, protein turnover, ubiquitination, oncogenesis, E3 ligase
The Myc transcription factor is a key regulator of both the G0 → G1 cell cycle entry and normal progression through the cell cycle. It is critical to control Myc levels within the cell to prevent abnormal cell growth. Since Myc mRNA and protein levels must be tightly regulated, it is not surprising that both mRNA and proteins exhibit rapid turnover. Particularly, Myc protein is highly unstable, with a typical half-life of <30 min (Hann and Eisenman 1984; Ikegaki et al. 1986). Considerable effort has been made toward investigating if reduced Myc protein turnover causes an increased net Myc protein level, which is commonly observed in cancers. For example, an extended half-life for Myc protein has been reported in neuroblastomas and lymphomas (Cohn et al. 1990; Gregory and Hann 2000; Malempati et al. 2006). So far, three different E3 ligase complexes have been implicated in Myc degradation: Skp2, Fbw7, and Huwe1 complexes (Kim et al. 2003; von der Lehr et al. 2003; Welcker et al. 2004; Yada et al. 2004; Zhao et al. 2008). In each case, either transient knockdown or genetic ablation of the E3 ligase increases Myc protein half-life and accumulation. These findings imply that each of these E3 ligases can be rate-limiting for Myc protein turnover in different cell backgrounds. The high levels of Myc protein found in the majority of cancer cells could be due to the impairment of Myc turnover pathways in addition to aberrant transcriptional activation of the myc genes. For instance, the Fbw7 gene is mutated in some cancers, but no genetic disruption or reduced expression levels of other E3 ligases associated with Myc turnover have been reported. Furthermore, there as no consistent changes in Myc phosphorylation in cancers with aberrant Myc accumulation but no Myc mutations. Thus, there may be other critical determinants that lead to differences in Myc protein destruction in different cancer cells.
Here, we identify a novel E3 ligase complex for both c-Myc and N-Myc that targets Myc proteins for proteasome-mediated degradation. This complex is a negative regulator of Myc function, and its consistent repression in cancer cells correlates with an extended Myc protein half-life.
Results and Discussion
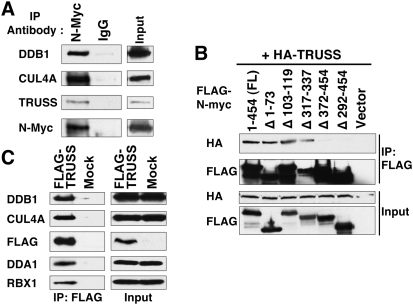
To identify Myc-interacting proteins that might regulate its functions, we affinity-purified complexes from HeLa cells that stably express Flag-tagged N-Myc. The eluant was subjected to mass spectrometry analysis to identify bound proteins (Supplemental Table 1). Mass spectrometric analysis revealed novel as well as known Myc-interacting proteins, such as TRRAP, TIP48/49, DMAP1, TIP60, and MAX (Cowling and Cole 2006). Among the novel Myc-interacting proteins, we decided to characterize the interaction between Myc and TRPC4AP (transient receptor potential cation channel, subfamily C, member 4-associated protein). TRPC4AP, also known as TRUSS (tumor necrosis factor receptor-associated ubiquitous scaffolding and signaling protein) (Soond et al. 2003, 2006). We verified the endogenous association by coimmunoprecipitation with anti-N-Myc and by Western blot with anti-TRUSS using IMR5 neuroblastoma cells (Fig. 1A). We next mapped the TRUSS-binding domain in N-Myc by coexpression of TRUSS with expression vectors containing various deletions of the N-Myc coding region. We found that the N-Myc mutant bearing a C-terminal deletion is unable to coimmunoprecipitate with TRUSS (Fig. 1B). Conversely, the other N-Myc mutants were able to bind TRUSS with equal affinity. This implies that TRUSS interacts with Myc via its C-terminal domain, which contains the basic helix–loop–heliz leucine zipper (B-HLH-LZ) motif. Since the B-HLH-LZ motif is also responsible for dimerization with Max, it was possible that TRUSS could prevent Myc dimerization with Max. However, we found no difference in Myc–Max dimerization with TRUSS overexpression (data not shown).
Figure 1.
Myc interacts with TRUSS and recruits the DDB1–CUL4 E3 ligase complex. (A) TRUSS forms a complex with native N-Myc. Endogenous N-Myc protein was immunoprecipitated from IMR-5 neuroblastoma cells with an anti-N-Myc monoclonal antibody, B8.4.B. Precipitating proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Inputs for immunoprecipitations are shown at the right. (B) Coimmunoprecipitation assay to locate the TRUSS-interacting region in N-Myc. HA-tagged full-length TRUSS-expressing vector (HA-TRUSS) was cotransfected with Flag-tagged N-Myc-expressing vectors containing the indicated deletions in N-Myc. Flag-N-Myc immunoprecipitates, together with inputs (5%), were probed for either HA (TRUSS) or Flag (N-Myc). (C) TRUSS associates with the DDB1–CUL4A complex. TRUSS was immunoprecipitated and immunoblotted from HeLa cells that stably expressed Flag-TRUSS. Antibodies used for immunoblots are described on the left. The input was split for each immunoprecipitation.
To understand the role of TRUSS in Myc function, we biochemically identified functional cofactors of TRUSS. We generated a HeLa cell line that stably expresses Flag-tagged TRUSS, which was then affinity-purified. Mass spectrometry analysis revealed that DDB1 (damage-specific DNA-binding protein 1), CUL4A (Cullin 4A), and DDA1 (DET1- and DDB1-associated 1) are strongly associated with TRUSS (Supplemental Table 2). This group of proteins precisely represents the DDB1–CUL4 E3 ubiquitin ligase complex, which belongs to a superfamily of cullin–RING finger ligase complexes that are found throughout eukaryotes (Petroski and Deshaies 2005). Notably, this is consistent with the mass spectrometry analysis of N-Myc complexes that also identified CUL4A and DDB1 (Supplemental Table 1). We used immunoblot to confirm that the TRUSS complex contains DDB1, CUL4A, DDA1, and the small RING finger protein RBX1 (RING box protein-1)/ROC1 (regulator of cullins-1) (Fig. 1C), which is essential for recruiting the E2 ubiquitin-conjugating enzymes (Jackson et al. 2000). We confirmed the endogenous interaction between N-Myc and the DDB1–CUL4 complex by coimmunoprecipitation analysis (Fig. 1A). In conclusion, we found that Myc interacts with TRUSS, and TRUSS is associated in the DDB1–CUL4 E3 ligase complex.
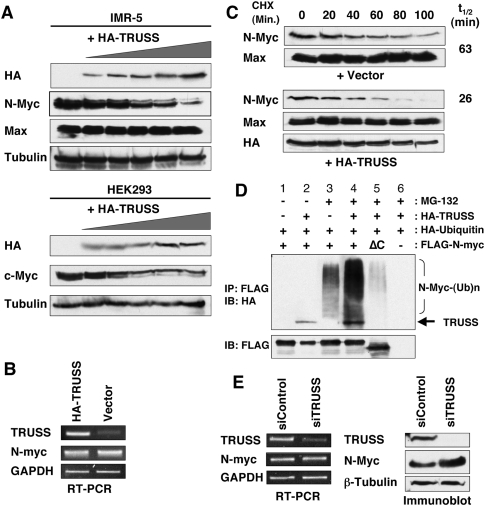
In the DDB1–CUL4 ubiquitin ligase system, substrate specificity is determined by DCAFs, which function as specific substrate receptors (Lee and Zhou 2007). Recently, 60 different putative DCAFs (DDB1- and CUL4-associated factors) were identified from human cells, including TRUSS/TRPC4AP (Lee and Zhou 2007 and references therein). Therefore, we explored the possibility that TRUSS is specific for the degradation of Myc by bridging it to the DDB1–CUL4 complex. To test this hypothesis, we ectopically expressed TRUSS in a dose-dependent manner in IMR-5 cells to determine if it could regulate the stability of endogenous N-Myc proteins. As expected, N-Myc protein levels decreased as TRUSS expression increased (Fig. 2A, top). The Myc-binding partner, Max, and β-Tubulin showed no change with increased expression of TRUSS, indicating that the decreased protein level is Myc-specific. We observed the same reduced protein levels for the c-Myc protein in the HEK 293 cell line (Fig. 2A, bottom). To test the possibility that TRUSS might change Myc mRNA levels, RT–PCRs were performed to show that endogenous N-myc mRNA levels were unchanged by ectopic TRUSS expression (Fig. 2B). Taken together, we conclude that TRUSS specifically regulates both c-Myc and N-Myc protein levels through a post-transcriptional mechanism, and not at the level of transcription or mRNA stability.
Figure 2.
TRUSS enhances Myc protein turnover. (A) TRUSS was expressed ectopically in IMR-5 cells (top panel) and HEK293 cells (bottom panel) at increasing dosages (0, 0.2, 0.5, 1, 2, and 3 μg, respectively, supplemented with an empty vector for constant DNA input). Total cell lysates were prepared after 36 h, and equal amounts of protein were loaded for immunoblot with indicated antibodies to detect native Myc protein levels. β-Tubulin was used as a loading control. (B) TRUSS does not affect Myc mRNA levels. TRUSS was expressed ectopically in IMR-5 cells (3 μg DNA), and total RNA was isolated and subjected to RT–PCR for N-myc mRNA after 36 h. GAPDH served as an input control. (C) Ectopic TRUSS expression shortens N-Myc protein half-life in IMR-5 cells. TRUSS or empty vector was expressed ectopically for 24 h, then cycloheximide (50 μg/mL) was added to the media to inhibit de novo protein synthesis. Whole-cell lysates were resolved by SDS-PAGE followed by immunoblotting. Max antibody was used as an input control. (D) TRUSS promotes N-Myc ubiquitination. Flag-N-myc, HA-TRUSS, and HA-Ubiquitin were cotransfected in HEK 293 cells ± MG-132. Flag-N-mycΔC was transfected in lane 5 instead of wild-type N-myc. N-Myc was immunoprecipitated with anti-Flag beads, and was immunoblotted with anti-HA antibody to detect TRUSS and high-molecular ubiquitin-conjugated N-Myc. In the absence of MG-132, N-Myc protein is degraded without accumulating ubiquitinated products. (E) Depletion of TRUSS leads to elevated N-Myc protein levels but no change in mRNA. siRNA against human TRUSS and control siRNA were transfected to IMR-5 cells to knock down TRUSS expression. (Left panel) After 36 h, RT–PCR was performed to evaluate the efficiency of siRNA. (Right panel) At the same time, total cell lysates were harvested to measure protein levels of TRUSS and N-Myc. β-Tubulin served as a loading control.
It is known that the Myc protein is extremely unstable, with a typical half-life of 20 min (Hann and Eisenman 1984; Ikegaki et al. 1986; Ingvarsson et al. 1988). To examine how TRUSS affects Myc stability, we measured the half-life of endogenous N-Myc protein in response to TRUSS overexpression. IMR-5 cells were transfected with a TRUSS expression vector, and de novo protein synthesis was blocked 24 h later with the translation inhibitor cycloheximide (Fig. 2C). Immunoblot analysis of N-Myc protein at different time points showed that N-Myc protein has a half-life of ∼66 min (Fig. 2C, top). With ectopic TRUSS expression, N-Myc protein half-life was reduced to 26 min (Fig. 2C, bottom), indicating that TRUSS accelerates the turnover of existing N-Myc protein, but not by slowing de novo synthesis. In contrast to the rapid turnover of Myc protein, Max is highly stable, indicating that Max is not a TRUSS target, although Myc–Max forms a heterodimer.
Myc protein is ubiquitinated and degraded by the proteasome (Bonvini et al. 1998; Flinn et al. 1998), and formation of a polyubiquitin chain is a hallmark for targeting a substrate toward the proteasome. Since TRUSS appears to be a Myc-specific receptor to mediate the interaction with the DDB1–CUL4 E3 ligase, TRUSS could facilitate Myc ubiquitination. To explore this, we analyzed the conjugation of ubiquitin onto N-Myc upon TRUSS overexpression in vivo. Ectopic TRUSS expression promoted the accumulation of higher-molecular-weight ubiquitin-conjugated Myc protein after MG-132 treatment to prevent protein degradation (Fig. 2D, lane 4). However, TRUSS fails to enhance Myc protein ubiquitination when their interaction is lost by the deletion of C-terminal Myc (Fig. 2D, lane 5). We also confirmed that TRUSS can ubiquitinate Myc in vitro (Supplemental Fig. 1). These data indicate that TRUSS facilitates ubiquitination of Myc through a direct interaction that targets Myc toward the proteasome for destruction. To further address if TRUSS is involved in the turnover of endogenous Myc, we generated an siRNA against TRUSS to knock down its expression, which was confirmed by RT–PCR after 24 h of transfection (Fig. 2E, left). Consistent with the overexpression studies above, TRUSS knockdown resulted in an increase in the level of N-Myc protein (Fig. 2E, right), without any change in its mRNA level (Fig. 2E, left). These data support that N-Myc protein accumulation is due to protein stabilization rather than transcriptional effects. Altogether, we conclude that TRUSS plays a key role as a Myc-specific receptor for the DDB1–CUL4 E3 ligase complex, and controls the turnover of Myc protein in a dose-dependent manner.
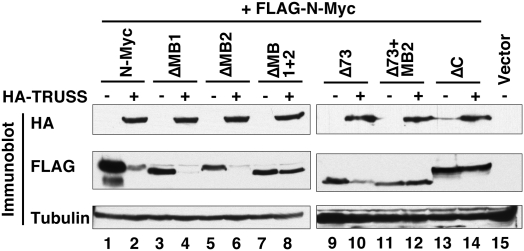
To better understand the mechanism by which TRUSS promotes Myc protein turnover, we mapped the Myc domains targeted by TRUSS. Since deletion mapping showed that the Myc C terminus was responsible for binding to TRUSS (Fig. 1), it was important to determine if this coincides with a domain responsible for enhanced turnover. A series of N-Myc deletion mutants were tested for TRUSS-mediated degradation by transient coexpression. Consistent with the findings above, coexpression of full-length N-Myc with TRUSS gave dramatically decreased protein levels (Fig. 3, lanes 1,2). In contrast, N-Myc with a C-terminal deletion showed no reduction with TRUSS overexpression (Fig. 3, lanes 13,14), confirming that TRUSS binding is necessary for enhanced turnover. N-Myc with deletion of either MB1 or MB2 was also susceptible to TRUSS overexpression (Fig. 3, lanes 3–6), whereas N-Myc with a dual MB1 and MB2 deletion was completely resistant (Fig. 3, lanes 7,8). N-Myc with a deletion of the N-terminal 73 amino acids also exhibited TRUSS-mediated degradation, but a further deletion of the MB2 domain was resistant (Fig. 3, lanes 9–12). Overall, these mapping data suggest that TRUSS recognizes Myc through the C terminus, but degradation is promoted by the molecular recognition of either the evolutionarily conserved MB1 or MB2 domains, which serve as apparently redundant determinants in the TRUSS pathway.
Figure 3.
TRUSS requires both N-terminal and C-terminal Myc domains for degradation. Full-length N-myc and various deletion mutants were expressed in HEK 293 cells in the absence or presence of TRUSS coexpression to map the TRUSS-dependent Myc degrons. Total lysates were immunoblotted to compare the net amount of Myc protein. ΔMB1, ΔMB2, and ΔC correspond to deletion of N-Myc amino acids 40–58, 103–119, and 372–454, respectively.
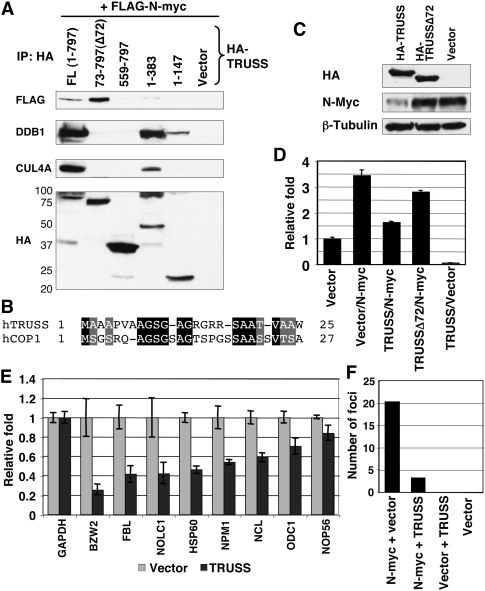
To further dissect TRUSS protein for domains responsible for Myc turnover, we generated a series of TRUSS deletion mutants. These mutants were coexpressed with N-Myc, and their interactions were assessed by coimmunoprecipitation, as well as with DDB1 and CUL4 (Fig. 4A). Full-length TRUSS interacts with Myc, DDB1, and CUL4A, consistent with the previous data (Fig. 1E). In contrast, deletion of the first 72 amino acids in TRUSS (Δ72) completely abolished the interaction with the DDB1–CUL4 complex. Notably, TRUSSΔ72 is still able to interact with N-Myc protein, implying that TRUSS recruits Myc independently of binding the DDB1–CUL4 complex. Amino acids 73–559 in TRUSS appear to be responsible for the interaction with Myc, since the C terminus (amino acids 559–797) does not interact with either Myc or the DDB1–CUL4 complex. We found that the first 72-amino-acid region of TRUSS is necessary but not sufficient to bind the DDB1–CUL4 complex because the 1–147 TRUSS mutant showed a much weaker interaction with the complex compared with the 1–383 TRUSS mutant (Fig. 4A). In addition, close examination of the TRUSS N terminus led to the identification of homology between amino acids 1–24 of TRUSS and 1–26 of hCOP1 (Fig. 4B), characterized previously as a DCAF for c-Jun destruction (Wertz et al. 2004). hCOP1 requires amino acids 1–26 to interact with DDB1–CUL4. We propose that both TRUSS and hCOP1 bind to DDB1–CUL4 complexes through this shared motif. To examine if the interaction between TRUSS and DDB1–CUL4 is necessary for TRUSS-mediated Myc degradation, full-length and mutant TRUSS were overexpressed in IMR-5 cells. Overexpression of full-length TRUSS reduced the Myc protein level compared with the control lysates, whereas there was no decrease in Myc level with TRUSSΔ72 transfection (Fig. 4C). In conclusion, the Myc-binding region in TRUSS is distinct from the domain required for the interaction with the DDB1–CUL4 complex, and this DDB1–CUL4 interaction with TRUSS is essential for the TRUSS-mediated degradation of Myc protein.
Figure 4.
The TRUSS N terminus is necessary for recruiting the DDB1–CUL4 complex and repressing transcription of Myc target genes. (A) HA-tagged full-length TRUSS or mutants containing different deletions were coexpressed with Flag-tagged N-myc. Expressed regions of TRUSS are indicated at the top. Immune complexes were resolved by SDS-PAGE and were blotted with the indicated antibodies. (B) Alignment between N-terminal regions of TRUSS and hCOP1. (C) Full-length and an N-terminal deletion (Δ72) of TRUSS were expressed in IMR-5 cells. Total-cell lysates were harvested and analyzed by immunoblotting. β-Tubulin is shown as a loading control. (D) Reporter construct harboring Myc-binding sites was analyzed in U2OS cells. Assays were performed 24 h after transfection. Luciferase activities were normalized to Renilla reporter activity as an internal control. Values were normalized to the vector control as 1. (E) RT–PCR analysis of Myc target genes in IMR-5 cells. A TRUSS expression vector was transfected to IMR-5 cells, and total RNAs were isolated for RT–PCR 36 h after transfection. GAPDH was used as an input control. (F) RK3E foci assay was carried out to measure the oncogenic potential of Myc in response to TRUSS. The numbers of foci from three independent transfections were averaged.
To begin to understand the in vivo role of TRUSS within the context of Myc function, we measured Myc transactivation in response to TRUSS using a luciferase reporter vector that harbors a tandem array of Myc/Max DNA-binding sites. In human U2OS osteosarcoma cells, N-Myc alone induced a 3.5-fold transactivation (Fig. 4D, lane 2). Coexpression of N-Myc with TRUSS compromised Myc-mediated transactivation, returning reporter expression to near basal levels (Fig. 4D, lane 3). Moreover, TRUSS expression alone reduced reporter activity below the basal level (Fig. 4D, lane 5), presumably by reducing endogenous Myc protein. Supportive of this, the expression of TRUSSΔ72, which is defective for binding to the DDB1–CUL4 complex, had little impact on reporter activity (Fig. 4D, lane 4). This result indicates that the TRUSS-mediated DDB1–CUL4 complex recruitment to Myc protein is crucial for the suppression of Myc transactivation. To verify this antagonistic effect in vivo, we measured Myc target gene expressions in IMR-5 cells. Ectopic expression of TRUSS reduced the mRNA level of known direct Myc target genes (Fig. 4E). We also expected that TRUSS is able to compromise cellular transformation induced by Myc. To test this, we assayed the transforming potential of Myc with TRUSS in E1A-immortalized rat kidney RK3E cells (Foster et al. 1999). We found that TRUSS coexpression significantly reduced the number of foci generated by Myc expression compared with Myc alone (Fig. 4F). Taken together, TRUSS can suppress Myc target gene transactivation and cellular transformation, presumably by reducing Myc protein levels.
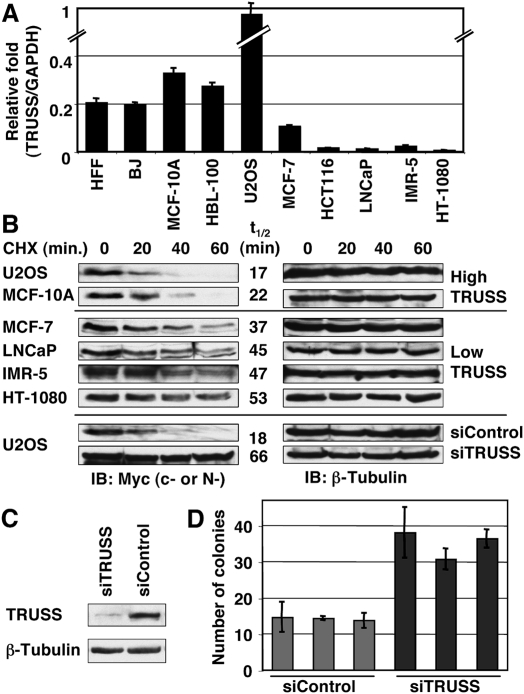
To explore if the relative level of endogenous TRUSS could be a significant determinant of Myc protein half-life, we compared TRUSS expression by RT–PCR across a panel of cell lines (Fig. 5A). We observed high TRUSS expression in all nonmalignant cell lines tested, including normal human foreskin fibroblast (HFF) cells, the HFF-derivative BJ cells, and nontumorigenic mammary epithelial cell lines MCF10A and HBL-100. In contrast, TRUSS expression was significantly lower to nearly undetectable in several cancer cell lines, including mammary carcinoma MCF-7, colorectal carcinoma HCT-116, prostate carcinoma LNCaP, neuroblastoma IMR-5, and fibrosarcoma HT-1080. One exception was the U2OS human osteosarcoma line, which had the highest TRUSS expression among the cell lines tested. If TRUSS promotes the instability of Myc proteins, then cell lines with high TRUSS expression might correlate with a short Myc half-life. Therefore, we measured the Myc protein half-life in all of the cell lines that had been assayed for TRUSS level (Fig. 5B). As anticipated, Myc protein turnover was particularly fast in high-TRUSS-expressing U2OS and MCF-10A cells (t1/2: 17 and 22 min, respectively), whereas the half-life was significantly longer in the TRUSS-deficient cell lines MCF-7, LNCaP, IMR-5, and HT-1080 (t1/2: 37, 45, 47, and 53 min, respectively). Myc protein levels were too low in HFF and BJ cells to determine an accurate half-life. To determine if TRUSS could be responsible for rapid Myc protein turnover, we studied U2OS cells that had the highest level of TRUSS expression and the shortest Myc half-life (Fig. 5A,B). siRNA transfection against TRUSS efficiently knocked down TRUSS expression within 48 h (Fig. 5C), and Myc protein half-life was significantly extended from 18 min to 66 min (3.7-fold) (Fig. 5B). The Myc protein half-life was unaffected with control siRNA transfection, and was similar to untransfected U2OS cells. TRUSS knockdown can also enhance the colony size of U2OS cells, presumably due to the increased Myc protein stability, which gives a growth advantage (Fig. 5D). These data demonstrate that TRUSS can control Myc protein half-life in various types of cells, and down-regulation of TRUSS expression is correlated with human malignancy and reduced Myc protein turnover.
Figure 5.
Down-regulation of TRUSS expression correlates with prolonged Myc half-life in cancer cells. (A) Total RNA from the indicated cell lines was analyzed by RT–PCR. TRUSS expression was normalized to GAPDH to compare the relative expression between lines. Data are the average of three independent assays. (B, top and middle) Half-life of Myc proteins from different human cell lines used in A are shown. Cycloheximide (CHX, 50 μg/mL) was added to the media, and whole-cell lysates were resolved by SDS-PAGE and were immunoblotted with anti-c-Myc antibody (anti-N-Myc antibody was used for IMR-5). After detection of Myc protein, the same blots were reprobed with anti-β-Tubulin antibody for a loading control. Myc half-lives calculated from image analysis are shown (t1/2). (Bottom) Knockdown of TRUSS expression prolongs c-Myc half-life in U2OS cells. c-Myc half-lives with t1/2 of 18 min in control cells and extended to 66 min after TRUSS knockdown. β-Tubulin served as a loading control. (C) Knockdown of TRUSS mRNA by siRNA in U2OS cells. (D) U2OS cells are embedded in soft-agar medium after siRNA transfection against TRUSS. After 20 d, colonies >150 μm are counted from three different areas. Three independent transfections were carried out.
Our results identify a novel Myc-specific E3 ligase complex containing TRUSS, DDB1, and CUL4, which can target both c-Myc and N-Myc. Among the different E3 ligase complexes implicated previously in Myc degradation, it has been proposed that Fbw7 is the primary mediator of Myc turnover, with the potential for its disruption in human cancer. Both the interaction and degradation of Myc by Fbw7 is dependent on phosphorylation of MycT58 by GSK3, and knockdown or genetic ablation of Fbw7 can prolong c-Myc half-life. However, stable expression of c-MycT58 mutants, which are defective for phosphorylation, or MycS, which lacks the Fbw7-binding domain, exhibits no net accumulation over c-MycWT (Chang et al. 2000; Gregory and Hann 2000; Hemann et al. 2005). Consistent with those observations, the TRUSS pathway is likely independent of GSK3 activity (Supplemental Fig. 2). Mutations in Fbw7 are found in some cancers, but cancers with wild-type Fbw7 exhibit similar Myc half-lives (O'Neil et al. 2007), making it unclear if Fbw7 mutations contribute directly to reduced Myc turnover. Fbw7 mutations can also activate Notch signaling, which enhances c-myc gene transcription (Thompson et al. 2007), making it important to separate transcriptional from post-transcriptional effects when assessing c-Myc protein levels.
The recognition of Myc by the TRUSS–DDB1–CUL4 complex appears to require multiple elements. For TRUSS, incorporation into the DDB1–CUL4 complex requires amino acids 1–72, similar to other DCAFs. For Myc, its recognition by TRUSS requires the B-HLH-LZ region, but Myc protein turnover is also dependent on redundant determinants within the N terminus involving the highly conserved MB1 and MB2 domains (Fig. 3). These data are consistent with previous mapping of determinants of c-Myc turnover in which MycS was unstable but MycSΔMB2 was stable (Gregory and Hann 2000). MycS is structurally and functionally similar to N-MycΔ1–73 (Cowling and Cole 2008), and the turnover of both proteins is dependent on MB2. The redundancy of Myc instability determinants and E3 ligases explains why mutation of the phosphorylation sites recognized by the Fbw7 E3 ligase complex or inactivation of Fbw7 itself does not always lead to elevated Myc protein levels (Chang et al. 2000; Gregory and Hann 2000; Hemann et al. 2005).
TRUSS levels are very low in all human tumor cells we surveyed, with the exception of U2OS. Coincidentally, this cell line has the shortest Myc protein half-life of any cancer cell line that we tested. Other normal cell lines also have short Myc protein half-lives, which correlate with higher TRUSS expression. Even small differences in Myc levels can affect transformation in response to several oncogenic pathways (Bazarov et al. 2001; Yekkala and Baudino 2007; Shachaf et al. 2008). Our data suggest that the down-regulation of TRUSS expression in cancer cells may play a significant role in tumor formation through enhanced Myc protein stability.
Materials and methods
Affinity purification of Flag-tagged protein complexes
Stable expression of Flag-N-myc or Flag-TRUSS was established in HeLa S3 cells. Cellular extracts were subjected to affinity purification using Flag-M2 beads (Sigma), and captured protein complexes were eluted using Flag peptide.
Immunoprecipitation and Western blot
Total cell lysates from IMR-5 and HeLa cells were prepared and immunoprecipitated with the indicated antibody. Western blots were carried out with antibodies detailed in the Supplemental Material.
Determination of protein turnover
Cells were plated, then transfected 24 h later with a TRUSS expression vector or empty vector. After 24 h, cycloheximide was added and lysates were analyzed by Western blot.
siRNA transfection and RT–PCR
Knockdown of TRUSS levels was achieved by transfection of an siRNA using standard protocols.
Luciferase reporter assay
Reporter assay was performed with Dual-Luciferase Reporter Kit with a 4× Myc/Max DNA-binding pGL3 reporter.
Transformation and soft agar assay
The RK3E transformation assay and soft agar assays were performed as described previously (Cowling et al. 2006).
Detailed information is available in the Supplemental Material.
Acknowledgments
This work was supported by a grant from the National Cancer Institute to M.D.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1920310.
Supplemental material is available at http://www.genesdev.org.
References
- Bazarov AV, Adachi S, Li SF, Mateyak MK, Wei S, Sedivy JM 2001. A modest reduction in c-myc expression has minimal effects on cell growth and apoptosis but dramatically reduces susceptibility to Ras and Raf transformation. Cancer Res 61: 1178–1186 [PubMed] [Google Scholar]
- Bonvini P, Nguyen P, Trepel J, Neckers LM 1998. In vivo degradation of N-myc in neuroblastoma cells is mediated by the 26S proteasome. Oncogene 16: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Chang DW, Claassen GF, Hann SR, Cole MD 2000. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol 20: 4309–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn SL, Salwen H, Quasney MW, Ikegaki N, Cowan JM, Herst CV, Kennett RH, Rosen ST, DiGiuseppe JA, Brodeur GM 1990. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene 5: 1821–1827 [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol 16: 242–252 [DOI] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2008. An N-Myc truncation analogous to c-Myc-S induces cell proliferation independently of transactivation but dependent on Myc homology box II. Oncogene 27: 1327–1332 [DOI] [PubMed] [Google Scholar]
- Cowling VH, Chandriani S, Whitfield ML, Cole MD 2006. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol Cell Biol 26: 4226–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn EM, Busch CM, Wright AP 1998. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol 18: 5961–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT, Ruppert JM 1999. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: Transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ 10: 423–434 [PubMed] [Google Scholar]
- Gregory MA, Hann SR 2000. c-Myc proteolysis by the ubiquitin–proteasome pathway: Stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 20: 2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, Eisenman RN 1984. Proteins encoded by the human c-myc oncogene: Differential expression in neoplastic cells. Mol Cell Biol 4: 2486–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436: 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegaki N, Bukovsky J, Kennett RH 1986. Identification and characterization of the NMYC gene product in human neuroblastoma cells by monoclonal antibodies with defined specificities. Proc Natl Acad Sci 83: 5929–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson S, Sundaresan S, Jin P, Francke U, Asker C, Sumegi J, Klein G, Sejersen T 1988. Chromosome localization and expression pattern of Lmyc and Bmyc in murine embryonal carcinoma cells. Oncogene 3: 679–685 [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD 2000. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10: 429–439 [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP 2003. Skp2 regulates myc protein stability and activity. Mol Cell 11: 1177–1188 [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P 2007. DCAFs, the missing link of the CUL4–DDB1 ubiquitin ligase. Mol Cell 26: 775–780 [DOI] [PubMed] [Google Scholar]
- Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, Sears RC 2006. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 20: 1572–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med 204: 1813–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Gentles AJ, Elchuri S, Sahoo D, Soen Y, Sharpe O, Perez OD, Chang M, Mitchel D, Robinson WH, et al. 2008. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res 68: 5132–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond SM, Terry JL, Colbert JD, Riches DW 2003. TRUSS, a novel tumor necrosis factor receptor 1 scaffolding protein that mediates activation of the transcription factor NF-κB. Mol Cell Biol 23: 8334–8344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond SM, Terry JL, Riches DW 2006. TRUSS, a tumor necrosis factor receptor-1-interacting protein, activates c-Jun NH(2)-terminal kinase and transcription factor AP-1. FEBS Lett 580: 4591–4596 [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. 2003. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci 101: 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23: 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekkala K, Baudino TA 2007. Inhibition of intestinal polyposis with reduced angiogenesis in ApcMin/+ mice due to decreases in c-Myc expression. Mol Cancer Res 5: 1296–1303 [DOI] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A 2008. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol 10: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]