Abstract
The N1-methyl-Adenosine (m1A58) modification at the conserved nucleotide 58 in the TΨC loop is present in most eukaryotic tRNAs. In yeast, m1A58 modification is essential for viability because it is required for the stability of the initiator-tRNAMet. However, m1A58 modification is not required for the stability of several other tRNAs in yeast. This differential m1A58 response for different tRNA species raises the question of whether some tRNAs are hypomodified at A58 in normal cells, and how hypomodification at A58 may affect the stability and function of tRNA. Here, we apply a genomic approach to determine the presence of m1A58 hypomodified tRNAs in human cell lines and show how A58 hypomodification affects stability and involvement of tRNAs in translation. Our microarray-based method detects the presence of m1A58 hypomodified tRNA species on the basis of their permissiveness in primer extension. Among five human cell lines examined, approximately one-quarter of all tRNA species are hypomodified in varying amounts, and the pattern of the hypomodified tRNAs is quite similar. In all cases, no hypomodified initiator-tRNAMet is detected, consistent with the requirement of this modification in stabilizing this tRNA in human cells. siRNA knockdown of either subunit of the m1A58-methyltransferase results in a slow-growth phenotype, and a marked increase in the amount of m1A58 hypomodified tRNAs. Most m1A58 hypomodified tRNAs can associate with polysomes in varying extents. Our results show a distinct pattern for m1A58 hypomodification in human tRNAs, and are consistent with the notion that this modification fine tunes tRNA functions in different contexts.
Keywords: N1-methyl-adenosine, tRNA, modification, microarray
INTRODUCTION
Over 100 types of RNA modifications have been identified in thousands of sites in tRNAs, rRNAs, mRNAs, snRNAs, and other RNAs (Bjork 1995; Maxwell and Fournier 1995; Rozenski et al. 1999; Ebhardt et al. 2005; Yu et al. 2005a,b). A human tRNA contains, on average, 13 modifications. A highly conserved modification in eukaryotic tRNA is N1-methyl-adenosine at position 58 (m1A58) (Fig. 1A). A58 forms a reverse Hoogsteen pair with conserved U54 in the tertiary structure of tRNA. Methylation at the N1 position of A58 introduces a positive charge at the elbow region of the tRNA tertiary structure without disrupting any hydrogen bonding interactions. This positive charge is thought to stabilize the structure of certain tRNAs on the basis of the crystal structure of yeast initiator-tRNAMet (Basavappa and Sigler 1991).
FIGURE 1.
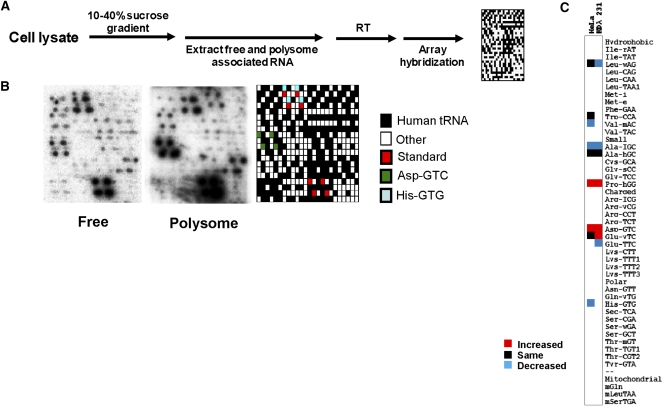
A microarray method to study m1A modifications in human tRNA. (A) Location of N1-methyl-A modification in tRNA. The RT primers are complementary to the region in red. (B) The amount of RT product is proportional to the amount of unmodified template tRNA. The primer is complementary to nucleotides 76–59, and all four dNTPs are included. The small amount of hypomodified yeast tRNAPhe at x = 0 is confirmed by another primer extension using the primer complementary to nucleotides 76–60 and a mixture of dATP, dTTP, dCTP, and ddGTP. A short exposure of the same lane on the left shows the separation of primer and primer stop at m1A58 (+1). (C) Scheme of a microarray experiment for m1A modification studies. (D) Detection of RT product using primers for individual human tRNA families. (E) Detection of RT products using a microarray. The array with a total human RNA sample (MCF10A cell line, left) shows much more products than the array with primers and standard tRNAs alone (middle). The array key is on the right. The array results correlates well with the gel analysis. For example, both methods show a very low amount of product for Met-i and a very large amount of product for Ala-hGC. (F) Specificity of the array method is indicated by the specific loss of the Ala-hGC signal when a large amount of the Ala-hGC oligo is added in array hybridization. The array key is on the right.
In yeast, inactivation of either subunit of the m1A58 methyltransferase (TRM6 and TRM61) results in growth arrest and cell death due to the rapid degradation of initiator tRNAMet lacking m1A58 modification (Anderson et al. 1998, 2000; Kadaba et al. 2004). Mutations of TRM6 and TRM61 cause derepression of GCN4 translation and its target genes because of the reduced level of initiator-tRNAMet (Cuesta et al. 1998; Calvo et al. 1999). The role of m1A58 in stabilizing the initiator-tRNAMet structure has been proposed to derive from its unique A54–A58 interaction (all other tRNAs in yeast have T54–A58 interaction) (Sigler 1975; Schevitz et al. 1979; Basavappa and Sigler 1991). Alternatively, the absence of m1A58 leads to the formation of a non-tRNA-like structure for the initiator-tRNAMet (Anderson and Droogmans 2005). Both explanations are consistent with the observation that three other tRNAs in yeast, 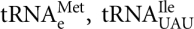 , and
, and  , were not destabilized by the lack of this modification (Anderson et al. 1998, 2000; Calvo et al. 1999).
, were not destabilized by the lack of this modification (Anderson et al. 1998, 2000; Calvo et al. 1999).
Most mammalian tRNAs contain m1A58 modification (Anderson and Droogmans 2005). Among the over 30 tRNAs sequenced, only two, rat tRNAAsp and tRNAGlu, have been reported to be unmodified at position 58 (Kuchino et al. 1981; Chan et al. 1982). The human homolog of the yeast tRNA m1A methyltransferase has been identified and it has been shown to have the same two subunit composition as the methyltransferase from yeast (Ozanick et al. 2005).
Very few reports exist, however, on whether the m1A58 modification is always complete for each tRNA species, or a fraction of some tRNA species may be hypomodified at this position inside cells. In principle, a varying level of m1A58 modification can be derived from low availability of cosubstrates (e.g., S-adenosyl-methionine used by the methyltransferase), regulated concentration or activity of the m1A58-modification enzyme, or an imbalance in the speed of tRNA folding and the modification reaction. Indeed, it has been reported that rat mammary adenocarninoma has reduced overall m1A58 levels in tRNA, and this reduction has been attributed to low methyltransferase levels in these tumors (Salas et al. 1982).
What are the pros and cons for cells to produce hypomodified tRNAs? On the neutral side, some modifications may be crucial only under certain conditions, so that the presence of hypomodified tRNAs is easily tolerated (Salas et al. 1982). On the negative side, hypomodified tRNAs may become dominant-negative inhibitors of tRNA aminoacylation or decoding of mRNA codons (Gustilo et al. 2008). On the positive side, hypomodified tRNAs represent multiple diverse tRNA species that may perform the same function at different efficiencies or may even perform distinct functions. Lack of a specific modification may weaken its interaction with the translation machinery, so that the hypomodified tRNA can be used more efficiently in cellular processes not directly involving ribosomes or elongation factors. For instance, hypomodified tRNAs may be more efficient activators of the GCN2 kinase in eukaryotes (Cuesta et al. 1998; Calvo et al. 1999).
Because hypomodified tRNAs may impact cell biology in many ways, it will be of interest to query which tRNAs are indeed hypomodified in the cell. Traditional methods involve the digestion of tRNA to fragments or mononucleosides, followed by chromatographic or mass spectrometric analysis (Kowalak et al. 1993; Yu et al. 1997; Qiu and McCloskey 1999; Zhao and Yu 2004). Although powerful, these methods generally require purification of individual tRNAs from a mixture of total tRNAs in some way, hence making it difficult to perform at the genomic scale. Furthermore, the presence or absence of certain modifications often impacts the purification procedure, complicating the interpretation for the analysis of hypomodification.
This work investigates m1A58 hypomodification in all human tRNAs and initial characterization of the functional significance of hypomodification in individual tRNA species. To simultaneously detect the presence of m1A58 hypomodified tRNA species in a total RNA mixture, we first perform primer extension using all primers complementary to the nucleotides 76–59 of all tRNAs. Because reverse transcriptase reads through m1A at a very low efficiency (Maden 1990; Maden et al. 1995; Kiss-Laszlo et al. 1996), only m1A58 hypomodified tRNAs can be extended at appreciable amounts. The primer extension products are then detected using a microarray method. We found that among the five cell lines examined, approximately one-quarter of human tRNA species contain hypomodified A58, and the pattern and the extent of hypomodified tRNAs are similar. We compare the hypomodified to m1A58-modified tRNAs in their relative association with polysome that approximates their involvement in translation. Our results indicate that cellular tRNAs are diverse in their modification status. The naturally present hypomodified tRNAs can have different efficiencies in polysome association, suggesting possible diversification in function.
RESULTS
A microarray method to analyze m1A58 hypomodification in tRNA
Our method makes use of the extremely low ability of reverse transcriptase (RT) to incorporate a base opposite to the N1-methyl-A modified nucleosides, thus leading to a stop during primer extension (Maden 1990; Maden et al. 1995; Kiss-Laszlo et al. 1996). Studies in Moloney murine Leukemia virus as well as HIV virus have shown that m1A plays a pivotal role in termination of reverse transcription in these viruses (Gilboa et al. 1979; Burnett and McHenry 1997; Renda et al. 2001). We utilized this inherent property of N1-methyl-A modification as the detection signal for hypomodification at A58 in human tRNA molecules.
To validate that reverse transcription can be used to quantitatively compare the extent of m1A58 in a tRNA, we performed primer extension using defined mixtures of purified yeast tRNAPhe, which contain this modification, and synthetic full-length yeast tRNA transcript, which is unmodified (Fig. 1B). The primer for tRNAPhe was designed to be complementary to the region encompassing the 3′ CCA to nucleotide 59 of the tRNA. This way, only tRNAs with unmodified A58 can be extended by RT. The reverse-transcription products range from RT stops at residue 37 where the wyobutosine (Y-base) is located to run-off reverse transcripts at the end of tRNA. Using 5′ 32P-labeled primers, all products from RT stops at Y37 to RT run-off were quantified. We found that the amount of product increases linearly from very low (x = 0, y = 0.04 when only yeast tRNAPhe isolated from the cell was present) to very high (x = 1, y = 0.88 when only tRNAPhe transcript was present). The small amount of product detected in the RT reaction using the yeast tRNAPhe isolated from the cell was derived from the presence of m1A58 hypomodified tRNAPhe, as validated using a different primer extension method (Fig. 1B). Detection of a small amount of m1A58 hypomodified tRNAPhe is also consistent with m1A58 being nonessential for elongator tRNAs in yeast (Anderson et al. 1998). This result indicates that reverse transcription is a useful strategy for comparative analysis of m1A58 modification in tRNAs.
We applied a multistep strategy to study m1A58 hypomodification at the genomic scale (Fig. 1C). First, total RNA was isolated from human cell lines using standard methods. Two bacterial tRNA standards (E. coli tRNALys, E. coli tRNATyr) were then added to the total RNA. These bacterial tRNAs do not contain m1A58 (Sprinzl and Vassilenko 2005), and hence serve as internal loading and array hybridization controls between microarray measurements. Second, a mixture of RT primers were added that are complementary to nucleotides 76–59 for 40 families of human nuclear-encoded tRNAs (Dittmar et al. 2006) and three human mitochondrial-encoded tRNAs that are known to contain m1A58 modifications (Florentz et al. 2003), making 45 primers in all, including those for the E. coli tRNA standards. Primer extension was carried out using a low amount of Avian Myeloblastosis Virus (AMV) RT. The RT reaction was performed with [α-32P]TTP to enable radioactive labeling of the RT products. After the RT reaction, the reaction mixture was treated with RNase H to remove the tRNA templates. The mixture was then filtered through size-exclusion columns to remove unincorporated [α-32P]TTP in order to reduce the background of array hybridization. Following phenol-chloroform extraction and ethanol precipitation, the RT product mixture was hybridized directly on a custom-printed tRNA microarray (Netzer et al. 2009). The hybridized array was exposed to a PhosphorImager in order to visualize the RT products.
The microarray was designed with DNA probes having the same sequence as tRNA, hence complementary to the RT products. A total of 96 probes repeated eight times each were printed on the array. We applied stringent hybridization conditions to ensure that only long transcripts resulting from primer extension would hybridize on the array. In general, these products range from RT stops at nucleotide 37 (∼40-nucleotides [nt] long) where many human tRNAs contain elaborate modifications to RT run-off products (75–90-nt long) (Fig. 1D). Excess primers themselves (∼18 mers) are not detected because the 32P-labeling requires primer extension. Using this design, numerous RT products were detected in a total RNA isolated from cultured human cells (Fig. 1E). In total, 136 spots showed signals above background, corresponding to 17 RT products.
We performed three control experiments to ensure that the majority of the signals on the arrays are derived from m1A58 hypomodified human tRNAs. First, we carried out the same experiment using just the primer sets plus the E. coli tRNA standards (Fig. 1E). In this case, only 40 spots corresponding to five RT products showed signals above background. As expected, the most intense signals (16 spots corresponding to two RT products) are derived from the two E. coli tRNAs. A total of 24 spots corresponding to three RT products can be attributed to products derived from the primers themselves; these products are not considered further in our subsequent analysis. Second, we tested the specificity of array hybridization by repeating the hybridization of the RT product in the presence of a molar excess of the Ala-hGC probe. The presence of this probe should compete only with the RT product derived from the Ala-hGC tRNAs. Indeed, signal from Ala-hGC tRNA is selectively diminished in the presence of this probe (Fig. 1F). Third, we carried out RT reaction with one primer at a time and analyzed the reaction product on denaturing PAGE (Fig. 1D). The reaction products detected in this way correspond nicely with those detected on the array. For example, high signal from Ala-hGC tRNA is detected by both array and gel methods. In contrast, signal from initiator-tRNA (Met-i) was at or below background in both array and gel experiments.
m1A58 hypomodification patterns in human cell lines
In order to obtain comprehensive information on m1A58 hypomodification patterns, we analyzed the total RNA extracted from five cell lines (Fig. 2). These cell lines represent a wide range of cell types: HeLa (cervical cancer), HEK293T (embryonic kidney), MDA-MB-231 (breast cancer, metastasized to lungs), MCF10A (human breast epithelium), and Neuroblastoma (adrenal gland).
FIGURE 2.
Profiling m1A58 hypomodification in human cell lines. (A) Hypomodified tRNA species in HeLa cells; the blue arrow indicates initiator-tRNAMet, which is completely modified at A58. The relative signal (y-axis) = 1 are those of the two tRNA transcripts that are unmodified at A58. For this work, we defined that y < 0.5 has low, y = 0.5-2 has medium, and y > 2 has high levels of hypomodified tRNA. (B) Array image from cell line HeLA showing the signals for Asp-GTC and Glu-yTC, and the absence of signal for Met-i. (C) Heat map showing ranges (high, medium, low) of m1A58 hypomodification in five human cell lines (HeLa, HEK 293T, MDA-MB-231, MCF-10A, and Neuroblastoma). The blue arrow indicates initiator-tRNAMet (negative control) and the orange arrows indicate tRNAAsp and tRNAGlu (positive controls). (D) Determination of hypomodification at m1G9 for tRNAGln,  and tRNAPro. The detection limit for hypomodification is ∼2% in this experiment. The gel shows the primer extension reaction for tRNAGln in the total RNA from MDA-MB-231.
and tRNAPro. The detection limit for hypomodification is ∼2% in this experiment. The gel shows the primer extension reaction for tRNAGln in the total RNA from MDA-MB-231.
tRNA from HeLa cells gave detectable RT signals for nine among 40 nuclear-encoded tRNA families and one among three mitochondrial-encoded tRNAs (Fig. 2A). Among the high signals were those obtained from tRNAAla(hGC, h = I/C/U) and tRNAPro(hGG). Both probes encompass several isoacceptors among Ala and Pro-tRNA families (IGC/CGC/UGC for Ala, and IGG/CGG/UGG for Pro). These isoacceptors have very similar sequences and cannot be separately detected in our array method (Dittmar et al. 2006). Three tRNAs with detectable signals are tRNAAsp and two isoacceptor families of tRNAGlu; these tRNAs have been shown previously to be undermodified at A58 in rat cells (Kuchino et al. 1981; Chan et al. 1982), and therefore likely to be hypomodified in human cells as well. On the other hand, no signal was detected for the initiator-tRNAMet (Fig. 2A,B). In yeast, m1A58 is essential for the stability of the initiator-tRNAMet, and this requirement seems to extend to human initiator-tRNA as well. These results further validate our microarray approach in the detection of specific m1A58 hypomodified tRNAs.
It is much more difficult to determine the absolute fraction of m1A58 hypomodification for each tRNA in the array experiment. Major variables include the differences in the efficiency of the RT reaction and array hybridization for each tRNA. For normalization purposes, we denoted y = 1.0 as the array signals detected from the two E. coli tRNA standards. This value corresponds to 0.066 pmol of each E. coli tRNA standard added to 1 μg of total human RNA (Fig. 2A). By this measure, we group the hypomodified tRNA species in three groups: high (y > 2), medium (y = 0.5–2), and low (y < 0.5). For HeLa cells, the high group includes one member (Ala-hGC), the medium group has seven members (Pro-hGG, Asp-GTC, Glu-yTC, Glu-TTC, Leu-wAG, Val-mAC, His-GTG), and the low group has two members (Trp-CCA, mitochondrial-Gln).
The pattern of the m1A58 hypomodified tRNAs is similar in five human cell lines (Fig. 2C). The same set of 10 tRNAs hypomodified in HeLa cells is hypomodified in all other cell lines as well, and the extent of hypomodification varies little. In all cases, no detectable signal was obtained for the initiator-tRNA (Fig. 2C, indicated by the blue arrow), and detectable signals were always obtained for Asp and Glu-tRNAs (Fig. 2C, indicated by the orange arrows). These results suggest that human cells have a basal pattern of m1A58 hypomodification, and this basal pattern includes ∼25% of human tRNA species.
In addition to these 10 common tRNAs, other cell lines appear to have one (HEK293, MCF10A), two (MDA-MB-231), and four (Neuroblastoma) more tRNAs that are hypomodified. Hypomodification signals from these noncommon tRNAs generally belong to the low group. They may play some unique roles that are specific for each cell type.
The MCF10A and MDA-MB-231 cell lines are often used for comparative studies between nontumorigenic versus tumorigenic breast cells (Chen et al. 1997; Gruber and Pauli 1999; Mehta et al. 2004). The nuclear-encoded tRNAs are overexpressed by approximately twofold, on average, in MDA-MB-231 over MCF10A cells (Pavon-Eternod et al. 2009). These cell lines have a similar pattern and extent for m1A58 hypomodification, therefore suggesting that the extent of hypomodification is maintained in breast cancer cells. Interestingly, MDA-MB-231 appears to contain 1.7–1.8 times more m1A58 methyltransferase subunits, TRM6 and TRM61, compared with MCF10A (Fig. 3; data not shown). This result suggests that tRNA overexpression is also accompanied by an increase in methyltransferase levels, presumably to ensure that no significant amount of m1A58 hypomodified tRNA accumulates in the cell.
FIGURE 3.
Effect of siRNA knockdown of m1A-methylase subunits, TRM6 and TRM61. (A) Western blot showing efficiency of siRNA treatment of MDA-MB231 cells. The siRNA treatment causes the decrease in protein level after 3 d of treatment. GAPDH level is used as a control. (B) Growth of control versus TRM6 and TRM61 siRNA-treated cells. (C) Representative array image from control and siTRM6-treated cells. (D) Heat map of hypomodification in control versus TRM6 and TRM61 knockdown cells. Purple arrows show tRNA species with significant increase in hypomodification in the siRNA-treated cells. Blue arrow indicates initiator-tRNAMet, which, as expected, is not hypomodified at all times. (E) Heat map of relative tRNA abundance of the control and siRNA-treated cells shows no change in tRNA abundance in all cases.
In order to determine whether human tRNAs are also hypomodified at other positions, we performed primer extension to examine the known 1-methyl-G modification at position 9 for three tRNAs (Bjork et al. 2001; Jackman et al. 2003) in three cell lines (Fig. 2D). For m1G9 modification, the primers are complementary to nucleotides 33–14 of these tRNAs. Using three dNTPs and one ddNTP, primer extension could generate two products, the shorter product corresponding to the reverse transcriptase stop at the m1G9 modification, and the longer product corresponding to the incorporation of the dideoxy-nucleotide (Sanger et al. 1977). The longer product is only observed when the tRNA is hypomodified at m1G9. Like m1A58, we found that m1G9 is also hypomodified at varying amounts for each tRNA, and each tRNA has a similar hypomodification pattern among the three cell lines. This result suggests that human cells contain distinct patterns of tRNA hypomodification at distinct modification sites.
siRNA knockdown of m1A58 methyltransferase subunits
To examine the consequence of m1A58 hypomodification and further establish our array method as a suitable tool for functional studies, we performed siRNA experiments on either of the human m1A58 methyltransferase subunits, TRM6 or TRM61 (Ozanick et al. 2005). We chose to carry out this experiment with MDA-MB-231 cells because tRNAs are known to be overexpressed in these cells (Pavon-Eternod et al. 2009), and this high amount of tRNA may be more sensitive to variations in this modification enzyme. siRNA treatment reduced the mRNA levels of both methyltransferase subunit genes by ∼10-fold compared with the siRNA control (data not shown); however, the reduction in protein level is less than twofold (Fig. 3A). This difference between changes in mRNA and protein levels of the m1A58 methyltransferase may be either derived from an unknown translational regulation for this multisubunit enzyme, or this enzyme may turn over very slowly. Nevertheless, this moderate reduction of the methyltransferase levels led to slower cell growth as compared with siRNA-control cells after 3 d (Fig. 3B), suggesting that this enzyme plays a role in cell proliferation. Observing the growth phenotype only after 3 d may be related to the longevity of tRNAs in mammalian cells, so that a time scale of days is needed to produce a significant change in the tRNA population.
We then determined the changes in the m1A58 modification levels in cells treated with siRNA targeted for either subunit of this methyltransferase (Fig. 3C,D). We found significantly higher levels of hypomodification for eight tRNAs that are already hypomodified in control cells. Again, initiator-tRNAMet remains fully modified in all cases. Furthermore, very little new hypomodified tRNA species are present significantly above background in the siRNA-treated cells. On the other hand, the amount of all tRNA species as measured by our standard tRNA microarray method using Cy3 and Cy5 labeled total tRNA samples (Dittmar et al. 2006; Pavon-Eternod et al. 2009, 2010; Zhou et al. 2009) in the siRNA-treated methyltransferase cells remains essentially unchanged as compared with siRNA-control cells after 3 d (Fig. 3E). A plausible explanation of the unchanging level of these tRNAs is that since the reduction of the methyltransferase level is less than twofold, a sufficient amount of the methyltransferase enzyme remains after siRNA treatment to still enable full modification of these tRNAs. Compared with the m1A58 hypomodified tRNAs, the fully modified tRNAs may be more efficient substrates for the methyltransferase enzyme.
There are several plausible explanations for the apparent slow growth phenotype. The phenotype may be derived from an overproduction of hypomodified tRNA species that is toxic to the cell. The phenotype may also be due to impairment of additional functions of the methyltransferase enzyme in the cell. Furthermore, the methyltransferase may act on yet-to-be-identified substrates that are not modified and are required for efficient cellular replication. Regardless, these results suggest that the presence of m1A58 hypomodified tRNAs in untreated cells are due to their inherent stability and not due to insufficient cellular tRNA degradation capacity.
Polysome association of m1A58 hypomodified tRNAs
Participation in translation for a tRNA requires its association with the polyribosomes. m1A58 modification could have a strong effect for polysome association because this region of tRNA interacts with the ribosome during translation (Ramakrishnan 2002; Korostelev and Noller 2007). In order to determine how m1A58 hypomodified tRNAs associate with polysomes, we extracted total RNA from free (low-sucrose density) and polysome-bound (high-sucrose density) fractions in a sucrose density gradient and performed RT reaction with these RNAs after the addition of E. coli tRNA standards (Fig. 4A).
FIGURE 4.
Effect of m1A hypomodification on polysome association. The analysis was performed with HeLa and MDA-MB231 cells. (A) Scheme of the procedure to measure polysome association. (B) Array images showing differences between free and polysome samples from HeLa cells. The array key is on the right. (C) Heat map of polysome association. Color coding shows the comparison of polysome association of m1A58 hypomodified tRNA with fully modified tRNA.
In HeLa cells, signals for eight of nine nuclear-encoded tRNAs are present in both free and polysome fractions (Fig. 4B,C). The mitochondrial tRNAGln was not detected in this experiment. We analyzed the array data by first dividing the amount of the RT product from polysome fractions by the free-tRNA fraction for each tRNA. This ratio was then normalized to the signals from the two E. coli tRNA standards. Among the nine tRNAs, the median value generated this way is 0.83 (mean value = 0.90 ± 0.59) which is close to the value of 1.0. Only four of the nine tRNAs show ratios within 1.5-fold of the median value generated in this way. Assuming the same experimental variation of 1.5-fold when comparing multiple microarray data, this result suggests that m1A58 hypomodification has a significant effect on ribosome association of these tRNAs. Two of the nine tRNAs, Pro-hGG and Asp-GTC, show ratios approximately twofold above median. For these two tRNAs, their m1A58 hypomodified species seem to be enriched in polysome. In contrast, three of the nine tRNAs, Val-mAC, Ala-IGC, and His-GTG, show ratios more than twofold below median. The hypomodified version of these three tRNAs seems to be depleted in polysomes.
In MDA-MB-231 cells, signals for seven out of 12 nuclear-encoded tRNAs are present in both free and polysome fractions (Fig. 4C). Polysome enrichments or depletion for these detectable hypomodified tRNAs are similar between these two cells lines. In particular, Pro-hGG and Asp-GTC also show high ratios in polysome-associated fractions in MDA-MB-231 cells, validating the similar result observed in HeLa cells.
DISCUSSION
Eukaryotic tRNAs contain a large number of post-transcriptional modifications. Most modifications are not essential for cell viability, and each modification can incrementally affect tRNA function at multiple levels, including maturation, aminoacylation, and ribosome utilization (Bjork 1995). Despite many years of studies, an open question remains on the identity and frequency of hypomodified tRNAs that are present in the cell. Using a functional genomics approach, we demonstrate in this work that ∼25% of human tRNA species are hypomodified at the conserved A58 nucleotide. The identity of m1A58 hypomodified tRNA species is similar in five human cell lines examined, suggesting that human cells share a common pattern of m1A58 hypomodification. As positive controls for our genomic approach, we identified that the initiator-tRNAMet is fully modified, and the tRNAAsp and tRNAGlu species are hypomodified in all five cell lines, in agreement with the literature results (Kuchino et al. 1981; Chan et al. 1982). It is an open question as to whether m1A58 modification is essential for the maturation of the majority of human tRNAs or whether fully modified tRNAs are simply more efficient substrates for the m1A58 methyltransferase.
The human tRNA species that are m1A58 hypomodified are distributed among distinct tRNA families. At this time, no obvious pattern derived from common sequence or alternate secondary structure motifs can be deduced for these tRNAs (data not shown). Nevertheless, the presence of m1A58 hypomodified tRNAs in human cells clearly indicate that this modification is not required for the maturation of all tRNAs. Three of the 22 mitochondrial-encoded tRNAs also undergo m1A58 modifications (Sprinzl and Vassilenko 2005), although, to our knowledge, the m1A58 methyltransferase in human mitochondria has not been identified. Here, we tested whether m1A58 hypomodification also occurs in these three mitochondrial tRNAs, two of which (mLeu-UAA; mGln) possess all features needed for a classical cloverleaf secondary structure (Helm et al. 1998; Florentz et al. 2003; Sprinzl and Vassilenko 2005). Only mitochondrial tRNAGln among these three tRNAs is hypomodified, and this hypomodification pattern is again conserved among the seven cell lines tested. The functional significance of the m1A58 hypomodificatioin in mito-tRNAGln is unclear.
Although m1A58 hypomodified tRNAs are inherently stable in human cells, in order to participate in translation, these tRNAs have to be aminoacylated. Because A58 is conserved in all tRNAs, this residue is not used directly as an identity element for aminoacyl-tRNA synthetases (Giege et al. 1998). Some synthetases, however, do use residues 59 and 60 as a part of their identity elements; for example, the E. coli aminoacyl-tRNAPhe synthetase. m1A58 modification with an extra positive charge and the presence of an extra methyl group may influence the charging efficiency of these synthetases.
We do not know at this time why human cells maintain a subset of tRNAs that are m1A58 hypomodified. However, several hypotheses can be proposed for their utilization.
Neutral/Null hypothesis: The hypomodified tRNAs are less-efficient substrates for the m1A58 methyltransferase enzyme compared with the other, fully modified tRNAs. Since most of the hypomodified tRNAs are still functional in polysome association, their presence is simply tolerated. This hypothesis would be consistent with the observed growth phenotype being derived from an unknown m1A58 methyltransferase function not related to tRNA methylation.
Nontranslation hypothesis: The hypomodified tRNAs are used for a function not involving mRNA decoding. Because N1-methylation at A58 introduces a positive charge in the elbow region of tRNA, the absence of this modification in some tRNAs may decrease its binding by the elongation factor eEF1α or by the ribosome. This decreased binding efficiency reduces competition by the major cellular components of translation, allowing the hypomodified tRNAs to be used as ligands for other proteins, e.g., the protein kinase GCN2 (Hinnebusch 2005).
Another possibility for keeping hypomodified tRNAs in the cell is to provide a reservoir for tRNAs that may be readily altered in stress response. For instance, many types of stresses in yeast and in mammalian cells generate tRNA fragments derived from cleavage of the anticodon loop (Thompson and Parker 2009a,b; Yamasaki et al. 2009). The amount of these tRNA fragments is small and does not affect the abundance of these tRNAs, yet these fragments alone can lead to translational repression in a mammalian cell (Yamasaki et al. 2009).
Speed-accuracy modulation hypothesis: Fully modified tRNAs often increase translation accuracy (Bjork 1995), but the increase in accuracy can reduce the overall speed of translation. The utilization of some hypomodified tRNAs may be useful to reduce the “bottleneck” in translation, allowing faster translation speed, and to balance that with sufficient accuracy (Drummond and Wilke 2008).
This brief discussion implies that the function of m1A58 modification may not be singular. It is plausible that m1A58 modification, even though occurring at the same position in all tRNAs, can confer different functions depending on the tRNA species. Additional possibilities for m1A58 function include dynamic changes of its hypomodification pattern as a function of cell cycle, cell density, or cell states. Furthermore, hypomodification patterns may also change in response to various types of stresses that cells may encounter. The ability to measure the hypomodification pattern at the genomic level as described here should provide a valuable tool to further examine dynamic variations and potential functions of this modification for individual tRNAs.
MATERIALS AND METHODS
RNA preparation
Total RNA from the HeLa, HEK293, MCF10A, MDA-MB-231, and Neuroblastoma cell lines were obtained using the mirVana miRNA Isolation Kit (Ambion, http:/www.ambion.com) according to manufacturer's manual. The two tRNA standards, E. coli tRNALys (No. R6018) and E. coli tRNATyr (No. R0258) and the yeast tRNAPhe (No. R4018) were purchased from Sigma-Aldrich (http:/www.sigmaaldrich.com) and used without further purification. The yeast tRNAPhe transcript was obtained by T7 RNA polymerase transcription. The DNA oligos for primer extension were synthesized by Integrated DNA Technologies (IDT, http:/www.idt.com). All DNA primers were purified by denaturing PAGE and stored in water.
Primer extension for array studies
The primer extension was carried out with 2 μg of total RNA, 1 pmol of each primer, 0.13 pmol each of E.coli tRNALys and tRNATyr in a total volume of 10 μL. The RNA and primers were annealed by heating at 90°C for 4 min with annealing buffer (50 mM Tris-HCl at pH 7.5 and 3 mM KCl) and the mixture was allowed to gradually cool to 40°C over a period of 30 min. The RT reaction was carried out at 43°C with 1 mM each dATP, dGTP, dCTP, 0.5 mM dTTP, and spiked with radioactive [32P]TTP and 2 U of AMV RT (USB, Inc.) in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.5 mM Spermidine, and 10 mM DTT for 20 min. The mixture was further treated with 0.1 U/μL RNase H (from Epicenter Technologies) at 43°C for 10 min, filtered though G-25 columns to remove excess [32P]TTP (GE Technologies), followed by phenol:chloroform extraction (to remove all enzymes) and ethanol precipitation. The pellet was dissolved in 10 μL of water, and 1 μL was loaded on a denaturing polyacrylamide gel containing 7 M urea to visualize RT products.
Primer extension for yeast tRNAPhe and m1G9 modification studies
The primer extension was carried out in a total volume of 10 μL with 1 μg of total RNA and 1 pmol of designated 5′ 32P-labeled primer. For yeast tRNAPhe, the primer was complementary to nucleotides 76–60; for human tRNAs, the primers were complementary to nucleotides 33–14 of the corresponding tRNA. The RNA and primers were annealed by heating at 90°C for 4 min with annealing buffer (50 mM Tris-HCl at pH 7.5 and 3 mM KCl) and the mixture was allowed to gradually cool to 40°C over a period of 30 min. Each RT reaction was carried out at 43°C with 1 mM each of three dNTPs (dA/C/TTP for yeast tRNAPhe and human tRNAGln; and dA/C/GTP for human initiator-tRNAMet and human tRNAPro), 1 mM of one ddNTP (ddGTP for yeast tRNAPhe and human tRNAGln; and ddTTP for human initiator-tRNAMet and human tRNAPro), and 1 U of AMV RT (USB, Inc.) in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.5 mM Spermidine, and 10 mM DTT for 20 min. The mixture was treated with 0.1 U/μL RNase H (Epicenter Technologies) at 43°C for 10 min. The reaction was quenched with the addition of an equal volume of 9 M urea/50 mM EDTA, and the RT product separated on denaturing polyacrylamide gels.
Polysome association measurements
To isolate RNA for polysome analysis, the cells with a confluence of 30%–50% were aspirated and washed twice with 1X PBS. Cells were scraped off the plates after adding lysis buffer containing a 2.5% (v/v) mixture of Nonidet P-40 in 0.2 M sucrose in 50 mM Tris (pH 7.5), 100 mM ammonium acetate, 5 mM MgCl2, and 3 mM β-mercaptoethanol was added to the plates. The cell lysate was centrifuged at 4°C for 30 min. The clear supernatant was collected and was loaded on top of a 10%–40% sucrose gradient. The gradient was prepared by dissolving ultra pure sucrose (Sigma Aldrich) in a gradient buffer consisting of the same buffer as above.
Hybridization and microarray analysis
The tRNA microarray for m1A58 modification contained a total of 95 oligonucleotide probes plus one buffer only slot repeated eight times each. The array contains 59 sense probes for human chromosomal-encoded tRNAs, 22 sense probes for human mitochondrial-encoded tRNAs, plus eight antisense tRNA probes from hybridization controls and six nonhuman tRNA probes as normalization controls. The oligonucleotides were printed manually to generate large array spots necessary for radioactive detection (Netzer et al. 2009). Array hybridization has been described previously for the analysis tRNA abundance and charging levels (Dittmar et al. 2005, 2006). A brief description follows. Immediately prior to hybridization, microarray slides were immersed in boiling water for 2 min to remove undesired particles and uncross-linked oligos. The reverse-transcribed RNA samples (corresponding to ∼2 μg of total RNA) were dissolved in microarray hybridization buffer (Sigma Aldrich) containing 20 μg of salmon sperm DNA and 10 μg of poly(A). The sample was applied to the Hybridization chamber of the GeneTAC Hyb4 station (Genomic solutions). The following program was used for hybridization: 75°C (2 min), 60°C (probe introduction), 90°C (5 min), 60°C (16 h). The slides were then washed on the Hyb4 station twice with 2XSSC, 0.1% (w/v) SDS at 50°C, twice with 0.1X SSC, 0.1% (w/v) SDS at 42°C, and twice with 0.1X SSC at 42°C. Slides were then removed from the station, rinsed with 0.1XSSC, and dried by centrifugation.
Microarray slides were imaged using a FUJI BAS scanner. 32P-intensities were quantified and background-subtracted using Fuji BAS software. Median values for eight replicate spots were used for further analysis.
Cell culture, siRNA transfection, and cell growth
MDA-MB-231 cells were obtained from the American Type Culture Collection and were maintained in RPMI 1640 (Mediatech) with 10% FBS (GIBCO), 100 U/mL Penicillin, and 100 μg/mL Streptomycin. Transfection of control siRNA (Dharmacon), TRM61, and TRMT6 siRNA (Santa Cruz Biotechnology) by Lipofectamine RNAiMAX (Invitrogen) was performed according to the manufacturer's protocol in 96-, 24-, and 6-well format. The cells were plated in RPMI 1640 with 10% FBS for 24 h. The cells with a confluence of 30%–50% were washed twice and changed to Opti-MEM (GIBCO) without serum before transfection was performed. For a 24-well format transfection, 12 nmol siRNA (final concentration of 20 nM) and 1.25 μL of RNAiMAX were used. The medium was changed back to RPMI 1640/10% FBS after 24 h. Cell growth curve was measured by CellTiter-Blue Cell Viability Assay (Promega) with an incubation time of 3 h at the time point of measurement.
qRT-PCR and Western blot analysis
Total RNA was isolated with the mirVana miRNA Isolation Kit (Ambion). qRT-PCR was carried out using the SuperScript III Platinum One-Step qRT-PCR System (Invitrogen) according to the manufacturer's protocol in the Applied Biosystems AB7300 system.
Western blot analysis was performed in a 4%–12% Tris-HCl gel, with a 1:400 dilution of TRMT6 (H-198), and TRM61 (C145I165) antibody (Santa Cruz Biotechnology) with the corresponding 1:1000 dilution of HRP conjugated secondary antibody (GE Healthcare).
ACKNOWLEDGMENTS
We thank Dr. J. Piccirilli for insightful discussions. We also thank all the reviewers for their suggestions and insightful comments. This work was supported by an NIH grant (GM88599 to T.P. and C.H.), and M.P.-E. was supported by a Ruth Kirschstein Pre-Doctoral Fellowship from the NIH (1F31CA139968).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2057810.
REFERENCES
- Anderson JT, Droogmans L 2005. Biosynthesis and function of 1-methyladenosine in transfer RNA. In Fine-tuning of RNA functions by modification and editing (ed. Grosjean H), pp. 121–140 Springer, Berlin [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB 1991. The 3 A crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J 10: 3105–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure, biosynthesis, and function (ed. Soll D, RajBhandary U), pp. 165–206 ASM Press, Washington, DC [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BP, McHenry CS 1997. Post-transcriptional modification of retroviral primers is required for late stages of DNA replication. Proc Natl Acad Sci 94: 7210–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio MT, Hinnebusch AG, Tamame M 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol 19: 4167–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JC, Yang JA, Dunn MJ, Agris PF, Wong TW 1982. The nucleotide sequence of a glutamate tRNA from rat liver. Nucleic Acids Res 10: 4605–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Guo X, Derguini F, Gudas LJ 1997. Human breast cancer cells and normal mammary epithelial cells: Retinol metabolism and growth inhibition by the retinol metabolite 4-oxoretinol. Cancer Res 57: 4642–4651 [PubMed] [Google Scholar]
- Cuesta R, Hinnebusch AG, Tamame M 1998. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 148: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T 2005. Selective charging of tRNA isoacceptors induced by amino acid starvation. EMBO Rep 6: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2: e221 doi: 10.1371/journal.pgen.0020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Thi EP, Wang MB, Unrau PJ 2005. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci 102: 13398–13403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M 2003. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci 60: 1356–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege R, Sissler M, Florentz C 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Pauli BU 1999. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res 59: 5488–5491 [PubMed] [Google Scholar]
- Gustilo EM, Vendeix FA, Agris PF 2008. tRNA's modifications bring order to gene expression. Curr Opin Microbiol 11: 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Brulé H, Degoul F, Cepanec C, Leroux J-P, Giegé R, Florentz C 1998. The presence ofmodified nucleotides is required for cloverleaf foldingof a human mitochondrial tRNA. Nucleic Acids Res 26: 1636–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Noller HF 2007. The ribosome in focus: New structures bring new insights. Trends Biochem Sci 32: 434–441 [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA 1993. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res 21: 4577–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Shindo-Okada N, Ando N, Watanabe S, Nishimura S 1981. Nucleotide sequences of two aspartic acid tRNAs from rat liver and rat ascites hepatoma. J Biol Chem 256: 9059–9062 [PubMed] [Google Scholar]
- Maden BE 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 39: 241–303 [DOI] [PubMed] [Google Scholar]
- Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77: 22–29 [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ 1995. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934 [DOI] [PubMed] [Google Scholar]
- Mehta K, Fok J, Miller FR, Koul D, Sahin AA 2004. Prognostic significance of tissue transglutaminase in drug resistant and metastatic breast cancer. Clin Cancer Res 10: 8068–8076 [DOI] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. 2009. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT 2005. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T 2009. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Wei M, Pan T, Kleiman L 2010. Profiling non-lysyl tRNAs in HIV-1. RNA 16: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, McCloskey JA 1999. Selective detection of ribose-methylated nucleotides in RNA by a mass spectrometry-based method. Nucleic Acids Res 27: e20 doi: 10.1093/nar/27.18.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V 2002. Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
-
Renda MJ, Rosenblatt JD, Klimatcheva E, Demeter LM, Bambara RA, Planelles V 2001. Mutation of the methylated
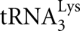 residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J Virol 75: 9671–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J Virol 75: 9671–9678 [DOI] [PMC free article] [PubMed] [Google Scholar] - Rozenski J, Crain PF, McCloskey JA 1999. The RNA Modification Database: 1999 update. Nucleic Acids Res 27: 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas CE, Uschmann BD, Leboy PS 1982. Methyl-accepting RNA in 13762 mammary adenocarcinoma correlated with low adenine methyltransferase levels. Cancer Res 42: 5004–5009 [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR 1977. DNA sequencing with chain-terminating inhibitors. Natl Acad Sci 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevitz RW, Podjarny AD, Krishnamachari N, Hughes JJ, Sigler PB, Sussman JL 1979. Crystal structure of a eukaryotic initiator tRNA. Nature 278: 188–190 [DOI] [PubMed] [Google Scholar]
- Sigler PB 1975. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng 4: 477–527 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009a. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009b. Stressing out over tRNA cleavage. Cell 138: 215–219 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA 1997. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3: 324–331 [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X 2005a. Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y-T, Terns RM, Terns MP 2005b. Mechanisms and functions of RNA-guided RNA modification. In Fine-tuning of RNA modifications by modification and editing (ed. Grosjean H), pp. 223–262 Springer, Göteborg, Sweden [Google Scholar]
- Zhao X, Yu YT 2004. Detection and quantitation of RNA base modifications. RNA 10: 996–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T 2009. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun 385: 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]