Abstract
Odor discrimination requires differential expression of odor detectors. In fact, olfactory input to the brain is organized in units (glomeruli) innervated only by olfactory sensory neurons that express the same odorant receptor (OR). Therefore, discriminatory capacity is maximized if each sensory neuron expresses only one allele of a single OR gene, a postulate sometimes canonized as the “one neuron–one receptor rule.” OR gene choice appears to result from a hierarchy of processes: differential availability of the alleles of each OR gene, zonal exclusion (or selection), OR gene switching during the initiation of OR gene transcription, and OR-dependent feedback to solidify the choice of one OR gene. The mechanisms underlying these processes are poorly understood, though a few elements are known or suspected. For example, the mechanism of activation of OR gene transcription appears to work in part through a few homeobox transcription factors (Emx2, and perhaps Lhx2) and the Ebf family of transcription factors. Further insights will probably come from several directions, but a promising hypothesis is that epigenetic mechanisms contribute to all levels of the hierarchical control of OR gene expression, especially the repressive events that seem to be necessary to achieve the singularity of OR gene choice.
Keywords: chromatin remodeling, epigenetics, olfaction, olfactory receptor, neural development
Introduction
A fundamental property of sensory systems is the elaboration of a sensor array in which the detector cells have distinct, but overlapping, receptive ranges or fields for each dimension of the stimulus. The differences allow distinct stimuli to be represented by unique patterns of sensor activity. The overlaps, which may in part be evolutionary conveniences, nevertheless ensure uninterrupted coverage of the biologically important fraction of the stimulus dimension. In mammalian olfactory systems, the establishment of distinct but overlapping sensors is accomplished by the property of singularity, the expression of a single odorant receptor (OR; also known as olfactory receptor) by each sensory neuron. Mammals typically have many millions of olfactory sensory neurons and ∼1000 OR genes, so a typical OR is expressed by a few thousand sensory neurons. These sensory neurons reside in the olfactory epithelium located in the back of the nasal cavity, from where they send their axons through the cribriform plate of the ethmoid bone to terminate in the glomerular layer of the olfactory bulb of the brain. The singularity of OR gene expression allows the projection of the sensor array onto the brain to be organized into a spatial map whereby convergence of all sensory neurons expressing the same OR establishes an array of units (glomeruli) that each represent the activity of one OR protein. Differences as small as one amino acid between ORs can result in segregation of axons to distinct glomeruli (Feinstein and Mombaerts 2004). Each OR can be activated by a subset of odorants, and each odorant appears to be detected by several ORs (Mombaerts 2004a). This organization provides an exquisite mechanism for the formation of neural activity patterns representing odor quality, in essence a spatial map of the activation of ORs. This map appears to be the primary basis of most odor discrimination (Fleischmann et al. 2008), a process capable of distinguishing odorants as similar as enantiomers. Obviously, this organization is wholly dependent on the regulation of OR gene expression, a property of olfactory sensory neurons that continues to be one of the great mysteries of the olfactory system.
ORs are the largest family of mammalian genes. Among sequenced mammalian genomes, the number of functional OR genes ranges from ∼384 in humans to ∼1284 in rats (Zhang and Firestein 2009). Mammals have two major types of OR genes. Class I OR genes are recognizably similar to fish ORs and are therefore thought to be of a more primitive origin than the larger group of Class II ORs genes, which appear to have evolved later. Though they have no sequence similarity, Class I and Class II ORs belong to the same subfamily of G protein–coupled receptors.
As is commonly the result of gene duplications giving rise to large gene families, the OR genes occur in clusters. Clustering of OR genes does not appear essential to the choice of which OR gene is expressed, though there are correlations between OR gene clusters on chromosomes and zonal expression patterns, such as the clustering of Class I OR genes and their nearly exclusive expression in the dorsal region of the olfactory epithelium (Zhang and Firestein 2007). Instead, OR gene choice appears to be driven primarily by mechanisms that treat each OR gene individually. For example, OR genes that are sensitive to the absence of the homeobox transcription factor Emx2 are found in the same chromosomal clusters as OR genes that are insensitive to the absence of Emx2 (McIntyre et al. 2008).
The phenomena that act at the level of individual OR genes and contribute to the singularity of OR gene expression include monoallelic expression, position within the epithelium (zonality), random switching during the initiation of OR expression, and feedback mediated by the appearance of a functional OR protein. Monoallelic expression is a random mechanism discovered soon after the first OR genes were identified (Chess et al. 1994). Similarly, zonality was apparent in the first in situ hybridization experiments, where expression of each OR was detected in zones restricted in the dorsoventral axis (Ressler et al. 1993; Vassar et al. 1993). Its consistency across individuals argues that overall it is a nonrandom, deterministic process. OR gene choice within a zone has a strong random component, however, because the initial selection from the available OR genes and the subsequent switching of the initiation of OR gene transcription are largely random processes (Shykind et al. 2004). Switching appears to be important for eliminating expression of OR pseudogenes encoding nonfunctional OR proteins. Finally, OR-dependent feedback locks in expression of one OR gene to the exclusion of all other OR genes (Serizawa et al. 2003; Feinstein et al. 2004; Lewcock and Reed 2004; Shykind et al. 2004).
The full diversity of the sensor array therefore appears to be achieved by a hierarchy of processes, some random and others deterministic, that result in the singularity of OR gene expression (Figure 1). The idea of hierarchical control was stimulated by the discovery of monoallelic expression of OR genes (Chess et al. 1994) but is supported by other arguments as well. For example, the deterministic framework of the zonal restriction of OR expression appears to constrain the random processes, such as OR gene switching (Shykind et al. 2004), that result in the singularity of OR gene expression. At all levels of this proposed hierarchy, the mechanisms involved are incompletely known, if identified at all. In this perspective review, I discuss results and make speculations relevant to understanding how an olfactory sensory neuron chooses to express a single allele of just one OR gene. For broader treatments of ORs, olfactory sensory neurons, the axonal wiring of the olfactory system, and the encoding of odor signals, I refer the reader to several recent reviews (Reed 2004; Mombaerts 2004a, 2004b, 2006; Imai and Sakano, 2008a, 2008b; Fleischer et al. 2009; Reisert and Restrepo 2009; Zhang and Firestein 2009).
Figure 1.
A hypothetical hierarchy of the regulation of OR gene expression. This diagram speculates an order of events that gradually restrict the OR genes available for expression. The earliest event appears to be differential marking of OR alleles, the random silencing of one allele of each OR gene, whereas the other allele is maintained in an available state. This would halve the number of OR alleles available for expression. Once the olfactory placode elaborates into the olfactory epithelium, zonal exclusion or selection might further restrict the number of OR loci available for expression. As the olfactory epithelium expands and begins to form immature neurons, transcriptional processes in the immature neurons select from the available OR loci and initiate OR gene transcription. OR gene selection is not yet fixed; random switching may still occur. If the OR protein made from the selected OR gene is not functional (or at least full length), this switching process continues. Whether further restriction of the available OR genes is continuing during this period is difficult to predict. At some point, the successful expression of an OR triggers a feedback process that results in the solidification of expression of this one OR allele and the apparent repression of all other OR gene loci. Dashed lines represent conditional events that may not occur in every developing olfactory sensory neuron.
Monoallelic expression
The first events that contribute to OR gene choice are probably those that underlie the monoallelic expression of ORs (Figure 1). The distinction of OR alleles appears to be established in early development, soon after the embryo passes through the morula and blastula stages, by differential epigenetic marking of the alleles (Takagi and Oshimura 1973; Kitsberg et al. 1993; Chess et al. 1994; Simon et al. 1999; Mostoslavsky et al. 2001). Although the details of the epigenetic mechanisms that allow monoallelic expression of autosomal monoallelically expressed genes are incompletely understood, all monoallelically expressed genes, including OR genes, are replicated asynchronously (Cedar and Bergman 2008). The allele that is more available for expression is replicated early, the less available allele is replicated late. The selection process that distinguishes the alleles is random and tends to act at the chromosome level. For example, the alleles of OR genes on the same chromosome show coordinated replication; all either early or late (Singh et al. 2003). Once established, asynchronous replication is maintained through cell divisions, arguing that the monoallelism of OR gene expression is established prior to the development of the olfactory epithelium, thereby silencing one allele of each OR gene so that the selection of an OR gene for expression results in the expression of just one allele. Alternative explanations where both alleles are equally available such that both the monoallelic and monogenic properties of OR gene choice are achieved in one step would require that each sensory neuron either silences or ignores one allele when it initiates transcription of the selected OR gene. These alternatives are also counter to the evidence, albeit circumstantial, that OR alleles are similar to other monoallelically expressed autosomal genes (Takagi and Oshimura 1973; Kitsberg et al. 1993; Chess et al. 1994; Simon et al. 1999; Mostoslavsky et al. 2001). Although it might also be possible that nascent sensory neurons or basal progenitor cells of the olfactory epithelium transiently relieve the early embryonic epigenetics believed to discriminate the alleles of monoallelically expressed genes (Cedar and Bergman 2008), parsimony argues that previously established allelic distinctions would be maintained. Therefore, each sensory neuron is probably born with one randomly determined allele of each OR gene that is more available for transcription than the other allele.
Note that though monoallelic expression accurately predicts asynchronous replication, the converse prediction is not reliable. Confirmation of monoallelic expression of OR genes predicted by asynchronous replication was therefore necessary and came first from reverse transcriptase–polymerase chain reaction of polymorphic alleles from limiting dilutions of isolated olfactory sensory neurons and subsequently from images of OR allele pairs that expressed different reporter genes (Chess et al. 1994; Strotmann et al. 2000; Shykind 2005).
Why is monoallelic expression fundamental to OR gene choice? If the preservation of unambiguous sensor types is the primary selective force on the mechanisms controlling OR gene expression, the likely answer is that monoallelic expression prevents olfactory sensory neurons from expressing two ORs that are very similar in sequence but might differ in their agonist profiles.
The mechanism underlying the monoallelism of OR gene expression is not known. However, monoallelic expression is common, with perhaps as many as 5–10% of all autosomal genes are monoallelically expressed (Gimelbrant and Chess 2006; Gimelbrant et al. 2007). Hypothesizing that ORs share a mechanism with other monoallelically expressed autosomal genes seems more reasonable than the alternative that an olfactory-specific mechanism exists. Though incompletely understood, several mechanisms are known to be important to one or more of the three types of monoallelic regulation (X-inactivation, imprinting, and random monoallelic expression of autosomal genes). These mechanisms include long noncoding RNAs that help discriminate the alleles, recruitment of Polycomb complexes to mark the chromatin of the inactive allele and DNA methylation to further repress transcription of the inactive allele (Keverne 2009, Zakharova et al. 2009). These general mechanisms are therefore candidate mechanisms for the monoallelic expression that is characteristic of OR genes.
OR zonality
The second level in the hierarchy of OR gene choice appears to be zonal exclusion, the restriction of OR expression to anatomical regions (zones) of the olfactory epithelium (Ressler et al. 1993; Vassar et al. 1993; Strotmann et al. 1994; Sullivan et al. 1995). The epithelium has two major regions, a large dorsal region that forms a single zone where all the Class I ORs are expressed along with a subset of Class II ORs, and a ventral region where other Class II ORs are expressed in zones that run the length of the anterior–posterior dimension. The selective forces behind OR zonality appear to be more than just a convenience to aid the singularity of OR gene expression. For example, the dorsal region is responsible for innate avoidance of predator odors and, therefore, the restriction of at least some ORs to this zone may be necessary to support this function (Kobayakawa et al. 2007).
For most ORs tested to date, OR expression zones span the anterior–posterior dimension of the epithelium but are only a fraction of the dorsoventral dimension. However, Class II ORs of the mOR262 subfamily are expressed in a patch that occupies portions of endoturbinates II and III plus part of ectoturbinate 3 (Strotmann et al. 1992, 1994). The available evidence indicates that the dorsoventral zones are the norm and the “patch” is the exception. The original interpretation that OR expression zones were four fixed regions, each circumscribing the expression of equivalent fractions of the OR gene repertoire, was subsequently revised by evidence that Class II OR genes are instead expressed in a dorsoventral continuum of overlapping zones (Iwema et al. 2004; Miyamichi et al. 2005).
The zonal and patch expression patterns of ORs imply that positional cues act to select a subset of OR genes as being available for expression. Whether these cues act by positive selection of the appropriate set of OR genes, by repression of all but a subset of OR genes, or by both positive and negative mechanisms working together, is unknown. The nature of the positional cues is also not known, but the continuum of overlapping zones seems most consistent with a dorsoventral gradient of some cue or cues. The continuous turnover of olfactory sensory neurons (OSNs), which appears to happen without altering OR zonality, argues either that the gradient is permanent or that a transient gradient laid down instructions that are permanent. If the latter is correct, then basal progenitor cells may be inherently biased to produce sensory neurons that will select from a zonal subset of OR genes. Experiments that test this idea, such as basal cell transplantation studies, have not yet been reported. In the absence of knowledge of the mechanism of zonality, alternatives to location-dependent processes remain possible. For example, to the extent that temporal patterns govern development along the dorsoventral axis of the olfactory epithelium, time could play as significant a role as location in OR gene choice.
Analysis of OR transgenes indicates that zonality depends on sequences within the OR gene locus itself, albeit interpreted within the genomic context in which the transgene resides (Vassalli et al. 2002). Like probable mechanisms of random monoallelic expression of autosomal genes, zonality might work through epigenetic mechanisms that regulate the availability of each OR gene in both positive (selection) and negative (exclusion) ways. Zonality might also be accomplished by, or might have significant contributions from, zonally restricted transcriptional regulators or zonal gradients of transcriptional regulators. In this scenario, zonal gradients of signals and transcription factors would act to select or repress those OR genes whose promoters were sensitive to them. No zone-specific transcription factors have yet been reported, however. Whether it be zonal exclusion or zonal selection, or a combination of the two, it is clear that location in the epithelium is deterministic, that is, location correlates with the expression of specific subsets of OR genes. Within the subset of OR genes that are available for expression at any given location, however, the choice process appears to be fundamentally random. Variation in the frequency of selection from among the available OR genes, which can be severalfold, is not necessarily inconsistent with the interpretation of randomness. Variation probably derives from biases in the events downstream of the random selection process. For example, the frequency of expression of the MOR28 cluster genes controlled by the H-region vary directly with proximity to the H-region (Fuss et al. 2007), a pattern that would occur if a random selection process acted through the biases of H-region enhancer activity.
OR expression singularity
Although evidence of occasional exceptions and the inherent difficulty of obtaining direct proof of the singularity of mammalian OR gene expression contribute to doubts about the universality of the “one neuron–one receptor rule,” the weight of the available evidence continues to favor this idea (Rawson et al. 2000; Serizawa et al. 2004, Mombaerts 2004b). Therefore, a reasonable working hypothesis is that once monoallelic and zonal mechanisms have restricted the OR gene choices available, an unknown mechanism with a random component completes the selection of one OR gene for expression. What then are the regulatory events that give rise to the singularity of expression? Numerous possibilities exist, ranging from a unique DNA enhancer element, to a protein complex so rare that only one OR gene is likely to bind it, to a unique DNA rearrangement, to a unique nuclear location (a “transcription factory”) for OR transcription, to rate-limiting kinetics that match a brief expression window period (Shykind 2005). Also possible is that the OR-dependent feedback mechanism that results in the repression of all but the active OR allele (described below) is the primary mechanism responsible for singularity.
An intriguing hypothesis is the idea that DNA rearrangement is responsible for the singularity of OR expression. Substantial tests of this hypothesis were achieved by generation of cloned mice using transfer of olfactory sensory neuron nuclei expressing a known OR (Eggan et al. 2004; Li et al. 2004). If DNA rearrangement is permanent then all olfactory sensory neurons of the cloned mice should express the OR expressed in the donor nucleus. Instead, OR expression patterns in the cloned mice were normal, arguing against the DNA rearrangement hypothesis. Of course, what cannot be completely excluded is the possibility that reprogramming of each donor nucleus reversed DNA rearrangement of the expressed OR gene. Nevertheless, these findings have cast significant doubt on this DNA rearrangement hypothesis.
Another appealing hypothesis is that a unique DNA element regulates not only the expression of nearby OR genes on the same chromosome (in cis) but also acts in trans to select for expression other OR genes on other chromosomes. A candidate for such a DNA element, the H-region, was originally discovered as an enhancer of expression of the OR genes in the MOR28 cluster, located 75 kbp away on mouse chromosome 14 (Serizawa et al. 2000). Evidence that the H-region could be trapped by the 4C chromosome conformation capture method along with DNA fragments containing ORs from other chromosomes (one of the earliest uses of the 4C method) supported the role of the H-region as the unique component necessary for OR gene choice (Lomvardas et al. 2006). However, targeted deletion of the H-region only reduced expression of the four MOR28 cluster OR genes that were already known to be regulated by the H-region (Fuss et al. 2007; Nishizumi et al. 2007). This finding argues strongly that the H-region cannot act in trans, or perhaps that any in trans activity is not necessary for OR gene choice. The trans-chromosomal association of OR loci with the H-region may have been a consequence of random captures exacerbated by the size of the OR gene family and the segregation of types of chromatin within nuclei (de Laat and Grosveld 2007; Simonis et al. 2007). The H-region, therefore, is unlikely to be the unique element responsible for the singularity of OR expression.
At present, the mechanism of OR gene choice remains a mystery. Some of the hypothesized mechanisms are much more difficult to test than DNA rearrangement or the role of the H-region. For example, the existence of a limiting protein, or perhaps more likely a complex of proteins, is particularly difficult to test in the absence of any clues about the nature of the proteins. The related idea that only a single transcription factory in a sensory neuron nucleus is capable of transcribing OR genes might be more easily approached, in part because transcription factories often occur at characteristic locations in nuclei (Carter et al. 2008; Sutherland and Bickmore 2009). Also difficult to test until more of the proteins involved are discovered is the hypothesis that feedback repression by one OR upon all others is the mechanism of singularity.
In considering mechanisms that produce singularity of OR expression, it seems unlikely that any mechanism would be perfect. Olfactory sensory neurons that express more than one OR, or even no ORs, might arise. That these neurons would then be selected for early death seems reasonable. Substantial evidence exists to suggest that olfactory sensory neurons lacking odor-stimulated activity or whose axons fail to coalesce into glomeruli have shortened life spans (Zheng et al. 2000; Zhao and Reed 2001; Feinstein and Mombaerts 2004; Feinstein et al. 2004; Zou et al. 2004). The increased rate of cell death of such sensory neurons may be sufficient for maintaining the specificity of olfactory sensory neuron input to olfactory bulb glomeruli. The life spans of sensory neurons expressing the “wrong” number of ORs might be even shorter. Apoptosis of immature olfactory sensory neurons, unlikely to have accumulated sufficient damage from exogenous stressors that seem to be the major cause of apoptosis of mature sensory neurons, does occur in normal, undamaged olfactory epithelium (Holcomb et al. 1995). Misexpression of ORs (too many or too few) is a potential cause of immature sensory neuron apoptosis.
A functional OR silences other OR genes?
Expression of a functional OR from an OR gene locus prevents expression of other OR genes, but expression of an unrelated protein or a nonfunctional OR from the same locus usually fails to prevent expression of another OR gene and does not prevent coexpression of the allele containing the nonfunctional OR (Serizawa et al. 2003; Feinstein et al. 2004; Lewcock and Reed 2004; Shykind et al. 2004). This effect extends even to OR transgenes randomly inserted into chromosomes or carried on artificial chromosomes (Serizawa et al. 2000), implying that the sequences responsible for the control of expression are contained within or very close to the transcribed regions of OR genes (Reed 2000). Some evidence suggests that the mechanism of feedback involves coupling to G proteins that stimulate the production of cyclic adenosine monophosphate such as the ability of the β2-adrenergic receptor expressed from an OR locus to behave just like an OR, causing homogeneous olfactory sensory neuron axon coalescence into glomeruli (Feinstein et al. 2004). Other evidence, particularly the ability of a full-length OR mutant that should be incapable of coupling with heterotrimeric G proteins to prevent coexpression of other ORs suggests that OR signaling through G proteins is not absolutely necessary either to drive feedback or to be a target of feedback (Nguyen et al. 2007). These data argue that a full-length OR may be sufficient for feedback.
While describing the feedback control of ORs as negative feedback conveniently describes the overall effect, the evidence does not rule out a positive feedback mechanism. OR gene loci may be susceptible to silencing by epigenetic mechanisms even in cells that do not express ORs (Miles et al. 2007; Hou et al. 2010), so the feedback mechanism might not need to enhance repression of OR genes, but instead might cooperate with positively acting factors on the active OR locus to protect it from repressive mechanisms that (presumably) become increasingly powerful during the differentiation of olfactory sensory neurons. Perhaps even more likely is that the feedback mechanism driven by the expressed OR contributes to both positive and negative regulation of OR gene loci.
If OR feedback involves powerful epigenetic mechanisms, might feedback be sufficient to explain the singularity of OR gene expression? The transient expression of several ORs in each immature sensory neuron as the ORs compete for dominance (termed “switching”) seems potentially consistent with this idea (Shykind et al. 2004). Testing how critical feedback is for OR singularity will require learning more about the feedback mechanism. At this point, whether the OR even needs to be functional is uncertain. Whatever the mechanism of feedback, however, the outcome seems likely to require some level of epigenetic control, a topic discussed further below.
OR gene promoters
OR genes are relatively simple, consisting of a single coding exon preceded by at most a few small noncoding exons. Transcriptional start sites can be a few hundred to several thousand bases upstream of the translational start site. A consensus TATA box may be present or (more commonly) an AT-rich region is found at the site where the TATA box would be expected (Hoppe et al. 2006; Michaloski et al. 2006). With the exception of the H-region enhancer that controls expression of four OR genes in the MOR28 cluster (Serizawa et al. 2000; Fuss et al. 2007; Nishizumi et al. 2007), the cis-elements necessary for normal expression of OR genes appear to be contained within small upstream regions of each OR gene (Qasba and Reed 1998; Rothman et al. 2005; Zhang et al. 2007; Vassalli et al. 2002). This upstream promoter region can be remarkably small; As little as ∼150 bp immediately upstream of the transcriptional start site of an OR promoter can be sufficient to drive zonal, singular expression of an OR transgene (Vassalli et al. 2002). A transgene driven by 358 bp of the promoter of an OR expressed in a patch pattern, Olfr157 (mOR262-12), also reproduced the normal expression pattern of the OR (Zhang et al. 2007). These data reinforce the conclusion that ORs are very compact genes whose essential control elements are maintained near the exons. The known conserved features of these minimal promoter sequences are at least one homeodomain-like site followed by at least one O/E (Olf1/early B-cell factor)-like site (Lane et al. 2001; Vassalli et al. 2002; Hoppe et al. 2006; Michaloski et al. 2006; Zhang et al. 2007). The O/E-like sites are typically found less than 200 bp upstream of the transcriptional start sites, whereas the homeodomain-like sites are distributed further upstream, frequency decreasing with distance (Michaloski et al. 2006). For ORs that have distinctive expression patterns, unique elements within the minimal promoters or in more distant enhancer regions would seem to be necessary. For the patch ORs of the mOR262 subfamily, candidate elements in their putative promoters have been identified via their conservation across the mOR262 subfamily (Hoppe et al. 2006; Zhang et al. 2007), but tests of these candidates have not been reported.
O/E-like sites, which are bound by the four members of the Ebf transcription factor family, are common to the promoters of ORs and other relatively olfactory-specific genes (Kudrycki et al. 1993; Wang and Reed 1993). O/E-like sites are therefore thought to help mediate the olfactory-specific expression of these genes in olfactory sensory neurons (Buiakova et al. 1996; Davis and Reed 1996). This has not been fully confirmed, in part due to difficulties in generating mice lacking all four Ebf family members (Wang et al. 2004). Sustained expression of the Ebf inhibitor, Zfp423 (OAZ), in olfactory sensory neurons suppresses expression of ORs and other olfactory-specific genes, but the near absence of olfactory sensory neuron maturation confounds the interpretation that loss of activity of Ebf transcription factors at O/E-like sites is responsible for reduced transcription of OR genes (Cheng and Reed 2007).
The homeodomain-like and O/E-like sites in putative OR promoters appear to work together. The expression of OR transgenes is geometrically reduced when both sites are mutated or deleted (Rothman et al. 2005). Similarly, targeted mutation of both these sites in the endogenous Olfr151 (M71) gene was required to produce even a 3-fold reduction in the frequency of neurons that express Olfr151 (Rothman et al. 2005). In addition to reinforcing the idea that the homeodomain-like and O/E-like sites work in concert, these data argue that other sites, as yet undefined, also regulate expression of endogenous OR gene loci. These sites could be additional homeodomain-like and O/E-like sites located more distantly from the transcriptional start site, or they may be sites bound by different types of transcriptional regulators.
Emx2 and Lhx2 act at the homeodomain-like site
Changes in the homeodomain-like site and the O/E-like site control the probability of OR gene choice rather than the level of expression per sensory neuron (Rothman et al. 2005). This suggests that OR gene choice mechanisms act through these sites, at least in part. It also predicts that the deletion or inhibition of the transcription factors acting at these sites will alter the frequency of gene choice. Although technical issues have prevented testing the Ebf transcription factors in this way, targeted deletion of the homeodomain transcription factor Emx2 did indeed alter the frequency of expression of many OR genes (McIntyre et al. 2008).
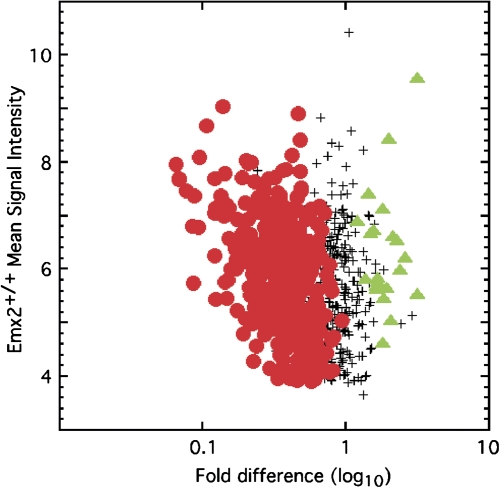
Emx2 is one of the 10 homeodomain transcription factors captured by the Olfr151 homedomain-like site or a promoter fragment of Olfr156 (mOR262-6) in yeast one-hybrid assays (Hoppe et al. 2003; Hirota and Mombaerts 2004). The others were Lhx2, Cart1 (Alx1), Dlx5, Dlx3, Prrx1 (Prx1), Prrx2 (Prx2), Alx3, Pitx1 (Ptx1) and Barx1. Emx2-deficient mice showed reduced amounts of mRNA for 365 OR genes, half of the OR mRNAs detected by the microarray platform used (Figure 2; McIntyre et al. 2008). These decreases were disproportionately greater than the 42% reduction in the number of mature sensory neurons in this mutant mouse and as predicted by the effects of mutating the homeodomain-like site, in situ hybridization revealed reduced frequencies of expression of OR genes but no evidence of reduced amounts of these mRNAs within the few sensory neurons that expressed them. The affected genes included both Class I and Class II OR genes. If OR genes are selected by a mechanism that involves switching and feedback to lock in the choice of a functional OR gene, then reduced expression of many, but not all, OR genes in Emx2-deficient mice should cause increased frequency of expression of other OR genes. As predicted, increased frequency of expression (2-fold to 6-fold) of 22 Class II OR genes was detected. These data argue that Emx2 is the homeobox transcription factor that acts most strongly at many of OR gene promotors. They also indicate that OR gene choice mechanisms act in concert with Emx2 for many ORs. Whether these mechanisms work through Emx2, or merely gate the ability of Emx2 to stimulate OR transcription, cannot yet be determined.
Figure 2.
Emx2 helps stimulate transcription for many OR genes. The fold differences of microarray signal intensities for all detected OR mRNAs in olfactory epithelium samples from Emx2−/− and Emx2+/+ mice are plotted against the mean signal intensities generated from the Emx2+/+ samples. Red circles are significant decreases in the Emx2−/− samples and green triangles are significant increases. Reprinted with permission from McIntyre et al. (2008). This figure appears in color in the online version of Chemical Senses.
Although Emx2 appears to be the predominant homeodomain transcription factor acting at the majority of OR genes, some OR genes clearly do not require Emx2. The hypothesis that many of these Emx2-insensitive OR genes might have promoters that are normally regulated by Emx2 but can be regulated by other homeodomain transcription factors in the absence of Emx2 has not yet been disproven. However, a more promising explanation is that at least one other homeodomain transcription factor helps activate these Emx2-insensitive OR genes even when Emx2 is present. Of the eight additional candidates known to bind an OR promoter, Lhx2 is the most promising because it is an abundant mRNA in olfactory sensory neurons and it was the most common binding partner detected in one-hybrid assays using OR promoter fragments (Hirota and Mombaerts 2004). The olfactory epithelia of mice lacking Lhx2 show little expression of any OR gene, but this is confounded by the fact that these mice have almost no mature OSNs and many fewer immature OSNs (Hirota and Mombaerts 2004; Kolterud et al. 2004; Hirota et al. 2007). As with mice lacking multiple Ebf transcription factors, the massive loss of OSNs prevents the interpretation that defects in OR gene expression, rather than OSN loss, are responsible for reduced abundance of OR mRNAs and the reduced number of cells detectably expressing ORs.
OR gene silencing and chromatin remodeling epigenetics
The flip side to the singularity of OR gene expression is the silencing of all other OR genes. Silencing, rather than expression, may therefore represent the most significant feature of OR gene choice. As yet, the little we understand about OR gene silencing is solely phenomenological. For example, separating the OR promoter and the OR coding sequence only partially relieves repression of OR coding region transgenes, indicating that the coding region itself is a sufficient signal for OR gene silencing (Nguyen et al. 2007). Even strong promoters such as the Omp promoter are much less active when placed in front of OR coding regions (Lane et al. 2005). These data suggest the hypothesis that epigenetic regulation plays critical roles in the negative control of OR gene expression.
When the ∼10,000 genes expressed by olfactory sensory neurons are assessed by functional bioinformatics, chromatin remodeling is one of the overrepresented biological processes (Sammeta et al. 2007). The Polycomb complex and other chromatin remodeling gene transcripts that comprise this category are expressed most abundantly in the immature sensory neurons, the stage where OR gene choice appears to be completed. Description of the types of chromatin marks at OR gene loci, especially the repressive marks that presumably predominate across the population of ORs and therefore would be the easiest to detect, should soon be forthcoming. Indeed, studies of the epigenetic regulation of the β-globin gene cluster in erythroid cells typically use a neighboring OR gene cluster as a control. These studies found that the OR gene cluster has histone modifications associated with silent genes, such as increased dimethylation of lysine 9 of histone-H3 (H3K9me2), and low levels of types of histone acetylation associated with actively transcribed regions of chromatin (Miles et al. 2007; Hou et al. 2010). By analogy to developmental processes in other tissues, we might find low levels of bivalent chromatin marks (a combination of active and repressive marks) on the histones at OR gene loci in basal progenitor cells followed by a shift to a predominance of repressive marks as the sensory neurons differentiate. How OR switching and feedback regulate chromatin modification at OR gene loci may be critical to understanding OR gene expression patterns. If this interpretation that epigenetic repression is a critical feature of OR gene expression is correct, then most intriguing of all is the question of how a single expressed OR allele manages to avoid repression mediated by chromatin remodeling. Answering this question may hold the key to understanding the singularity of OR gene choice.
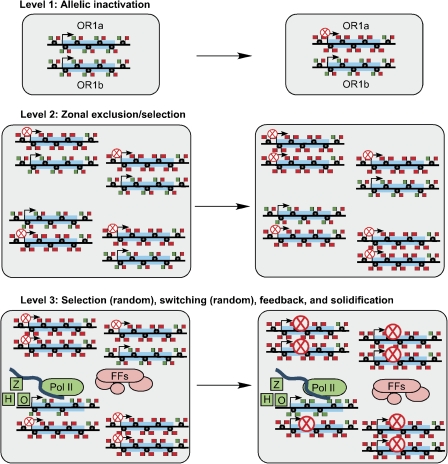
A speculative view of the regulation of OR gene expression that emphasizes the role of chromatin remodeling is given in Figure 3. OR gene loci are probably equivalent or nearly so until early in embryonic development when one allele of each OR locus is marked, probably by accumulating more repressive chromatin modifications (level 1). This allele henceforth would have a low probability of transcription. The other allele has a high probability of transcription and might be sparsely marked with a combination of active and repressive histone modifications if it shares properties thought to be common to other developmentally regulated genes (Mikkelsen et al. 2007; Heintzman et al. 2009). As development proceeds and the olfactory epithelium is laid down, zonal selection of OR gene loci must occur (level 2). Whether this occurs in basal progenitor cells or in immature sensory neurons is unknown. Given the propensity of OR gene loci to be repressed, the mechanism of zonal selection may be one of exclusion mediated by a predominance of repressive chromatin modifications. This hypothetical mechanism of zonality does not preclude roles for zone-specific signaling or transcription factors, in part because these signals might work by controlling chromatin modification. At this point in development, each nascent sensory neuron would be in a state where repressive marks dominate at the vast majority of OR loci. Via a random process, at least one OR locus captures transcriptional machinery that includes a homeobox transcription factor (most commonly Emx2), an Ebf family transcription factor, and other factors as yet unknown but sufficient to initiate the synthesis of an OR mRNA and translation into protein (level 3). This situation is not stable and although the locus (or loci) being expressed would now have a greatly increased probability of becoming the locus expressed in the mature neuron, switching can still occur. At this immature neuron stage in the olfactory sensory neuron cell lineage, expression of many chromatin-modifying enzymes and associated genes is peaking so the immature neurons appear to be primed to solidify the gene expression patterns required for the mature olfactory sensory neuron phenotype (Sammeta et al. 2007). Mechanisms of solidification might include DNA methylation, further increases in repressive histone modification, and incorporation of nonconventional histones into the nucleosomes at OR genes. One component of these epigenetic changes is the role of feedback from an expressed OR to solidify the singularity of OR gene choice. Feedback may act to shield one active OR locus from epigenetic repression, or to enhance epigenetic repression of all other OR loci, or both. The solidification process is probably completed before the sensory neuron reaches maturity given the role of the OR protein in the specific convergence of sensory neuron axons into a glomerulus (Mombaerts et al. 1996; Mombaerts 2006).
Figure 3.
Speculations on the epigenetic control of OR gene expression. In this figure, pairs of DNA strands depict the two alleles of an OR gene (transcribed region highlighted in light blue) on paired chromosomes. The flags at each nucleosome (small black circles) are coded red for repressive marks and green for active or permissive marks. Nucleosomes showing both red and green flags indicate bivalently marked chromosomal regions that are available to be further modified into repressed chromatin (all red flags) or actively transcribed chromatin (all green flags). Each circled red X emphasizes repressed OR alleles, and a larger circled red X depicts repression that has been solidified (as might happen when the sensory neurons reach their mature state). H represents homeobox transcription factors such as Emx2 and O represents the O/E-like (Ebf) family of transcription factors that are necessary for transcription of most, perhaps all, OR genes. Z represents as yet unknown factors that also contribute to OR transcription, factors that may be responsible for the singularity of OR expression. Pol II, RNA polymerase II; FFs, feedback factors whose identities are unknown. The thick blue strand depicts newly transcribed RNA.
This model is certainly wrong, perhaps only in its minor details, perhaps completely. Alternative mechanisms that might be able to discriminate OR genes and alter their probability of expression without involving common mechanisms of chromatin remodeling include noncoding RNAs of several types and novel proteins that specifically bind OR coding regions. However, chromatin remodeling is already known to be fundamental to the regulation of gene expression. It is flexible, has graduated levels in both positive and negative directions, is able to integrate signals of many types, and is often reversible (Kouzarides 2007; Mikkelsen et al. 2007; Cairns 2009; Heintzman et al. 2009). For these reasons, it is easy to imagine that it contributes to OR gene regulation, perhaps even to the extent that mechanisms of OR gene choice work through it.
Conclusion
A mechanistic understanding of how OR genes are selected for expression is still lacking. However, significant progress has been made since the 2001 Banbury Conference on the molecular biology of chemosensory receptors where Richard Axel was asked to comment on the mechanism of OR gene choice. He replied, “I don't even know how to think about that problem yet.” While my interpretations of the clues discussed in this review will prove mistaken to a greater or lesser extent, the mere existence of the clues allows testable hypotheses to be generated. There is reason to be optimistic that these clues, combined with the rapidly evolving understanding of epigenetic regulation of gene expression in general, will soon lead to the discovery of mechanisms that cause the robust transcription of one allele of one OR gene and the repression of all other OR genes in each olfactory sensory neuron.
Funding
National Institutes of Health (R01 DC002736); National Institutes of Health (R01 DC007194).
Acknowledgments
For stimulating discussions relevant to this review, I thank Neeraja Sammeta, Guangfan Zhang, Randy Reed, Stuart Firestein, Paul Feinstein, Tom Bozza, Anne Vassalli, Andrea Rothman, Andreas Walz, Junji Hirota, Stefan Fuss, Tomohiro Ishii, and Masayo Omura.
References
- Buiakova OI, Baker H, Scott JW, Farbman A, Kream R, Grillo M, Franzen L, Richman M, Davis LM, Abbondanzo S, et al. Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proc Natl Acad Sci U S A. 1996;93:9858–9863. doi: 10.1073/pnas.93.18.9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem Soc Trans. 2008;36:585–589. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Choreography of Ig allelic exclusion. Curr Opin Immunol. 2008;20:308–317. doi: 10.1016/j.coi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54:547–557. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev. 2007;17:456–464. doi: 10.1016/j.gde.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Fleischer J, Breer H, Strotmann J. Mammalian olfactory receptors. Front Cell Neurosci. 2009;3:9. doi: 10.3389/neuro.03.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Shykind BM, Sosulski DL, Franks KM, Glinka ME, Mei DF, Sun Y, Kirkland J, Mendelsohn M, Albers MW, et al. Mice with a “monoclonal nose”: perturbations in an olfactory map impair odor discrimination. Neuron. 2008;60:1068–1081. doi: 10.1016/j.neuron.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Chess A. An epigenetic state associated with areas of gene duplication. Genome Res. 2006;16:723–729. doi: 10.1101/gr.5023706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci U S A. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol Cell Neurosci. 2007;34:679–688. doi: 10.1016/j.mcn.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Holcomb JD, Mumm JS, Calof AL. Apoptosis in the neuronal lineage of the mouse olfactory epithelium: regulation in vivo and in vitro. Dev Biol. 1995;172:307–323. doi: 10.1006/dbio.1995.0025. [DOI] [PubMed] [Google Scholar]
- Hoppe R, Breer H, Strotmann J. Promoter motifs of olfactory receptor genes expressed in distinct topographic patterns. Genomics. 2006;87:711–723. doi: 10.1016/j.ygeno.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Hoppe R, Frank H, Breer H, Strotmann J. The clustered olfactory receptor gene family 262: genomic organization, promotor elements, and interacting transcription factors. Genome Res. 2003;13:2674–2685. doi: 10.1101/gr.1372203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Sakano H. Interhemispheric olfactory circuit and the memory beyond. Neuron. 2008a;58:465–467. doi: 10.1016/j.neuron.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H. Odorant receptor-mediated signaling in the mouse. Curr Opin Neurobiol. 2008b;18:251–260. doi: 10.1016/j.conb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J Neurosci. 2004;24:356–369. doi: 10.1523/JNEUROSCI.1219-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keverne B. Monoallelic gene expression and mammalian evolution. Bioessays. 2009;31:1318–1326. doi: 10.1002/bies.200900074. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131:5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kudrycki K, Stein-Izsak C, Behn C, Grillo M, Akeson R, Margolis FL. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol Cell Biol. 1993;13:3002–3014. doi: 10.1128/mcb.13.5.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AP, Zhao H, Reed RR. Development of transgenic mouse models for the study of human olfactory dysfunction. Am J Rhinol. 2005;19:229–235. [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Young J, Athanasiou M, Friedman C, Rowen L, Evans G, Axel R, Hood L, Trask BJ. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc Natl Acad Sci U S A. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428:393–399. doi: 10.1038/nature02433. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chem Senses. 2008;33:825–837. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PA, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Res. 2006;16:1091–1098. doi: 10.1101/gr.5185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J, Mitchell JA, Chakalova L, Goyenechea B, Osborne CS, O'Neill L, Tanimoto K, Engel JD, Fraser P. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human beta-globin locus. PLoS One. 2007;2:e630. doi: 10.1371/journal.pone.0000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004a;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004b;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizumi H, Kumasaka K, Inoue N, Nakashima A, Sakano H. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc Natl Acad Sci U S A. 2007;104:20067–20072. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba P, Reed RR. Tissue and zonal-specific expression of an olfactory receptor transgene. J Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson NE, Eberwine J, Dotson R, Jackson J, Ulrich P, Restrepo D. Expression of mRNAs encoding for two different olfactory receptors in a subset of olfactory receptor neurons. J Neurochem. 2000;75:185–195. doi: 10.1046/j.1471-4159.2000.0750185.x. [DOI] [PubMed] [Google Scholar]
- Reed RR. Regulating olfactory receptor expression: controlling globally, acting locally. Nat Neurosci. 2000;3:638–639. doi: 10.1038/76584. [DOI] [PubMed] [Google Scholar]
- Reed RR. After the holy grail: establishing a molecular basis for mammalian olfaction. Cell. 2004;116:329–336. doi: 10.1016/s0092-8674(04)00047-9. [DOI] [PubMed] [Google Scholar]
- Reisert J, Restrepo D. Molecular tuning of odorant receptors and its implication for odor signal processing. Chem Senses. 2009;34:535–545. doi: 10.1093/chemse/bjp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol. 2007;502:1138–1156. doi: 10.1002/cne.21365. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Shykind BM. Regulation of odorant receptors: one allele at a time. Hum Mol Genet. 2005;14(1):R33–R39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A. Coordination of the random asynchronous replication of autosomal loci. Nat Genet. 2003;33:339–341. doi: 10.1038/ng1102. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Conzelmann S, Beck A, Feinstein P, Breer H, Mombaerts P. Local permutations in the glomerular array of the mouse olfactory bulb. J Neurosci. 2000;20:6927–6938. doi: 10.1523/JNEUROSCI.20-18-06927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Helfrich T, Beck A, Breer H. Rostro-caudal patterning of receptor-expressing olfactory neurones in the rat nasal cavity. Cell Tissue Res. 1994;278:11–20. doi: 10.1007/BF00305773. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Krieger J, Raming K, Breer H. Expression of odorant receptors in spatially restricted subsets of chemosensory neurones. Neuroreport. 1992;3:1053–1056. doi: 10.1097/00001756-199212000-00005. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Bohm S, Ressler KJ, Horowitz LF, Buck LB. Target-independent pattern specification in the olfactory epithelium. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Takagi N, Oshimura M. Fluorescence and Giemsa banding studies of the allocyclic X chromosome in embryonic and adult mouse cells. Exp Cell Res. 1973;78:127–135. doi: 10.1016/0014-4827(73)90046-3. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Zakharova IS, Shevchenko AI, Zakian SM. Monoallelic gene expression in mammals. Chromosoma. 2009;118:279–290. doi: 10.1007/s00412-009-0206-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics. 2007;89:441–450. doi: 10.1016/j.ygeno.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. Genomics of olfactory receptors. Results Probl Cell Differ. 2009;47:25–36. doi: 10.1007/400_2008_28. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Breer H, Strotmann J. Promotor elements governing the clustered expression pattern of odorant receptor genes. Mol Cell Neurosci. 2007;36:95–107. doi: 10.1016/j.mcn.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Zhao H, Reed RR. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 2001;104:651–660. doi: 10.1016/s0092-8674(01)00262-8. [DOI] [PubMed] [Google Scholar]
- Zheng C, Feinstein P, Bozza T, Rodriguez I, Mombaerts P. Peripheral olfactory projections are differentially affected in mice deficient in a cyclic nucleotide-gated channel subunit. Neuron. 2000;26:81–91. doi: 10.1016/s0896-6273(00)81140-x. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Feinstein P, Rivers AL, Mathews GA, Kim A, Greer CA, Mombaerts P, Firestein S. Postnatal refinement of peripheral olfactory projections. Science. 2004;304:1976–1979. doi: 10.1126/science.1093468. [DOI] [PubMed] [Google Scholar]