Abstract
A fundamental and shared process in all forms of life is the use of DNA glycosylase enzymes to excise rare damaged bases from genomic DNA. Without such enzymes, the highly-ordered primary sequences of genes would rapidly deteriorate. Recent structural and biophysical studies are beginning to reveal a fascinating multistep mechanism for damaged base detection that begins with short-range sliding of the glycosylase along the DNA chain in a distinct conformation we refer to as the search complex (SC). Sliding is frequently punctuated by the formation of a transient “interrogation” complex (IC) where the enzyme extrahelically inspects both normal and damaged bases in an exosite pocket that is distant from the active site. When normal bases are presented in the exosite, the IC rapidly collapses back to the SC, while a damaged base will efficiently partition forward into the active site to form the catalytically competent excision complex (EC). Here we review the unique problems associated with enzymatic detection of rare damaged DNA bases in the genome, and emphasize how each complex must have specific dynamic properties that are tuned to optimize the rate and efficiency of damage site location.
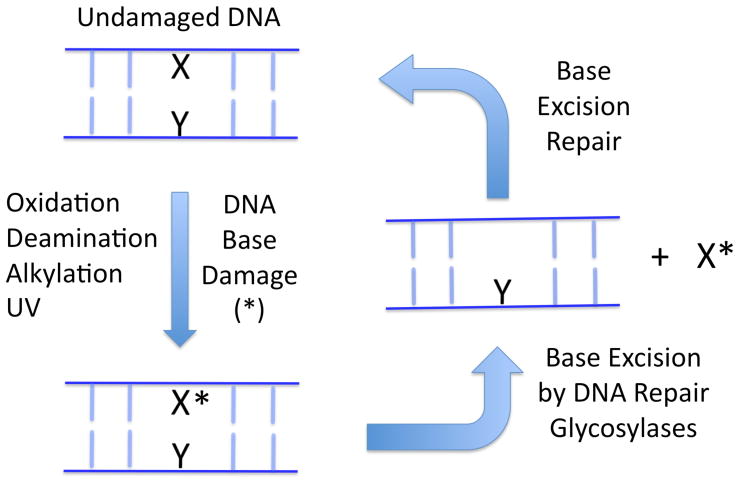
The problem of enzymatic detection of a single damaged base in the context of a vast genome of nearly isomorphous undamaged bases has intrigued the DNA repair community virtually since the discovery of the DNA base excision repair pathway (1). The initiating step in this pathway begins with the enzymatic hydrolysis of the glycosidic bond that attaches the damaged base to the deoxyribose phosphate DNA backbone, setting the stage for the multistep base excision repair process to begin (Fig. 1). The cellular sentinels at this first step are the remarkable DNA glycosylase enzymes (2). Although these enzymes fall into different structural classes, and each is specialized for the detection and removal of different types of damaged bases (Table 1), with the sole exception of pyrimidine dimer DNA glycosylase (3), these enzymes have converged on a single mechanistic solution for damaged base recognition and excision: rotation of the damaged base from the DNA base stack into a sequestered active site pocket where chemistry occurs (Fig. 2). This process has been called either base or nucleotide “flipping” by various investigators (4, 5), and connects the damage encounter event with the catalytic step of bond scission.
Figure 1.
The DNA base excision repair pathway begins with the encounter of a rare genomic damaged base (X*) by a damage specific DNA glycosylase. These enzymes hydrolytically excise the damaged base and begin the extended process of replacing it with the original undamaged base (X).
Table 1.
Summary of substrate specificities, structural characterizations with bound DNA, and kinetic studies on representative DNA repair glycosylases
| Enzyme | Primary Substratesa | Enzyme Fold Family | IC Structureb | EC Structureb | Transition State Structure | Rapid Kinetic Studiesc | Facilitated Diffusion Studiesc |
|---|---|---|---|---|---|---|---|
| UNG | U/A, U/G, ssU | UNG | 2OXM (6) | 1EMH (7) | 1Q3F (8, 9) | (10–13) | (14–17) |
| SMUG | U/G, ssU | UNG | 1OE4d (18) | 1OE5d (18) | |||
| TDG | T/G, U/G | UNG | 2RBAe (19) | 2RBAe (19) | |||
| hOGG1 | °G/C | HhH | 2I5W, 1YQK (20, 21) | 1EBM (22) | (23, 24) | (25) | |
| MutM | °G/C | MutM | 2F5O 3GO8, 3JR4 (26, 27) | 1R2Y (28) | (29, 30) | (25) | |
| MutY | A/°G | HhH | 3G0Q (31) | 3FSP (32) | (33) | (34) | |
| AlkA | m3A, m2T, m2C | HhH | 3D4V (35) | ||||
| AAG | hyX, m3A, εA, | AAG | 1F6O (36) | 1BNK (37) | (38) | ||
| TAG | m3A, m3G | HhH | 2OFI (39) | ||||
| EndoIII | Tg, urea, ho5C, hU | HhH | 1P59f (40) | ||||
| Pyrimidine dimer DNA glycosylase | T<>T photo dimers | T4 pyrimidine dimer glycosylase | 1VAS, 2FCC | (75) | |||
The base that is excised is shown in bold, and the opposing base is also shown if this base is also important in recognition.
IC, interrogation complex; EC, excision complex
Studies are listed that focus on the kinetics of base flipping and facilitated diffusion on DNA.
IC is with partially extrahelical uracil, EC is approximated by a product complex with abasic DNA mimic and uracil.
The EC and IC structures were observed simultaneously in the same crystal. The EC is approximated with an abasic product mimic.
Trapped Schiff base covalent intermediate.
Figure 2.
The extended reaction coordinate for extrahelical recognition of undamaged and damaged bases by uracil DNA glycosylase (UNG), and the bacterial and human 8-oxoguanine DNA glycosylases (MutM and hOGG1). Each of these enzymes excises its cognate damaged base through a multistep base flipping reaction coordinate beginning with one or more early interrogation complexes (IC) where normal and damaged bases are inspected, and ending with a fully extrahelical excision complex (EC) poised for glycosidic bond cleavage of the damaged base. The trajectory for base flipping is shown viewing down the helical axis (top) and from an orthogonal perspective in which the enzyme has been removed for clarity (bottom).
Base flipping involves one of the most extended and improbable reaction trajectories in biology. The overall reaction is driven forward solely by the use of enzyme binding energy for DNA, which is used to pay for the significant energetic costs of extracting a base from the DNA base stack (11). These costs include breaking of Watson-Crick hydrogen bonds, the disruption of aromatic stacking interactions with adjacent bases, and large perturbations in the phosphate torsion angles around the flipped base. Initial structural investigations into enzymatic base flipping focused on elucidating the final extrahelical state, where the damaged base was rotated fully into the enzyme active site and poised for glycosidic bond cleavage. Such structural efforts have been extraordinarily successful, and coordinates for over 21 enzyme-DNA complexes have been deposited in the protein data bank (Table 1). Notwithstanding with these successes, a mechanistic understanding of the extended reaction coordinate for base flipping has not been forthcoming by documenting interactions present in structural snapshots of the reaction endpoint. This realization has been the driving force for the development of new structural and biophysical methods capable of elucidating transient intermediates that occur very early in the pathway, and for understanding the important mechanism of diffusional encounter with a damage site. These intriguing aspects of damage detection, and the mechanistic insights provided by these new approaches, are the primary focus of this review.
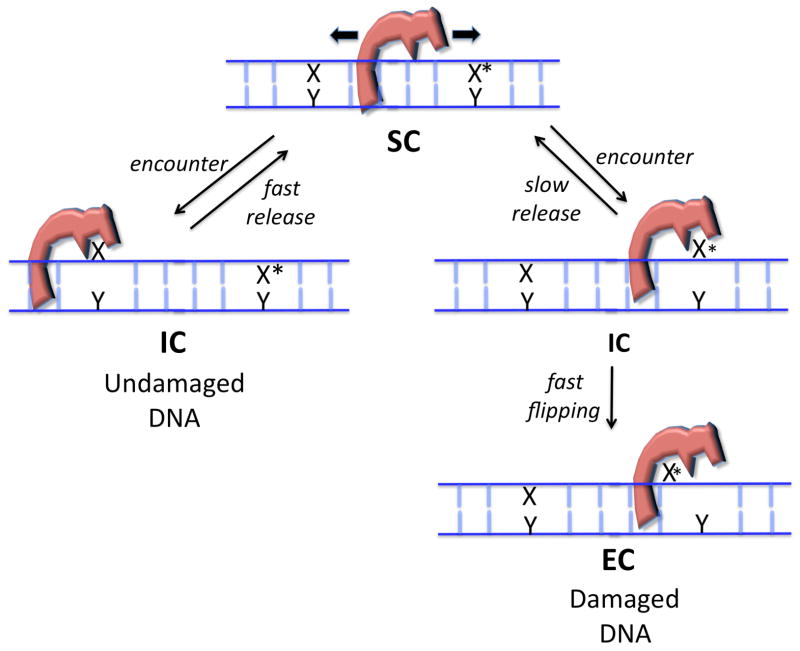
Here we highlight recent studies that are beginning to reveal the basic mechanistic elements in damaged base detection by several DNA glycosylases. We first discuss the fundamental physical problems associated with locating rare damaged bases in DNA so that the mechanistic solutions taken by these enzymes can be placed in a contextual framework. The first and most fundamental problem is genome coverage. In other words, what mechanisms are used to insure that these enzymes come in contact with three billion individual base pairs in the human genome such that an extremely rare (~1/106 bp) site of damage may be discovered before it permanently alters the genetic code? This question requires consideration of various possible mechanisms for diffusional encounter and their contribution to both speed and coverage. We refer to this as the enzymatic “search” problem and requires a special form of the enzyme we refer to as the search complex (SC, Fig. 3). A second problem is the initiating event that moves the enzyme from its search mode into an “interrogation” mode, where the enzyme can actually discern a structural or dynamic property of the damaged base that distinguishes it from a normal base. This key transition from the search complex to the interrogation complex (IC, Fig. 3) must be guided by a structural or dynamic property of the DNA, or alternatively, the enzyme. For many enzymes this step is especially enigmatic because the target base lesion neither structurally perturbs the average structure of the DNA duplex, nor does it provides a significant “handle” for detection while it is buried in the DNA base stack. Finally, the transition from an interrogation complex to a catalytically productive “excision” complex (EC, Fig. 3) involves a complex reaction coordinate for base flipping, which has been characterized in great detail for several enzymes. Our emphasis will be weighted towards three well-studied DNA glycosylases that recognize the pyrimidine base uracil (uracil DNA glycosylase, UNG), or the oxidized purine base 8-oxoguanine (the human or bacterial 8-oxo-guanine DNA glycosylases, hOGG1 and MutM). Although we focus on three enzymes where the experimental light now shines the brightest, we will attempt make connections to other related enzymes wherever possible.
Figure 3.
Damaged base recognition involves three discrete enzyme DNA complexes. The search complex (SC) is a loose nonspecific complex that is competent for hopping and sliding along the DNA chain. The interrogation complex (IC) is competent for extrahelical or intrahelical interrogation of damaged and undamaged basepairs. The excision complex (EC) is specific for binding of an extrahelical damaged base.
THE MAGNITUDE OF THE SEARCH PROBLEM
It may be estimated that during each day of our lives about 104 base lesions arise in each cell of our bodies (41). Since mutations do not also arise with such alarming frequency, it must be concluded that base excision repair efficiently corrects these lesions, providing a lower limit to the efficiency by which DNA glycosylases locate these sites and initiate repair. In a genome size of ~7 × 109 bp and given the relatively high abundance of DNA repair glycosylases (~105 per nucleus)(42), it may be estimated that an individual glycosylase must survey about 70,000 bps of DNA. Of course this calculation assumes that all the base pairs of DNA are accessible to the glycosylase, which may not be true given the highly packed structure of chromatin, but it does give one measure of the magnitude of the problem.
It is of interest to ask how long the target search takes in a human nucleus? As a first approximation of the problem, one may use the Smoluchowski diffusion equation to calculate the average time for encounter of a single enzyme molecule with a target base pair in the absence of interactions with the rest of the DNA chain (eq 1) (43). Equation 1 simply states that the search time (tsearch, 3D) for diffusional encounter with a damaged base pair
| (1) |
increases in direct proportion to the volume of solution that must be searched (in this case the nuclear volume), and shortens with increasing rates of three-dimensional diffusion (D3), and as the target radius (r) increases (big targets are more likely to be encountered than small targets). Using reasonable values for D3 = 108 nm2/s for a typical enzyme with a diameter of 5 nm, a nuclear volume of ~1011 nm3, and a target radius of 0.34 nm for a single base pair, a search time of less than one hour may be calculated, which seems rapid enough compared to the average time for DNA replication in human cells. Although diffusion constants may be smaller in the nucleus, direct measurements indicate that this diminishment is less than 10-fold (44)*, and the search time would still smaller than the typical dividing time of a cell. Moreover, the time that is required for target location by simple diffusion is decreased in direct proportion to the number enzyme molecules present in the nucleus (~105), which when considered, pushes the calculated search time down to a very short time scale indeed.
However, this most simple of diffusional mechanisms becomes unsatisfactory when one considers that each enzyme molecule must locate a single damaged base pair that exists in a 30,000-fold excess of nonspecific DNA binding sites. Thus, it is 30,000 fold more probable that the enzyme will encounter undamaged DNA rather the target site. If no pathway existed for the transfer of the enzyme from a nonspecific site to a damage site, then undamaged DNA would serve as a potent competitive inhibitor of damage site repair. Fortunately, these enzymes have evolved a capacity to use nonspecific binding interactions with the DNA chain in a discrete search mode that allows efficient intramolecular transfer from an initial non-specific binding site to the site of damage (45). As we further develop below, intramolecular transfer, even over short segment lengths, serves another important purpose beyond providing a pathway to the target site: the enzyme remains in contact with the DNA, which is a fundamental requirement if rare sites of damage are to be detected.
MOLECULAR SPEED LIMITS IN THE DAMAGE SEARCH
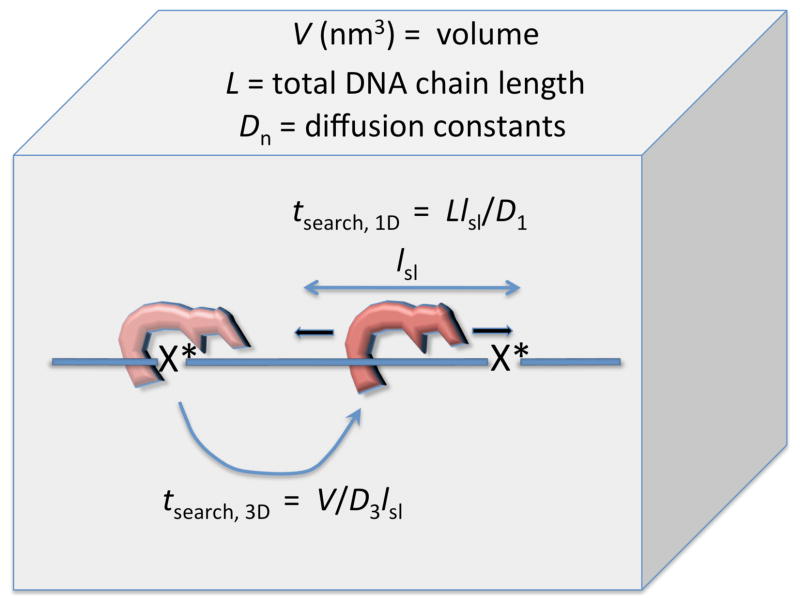
The above discussion reveals that three-dimensional (3D) diffusion and the relative abundance of glycosylase enzymes in the nucleus is sufficeint to lead to rapid encounters with a DNA chain in the nucleus, and is itself not a speed limit that would prevent timely repair. However, rapid encounter with single base pairs in DNA followed by dissociation to bulk solution provides exceedingly inefficient coverage because the enzyme must repeatedly diffuse to new landing sites, and it is exceedingly unlikely that a given association event will place the enzyme in perfect register with a damaged base if it is present. Thus it would be advantageous if the enzyme could use the DNA chain as a conduit for 1D sliding to the target site (Fig. 4). The average length of the DNA that can be sampled in a single stochastic sliding event (lsl) will depend on the 1D diffusion constant (D1) and the lifetime (tbound = 1/koff) of the enzyme on the DNA (lsl ~ √D1tbound)(43). In the case of a search that uses sequential 3D and 1D steps, the total search time (tsearch) to find a damage site embedded in DNA chain of total length L will be equal to the sum of the 3D and 1D search times (eq 2 and 3).
Figure 4.
Damaged base recognition involves both three dimensional diffusion and one dimensional DNA sliding (see text and eqs 2 and 3 for definitions).
| (2) |
| (3) |
The implications of eq 3 may be appreciated in straightforward way. The first 3D term reflects the time needed to encounter a target region of the DNA chain of size lsl nm. Note that this target size will be much larger than that of a single base pair (eq 1), and reflects the fact that if the enzyme lands within lsl nm of a damage site it will find it by sliding (Fig. 4). The second 1D term reflects the time it takes to search an entire DNA chain of length L using a 1D search velocity of D1/lsl (this 1D search requires many repeated 3D encounter events). An important aspect of tsearch is that increasing values of lsl decrease the tsearch, 3D term and increase the tsearch, 1D term. The decreasing efficiency of 1D intramolecular transfer with increasing sliding lengths reflects the intrinsic limitation of 1D stochastic motion. That is, the probability of taking a step towards a target site is exactly the same as that for moving away from the site, resulting in the statistical outcome that n2 steps need to be taken to move n basepairs towards a site (46). Theoretical analyses have concluded that sliding lengths in the range 10 to 100 bps (3.4 nm to 34 nm) will optimize the total search time in the human nucleus as described by eq 3 (43). In conclusion, these considerations of 1D and 3D pathways suggest that efficient damage site location would involve frequent enzyme dissociation events. These dissociation events provide the opportunity for the enzyme to reassociate nearby to its site of dissociation, or alternatively, to a distant site in the linear chain, which would be a likely outcome in the compact DNA context of the human nucleus. These new landing sites could then be interrogated using one base pair sliding steps over a 10–100 bp total sliding length. Such a mechanism encompasses the most efficient features of the 3D and 1D processes (Fig. 4).
GENERAL MECHANISTIC CONSIDERATIONS
Kinetic Aspects of the Search Process
The above theoretical considerations on diffusive pathways for damage site location, although highly instructive, treat the enzyme as a diffusing particle, and thus do not provide any mechanistic or physical basis for how an enzyme might interact with nonspecific DNA in a sliding mode, or what properties of the enzyme or DNA result in engagement of a damaged base when it is encountered. Although the process of DNA sliding is poorly understood both structurally and mechanistically (47)(48), it is possible to envision the central kinetic properties that would comprise a productive sliding mode. First, the enzyme must form sequence independent bindings interactions with the DNA that are neither too tight nor too loose. The simple rational behind this statement is that very tight interactions lead to a lack of mobility (small D1), and an increase in tsearch (eq 3). Alternatively, very loose interactions decrease the lifetime (1/koff) of the enzyme on the DNA, resulting in very short sliding lengths (lsl). Second, when the damaged site is encountered it is essential that a property of the site, or the enzyme, must somehow efficiently lead to a halt in the sliding process, and shift the enzyme into the interrogation conformation where it may begin the process of specifically engaging the site. If pausing at the site did not occur efficiently, leading to forward commitment along the base flipping pathway, then sliding would be an ineffective process. A closely related and essential question is whether pausing and interrogation of normal base pairs occurs during the sliding process. If such interrogation events occur (and several lines of experimental evidence suggest that they do), then these must be extraordinarily dynamic events that do not result in time consuming and unproductive pausing at undamaged base pairs. Consideration of these essential elements of sliding leads to the conclusion that each enzyme must have evolved a balance between its nonspecific binding lifetime on the DNA chain, its rate of sliding, and the rate of base pair interrogation. Although these individual rates may differ by orders of magnitude between different enzymes (see below), they must still be tuned within a given system to allow effective recognition. Thus, the relative magnitudes of these individual rates are key unknowns in developing an understanding of the search mode for each DNA glycosylase, and any experimental measurements of these individual rates (sliding, dissociation and the rate of recognition) must be interpreted in the context of the other rates to achieve a satisfying mechanistic model for the process.
What does a Sliding Enzyme Look Like?
What interactions between the enzyme and DNA are important for sliding? The answer to this question is paramount, yet little experimental data exists to answer it. In lieu of hard measurements, models have been proposed for phosphate backbone tracking, as well as minor and major groove tracking (49, 50). Indeed it is possible that different enzymes may employ different tracking mechanisms depending on the individual features of the damaged base that must be detected. For instance, detection of a minor groove perturbing lesion such as N3-methyladenine may be enhanced by minor groove tracking, whereas it is difficult to envision how uracil would be detected by such a mechanism. Tracking mechanisms invariably bring forth visions of these enzymes spiraling around the DNA helix as they move from one site to the next (50), but it cannot be excluded that more complicated trajectories may be followed where the enzyme diffuses or tumbles on the DNA surface (51)(15), rapidly reorientating itself with respect to the DNA strand polarity (see examples below). The most likely interactive force that would allow rapid sliding on the DNA, irrespective of DNA sequence, would be the comparatively weak electrostatic force between the enzyme and the DNA phosphate backbone. The electrostatic force has the virtue of being active over comparatively large distances (the energy of interaction between two point charges is proportional to 1/r, where r is their separation), which would serve to keep the enzyme in the vicinity of the DNA. In addition, the dense linear array of negatively charged phosphate atoms of the DNA backbone is ideally suited for intramolecular migration because the enzyme is always in close proximity to another set of phosphate interactions that would serve to pull it another step along the DNA chain. Electrostatically guided migration along the DNA backbone, perhaps transiently utilizing different regions of the protein surface distinct from the primary DNA binding mode, provides a plausible mechanism for rapid intramolecular transfer, and is consistent with the observation that facilitated diffusion is dramatically reduced in the presence of physiological concentrations of salt in all systems that have been investigated (52).
Base Flipping
After the enzyme has contacted the damaged site, the reaction coordinate for base flipping begins (Fig 2). One of the most controversial aspects of base flipping is whether the enzyme uses binding energy to perturb the DNA structure and facilitate base pair opening, or alternatively, whether the dynamic properties of DNA base pairs themselves are sufficiently rapid, and along the correct trajectory, to initiate the process of base flipping (53). These two mechanisms have been historically labeled “active” and “passive” with respect to the role of the enzyme, but this terminology is mechanistically ambiguous because it fails to address the essential aspects of the problem. First, the term active is poorly descriptive because base flipping always involves active participation of these enzymes. This is obvious from inspection of large structural distortions in the DNA (Fig. 2), which requires the utilization of enzyme binding energy to drive the base along the flipping reaction coordinate. However, structural observations of an apparently “active” mechanism, do not in any way address the elusive question of whether dynamic motions of the DNA served to initiate the process. The passive term is inscrutable because it also obscures the intrinsic dynamic nature of macromolecules, where thermally induced dynamic motions are often used to generate reactive conformers for a given biological process (54). Thus, a more useful way of framing the question is to ask whether damaged and undamaged DNA base pairs have kinetically competent motions along a coordinate that could serve to initiate the base flipping process, and whether differences in the dynamics of damaged and normal base pairs give rise to specificity.
Base flipping can be separated into two reaction coordinates that are distinct yet coupled (Fig. 5). The first describes the rotation of the base from the DNA base stack, which may be quantified in different ways, but is usually described by a pseudodihedral angle defined by the base, sugar and 3′ and 5′ phosphates of the flipped nucleotide (55). The second coordinate describes the extent of DNA bending, which can be quite severe (Fig. 2). The act of DNA bending likely promotes the flipping process by disrupting the duplex structure, and providing an unhindered path of the base out of the base stack. When intermediates during base flipping have been structurally characterized (Fig. 2), various enzymes show different degrees of progress along each reaction coordinate, possibly reflecting different structural requirements for flipping purine and pyrimidine bases. For instance, the structure of the IC for human UNG shows little bending, yet base rotation has proceeded to 20 % of the full trajectory (Fig. 2). In contrast, the IC structures of hOGG1 and MutM show extreme amounts of bending before or during rotation of guanine or 8-oxoguanine from the duplex (see discussion below).
Figure 5.
The free energy surface for enzymatic base flipping involves a DNA bending and base rotation reaction coordinate. Different enzymes show different degrees of progression along each axis during the process of flipping (Fig. 2). The progression of hOGG1 along both coordinates is qualitatively depicted in the free energy surface shown. The trajectory followed by UNG involves a greater progression along the rotation coordinate as compared to bending, while that of MutM may involve full bending before any rotation occurs (Fig. 2).
SPECIFIC EXAMPLES
There are a handful of DNA glycosylases for which mechanistic and structural studies have reached a high level of detail. In these cases, sufficient experimental observations have been made so as to allow the construction of reasonable models for the search, interrogation and excision stages of damaged base repair. These enzymes serve as paradigms for other glycosylases where less experimental data is currently available. The first enzyme that will be considered below is uracil DNA glycosylase (UNG), which locates and excises the pyrimidine base uracil from U/G and U/A base pairs, and also from single stranded DNA. The enzyme falls in its own structural class (Table 1), and is the most catalytically powerful of all the DNA repair glycosylases (2)(56). Unlike the other enzymes considered here, UNG acts equally well on double stranded and single stranded DNA substrates, and for this reason, the search and interrogation mechanisms of UNG may differ from other glycosylases listed in Table 1 that require duplex DNA for maximal activity. The remaining two enzymes that we consider recognize the oxidized guanine base, 8-oxoguanine (°G), in the context of °G/C base pairs (57). Human 8-oxoguanine DNA glycosylase (hOGG1) is a representative member of the largest structural superfamily of DNA glycosylases. These enzymes are characterized by a “helix-hairpin-helix” (HhH) structural motif that is associated with their DNA binding mode, making hOGG1 an important reference point for understanding the recognition mechanism of this entire superfamily. Bacterial 8-oxoguanine DNA glycosylase (MutM), has an entirely different protein fold from hOGG1, providing a nice comparative example for °G detection. In each case, we try to link the experimental observations with the intrinsic problems associated with searching the genome, pausing to interrogate base pairs, and moving damaged bases along the base flipping reaction coordinate.
The Case of Uracil and Uracil DNA Glycosylase (UNG)
The Search for Uracil
Uracil in DNA is generated from the deamination of cytosine or through the incorporation of dUTP by DNA polymerases, resulting in U/G or U/A base pairs that are substrates for UNG, as well as uracil in ssDNA (2). The DNA damage search mechanism of UNG has been investigated independently by several groups (15, 16, 58). The most extensive study employed an approach where two uracil sites were incorporated into a single linear DNA chain, and spacing between sites was varied over the range 20 to 800 base pairs (15). Under appropriate initial rate conditions, it is possible to quantify what fraction of single site excision events are then followed by intramolecular translocation and excision of the second uracil site in the same DNA chain, without dissociation of the enzyme to bulk solution. The efficiency of second site excision is referred to as the intramolecular transfer efficiency (fintra = T × E), and is determined by the probability that intramolecular transfer between sites occurs successfully (T), and the probability that once the second site is reached, the uracil is excised before the enzyme falls off the DNA (E). By varying the spacing between the two sites it is possible to evaluate if T diminishes with an n2 dependence on base pair (n) separation, as expected for a pure sliding mechanism (see above), or a 1/r dependence as expected from a hopping mechanism (where r is the distance in nm between the two sites) (46). Such data for Escherichia coli UNG at low salt concentration clearly followed a 1/r dependence(15), indicating that intramolecular transfer involved predominantly hopping, rather than sliding. A key finding was that only about 40% of UNG molecules that cleave one site ever make it to a second site located 20 bps away. The landing site size that could be extracted from this data was about one turn of the DNA helix, suggesting that when the enzyme lands within ~10 bp of a uracil site it remains in contact with the DNA, and uses short range sliding as the final step of the search process (lsl ~ 10 bp). By expanding the target region by a factor of ten over that for a single base pair, UNG has greatly increased its coverage of the genome per binding event. These findings are entirely consistent with the expected elements of a highly efficient search process as discussed above (eq 3).
A very interesting finding that arose from the two site intramolecular transfer studies of UNG was that fintra was the same regardless of whether the two uracils were on the same or opposite strands of the DNA, even for site spacings as small as 20 bps (15). As originally noted by Halford and coworkers (51), when two target sites are located on opposite DNA strands, successful intramolecular transfer must require at least one microscopic “dissociation” event that allows the enzyme to reorient its binding site with respect to the DNA strand polarity. Such a dissociation event does not result in loss of the enzyme from the vicinity of the DNA chain, but is sufficient to allow rapid rotation of the enzyme molecule with respect to the DNA axis. The importance of this observation with respect to the mechanism of intramolecular transfer by UNG cannot be overstated: UNG cannot simply track along a DNA groove or the DNA backbone. If such 1D tracking occurs, it must be frequently punctuated by rotation events.
Much is known about the kinetic properties of UNG, allowing a rough reconstruction of the temporal events involved in DNA association and scanning. UNG associates with short DNA oligonucleotides with a diffusion-controlled rate constant in the range 2–4 × 108 M−1 s−1 (D3 ~108 nm2 s−1) as determined in stopped-flow association measurements at room temperature in the presence of 10 mM NaCl (11). Thus, UNG has optimized tsearch, 3D (eq 2). With respect to the 1D search time via sliding (tsearch, 1D, eq 2), an estimated sliding length of lsl ~ 10 bp can be obtained from the target size measured in intramolecular transfer studies of UNG discussed above. Combining this estimate with measurements of the binding lifetime of UNG to undamaged DNA (tbound ~ 5 ms under the same conditions)(15), and the assumption that each sliding event results in stochastic sampling of each base pair over a sliding distance of lsl, a sliding diffusion constant of D1 ~ 104 bp2 s−1 can be calculated (D1 = lsl2/2tbound)(43). Although these are only rough magnitude estimates, they are instructive: over its relatively short ten base pair (n) sliding distance UNG will make n2 = 100 random walk steps, providing great redundancy for the search process without the time consuming oversampling of longer DNA stretches.
Dynamic Properties of the UNG Search Complex
Human UNG is amenable to modern heteronuclear NMR studies both in its free form and while bound to DNA (6, 59). Recent NMR studies focused on the nanosecond-millisecond timescale dynamic motions of the peptide backbone UNG, and revealed that the free enzyme has very little measureable dynamics over this broad timescale range (59). In contrast, when UNG binds to an undamaged 10 mer DNA duplex, dynamic motions were induced on a millisecond and microsecond timescale, suggesting that nonspecific DNA binding loosens the structure of UNG. The regions of the enzyme that displayed enhanced dynamics mapped to the known DNA binding surface that includes a minor groove intercalating residue (Leu272) and important serines that interact with the phosphate backbone around the flipped base (60). A normal mode analysis was performed to provide a structural interpretation for these dynamic motions. These computations revealed that the lowest energy motions available to the enzyme involved an open-to-closed structural transition that resembled the structural changes that occur when UNG binds to DNA (Fig. 6a). A rapid oscillation of UNG between an open form that interacts weakly with DNA and is competent for sliding, and a closed form that interrogates base pairs, is consistent with the dynamics of the sliding step and also the dynamics of base pair opening (see below). Thus, such dynamics may serve as the bridge that links the open search conformation to the closed interrogation conformation allowing the enzyme to pause and inspect individual base pairs.
Figure 6.
Normal mode analysis and NMR dynamic data suggest an open (left) to closed (right) transition in UNG during the interrogation of DNA for damage (see text). The open state is envisioned as competent for DNA sliding and the closed conformation (1OXM, Fig. 2) is competent for base interrogation.
Spontaneous Uracil Dynamics Initiates Formation of the Interrogation Complex (IC)
Duplex DNA is a very stable molecule held together by the synergistic effects of nucleobase π-stacking, hydrogen bonding, solvent exclusion and steric interactions. However, on a per residue basis, those forces favoring the canonical B-form structure are relatively weak, and bases are surprisingly free to sample conformations outside the DNA helix driven solely by thermal background energy (61). These spontaneous opening events are of significant interest as they seem to mirror the early stages of the base flipping pathway utilized by DNA glycosylases, and suggest that the ubiquity of the base flipping strategy may stem from exploitation of the intrinsic dynamic properties of the DNA molecule itself. It has been stated that spontaneous base pair opening events cannot serve as a viable mechanism for the initiation of the base flipping pathway because the concentration of the extrahelical conformation is vanishingly small (Kop = [out]/[in] < 10−5)(62). However, for a sliding enzyme the problem is not the unfavorable equilibrium thermodynamics, the issue is entirely kinetic (63). Thus, two conditions must be satisfied for a sliding enzyme to capture a spontaneously emerging base. First, the spontaneous breathing motions must be fast enough for the bound enzyme to sample, on average, at least one opening event during its lifetime on each base pair, and secondly, the motions must move the base into a pose that is recognized by the interrogation conformation of the enzyme (i.e. the motion must place the base on the flipping pathway).
Do spontaneous DNA base pair dynamic motions meet these two criteria? The fastest forward kinetic step that has been detected during rapid kinetic studies of multistep enzymatic base flipping by UNG is about k = 700 s−1 at room temperature, and this step has been assigned to exit of the uracil base from the base stack (64)(65). Therefore, if spontaneous base pair breathing motions are involved in initiating the whole process, these motions must be at least as fast as this reference rate to be kinetically competent. The spontaneous opening rates (kop) of T/A (30 s−1) and U/A (200 s−1)2 DNA base pairs in matched duplexes have been directly measured by NMR imino proton solvent exchange methods at 10 °C (63). After normalization to room temperature to match the kinetic conditions (66), the rates of U/A opening exceed several thousand per second, and easily meet the kinetic competence requirement. The 7-fold faster rates of U/A opening as compared to T/A also suggests that the intrinsic dynamic differences between these base pairs may facilitate specific damage recognition (U/G base pairs are expected to be even more dynamic)(66). Finally, the evidence that spontaneous breathing motions generate a conformer that is on the enzymatic base flipping pathway is compelling. Computer simulations indicate that imino exchange requires an ~50° rotation of the base from the DNA stack (67), which results in breaking of the interstrand hydrogen bonds but only partial unstacking of the flipped base. This computational view of extrahelical DNA bases is very similar to the actual crystal structure of extrahelical T captured in the exosite of UNG (Fig. 6b) (6). We have concluded from these comparisons that spontaneous opening events also meet the criterion of generating on-pathway conformers.
Finally, if spontaneous base pair opening motions are the initiating event in recognition, it follows that the rate of these motions should not be altered by the enzyme. Indeed, comprehensive NMR studies of the opening rate of T:X base pairs in the presence and absence of UNG have shown that the enzyme does not significantly alter the opening rate (kop)(6, 63, 68). Instead, UNG only acts to increase the open lifetime of T and U by transiently holding these bases in the exosite. The lifetime of thymine in the exosite is vanishingly small (~0.1 ms), which is consistent with the expectation that an efficient enzyme should not spend an undue length of time interrogating normal base pairs. In this regard, UNG does not appear to spend time interrogating normal G/C base pairs because the intrinsic stability of this pair does not allow frequent enough opening events for capture by the enzyme, and the exosite is not compatible with binding of C, G or A bases (6, 63).
Late Events on the Reaction Coordinate for Uracil Flipping
Once base pair dynamic motions have ejected uracil or thymine, placing the Watson-Crick edge of these bases in the U and T specific exosite, binding energy of the enzyme is used to selectively guide uracil the rest of the way into the active site. Rapid kinetic studies have revealed the presence of at least one more kinetic intermediate between the initial exosite complex and the final catalytically competent Michaelis complex (64). Although a structure of this intermediate has not been obtained, indirect evidence suggests that the DNA is more severely bent than in the exosite structure (11). The immense specificity for uracil over thymine may be achieved at this intermediate, or alternatively, at the subsequent Michaelis complex. The net effect is that thymine is excised 108 times slower than uracil because of the combined kinetic and thermodynamic barriers that select against this normal base. The enzymatic strategy during the late stages of base flipping is to break the process into discrete motional segments, punctuated by one or more base docking events that specifically guide uracil down a thermodynamic gradient into the active site pocket.
The Case of 8-oxoguanine Detection
The Search for 8-oxoguanine
Oxidized guanine bases are a prevalent form of DNA damage caused by reactive oxygen species and are removed by two structurally distinct enzymes, MutM in bacteria, and hOGG1 in humans (57). Similar to UNG, MutM and hOGG1 have diffusion-controlled association rates with duplex DNA oligonucleotides containing 8-oxoG (~3 × 108 M−1 s−1)(23, 30), and therefore, both enzymes have optimized tsearch, 3D. The ability of both enzymes to track along the DNA chain has been investigated using DNA constructs with two substrate sites incorporated into a single chain (see above for UNG)(25), and also, by direct observation using high-speed imaging of single-molecules (50, 69). The efficiency by which hOGG1 and MutM translocate and cleave a second 8-oxoG:C site spaced 19 bps away from another site was determined as ~0.65 – 0.8 in the presence of 0–10 mM KCl. However, these efficiencies fell to around 0.2 at a physiological concentration of salt ([KCl] = 150 mM). Although both enzymes have higher transfer efficiencies than UNG under similar conditions, the distance that is being interrogated is quite short, and it is important to realize that about 80 % of the molecules dissociate before translocating even 19 bps at physiological salt concentrations. Thus, the picture that emerges from these findings with MutM and hOGG1 is one of rapid and efficient three-dimensional diffusion to the DNA chain, and relatively short range sliding. These features are mechanistically similar to UNG.
Single-molecule imaging of hOGG1 and MutM sliding on undamaged, flow-stretched lambda DNA presents a somewhat different picture compared with the ensemble measurements (69). First, since flow methods efficiently wash away dissociated enzyme molecules, only sliding events can be measured using this approach. (However, single-molecule hopping has been observed using non-flow conditions, as recently demonstrated with nucleotide excision repair enzymes.) (70) At a near physiological salt concentration ([NaCl] = 100 mM), a mean sliding distance of 440 bp was reported for hOGG1. This mean sliding distance occurred during a mean binding lifetime of 25 ms, and thus required an extremely rapid 1D diffusion constant of D1 = 5 × 106 bp2 s−1, corresponding to a near barrierless activation energy for sliding of 0.5 kcal/mol. Similar studies with MutM revealed a 10-fold slower diffusion constant than hOGG1 (D1 = 4 × 105 bp2 s−1), which was rationalized with respect to the decreased requirement for scanning a bacterial genome that is has 1/1,000th the number of base pairs. Since 8-oxoG damaged sites were not present in the lambda DNA, it is not known how efficiently such rapid sliding events may be paused in order to interrogate base pairs for damage.
A second aspect of the search mechanism of hOGG1 and MutM has been indirectly revealed by single molecule imaging: these proteins move by spinning around the DNA helix (50). This interesting study varied each protein’s radius (r) through conjugation of strepavidin, and then measured the 1D diffusion constants (D1) of the native and conjugated forms of the enzymes. Although the limited spatial resolution of single molecule imaging cannot directly detect spinning of individual enzyme molecules on DNA, it can test the expected effect of a given change in molecular radius on D1. If only linear motion along the DNA chain axis is occurring, then D1 should show a 1/r dependence, if coupled rotation is also occurring, a dependence that is proportional to 1/r3 would be expected. The data for both hOGG1 and MutM were best fitted with the 1/r3 dependence on protein radius, leading the authors to conclude that these proteins track along the major or minor groove, or the backbone of the DNA chain. It will be of interest to uncover whether this apparent spinning behavior also involves rapid molecular rotation of the enzymes with respect to the strand polarity, as required in strand transfer by UNG (see above).
Transitioning to the Interrogation Complex
A key question raised by the rapid sliding rates of both MutM and hOGG1 is how these enzymes know when to stop moving like race cars down the track? A further enigma is that hOGG1 is known to dramatically kink DNA both in the absence and presence of an 8-oxoG/C lesion, and the kinking appears to be required to present both G and 8-oxoG in the exosite (21, 22, 71). Since it seems implausible that the kinked complex is competent for the observed rapid sliding, let alone spinning, there must be an essential dynamic transition between a loose sliding conformation of hOGG1 and the conformation in which the DNA is bent. Achievement of the MutM or hOGG1 complexes in which the DNA is bent and destabilized occurs on a time frame of less than 10 ms as judged by rapid kinetic measurements (23, 30), and the initial nonspecific encounter complexes dissociate rapidly (bound lifetimes in range 1 to 5 ms at [KCl] = 50 mM) )(23, 30). The shorter lifetimes of the encounter complexes in these ensemble kinetic measurements, as compared to the single molecule studies described above, may arise from the short DNA sequences that were used (12 mers), or may reflect that the single molecule studies observe a small fraction of the binding events due to the time resolution limitations of the imaging and flow methods. Regardless, the similarities between the interrogation mechanisms of MutM, hOGG1 and UNG are abundant, with rapid dynamic motions leading to efficient interrogation of base pairs without undue pausing at undamaged sites. The most significant mechanistic difference between these 8-oxo-G detecting enzymes and UNG is that they severely bend DNA, even in the absence of damage. Thus on the basis of these limited examples, there appears to be a divergence in the enzymatic strategy for extrahelical inspection of small pyrimidine bases such as thymine and uracil, and bulky purine bases such as G and 8-oxo-G. It is possible that the kinetic barriers for spontaneous exposure of G and 8-oxo-G are not kinetically competent for efficient recognition. [Spontaneous G/C base pair opening is typically much slower than T/A opening (62), but 8-oxo-G has been suggested by computation to have a lower barrier to base extrusion as compared to G (72)]. Alternatively, some other structural requirement for recognition may not be satisfied by a spontaneous base pair opening process, forcing these enzymes to evolve a bend and flip strategy for extrahelical recognition of G and 8-oxo-G. If this is true, it may be generally found that extrahelical recognition of purine bases follows such a strategy.
Events on the Reaction Coordinate for 8-oxoG Flipping by MutM and hOGG1
The strategy of bending both damaged and undamaged DNA (at least in the case of hOGG1) likely comprises the earliest event in G and 8-oxoG flipping by these enzymes. Once DNA is bent, its dynamic properties are no longer that of naked B DNA, but instead, that of the new enzyme induced conformation. Thus, if spontaneous thermal motions of G and 8-oxo-G are important in moving these bases onto the flipping reaction coordinate, enzyme binding energy may be used to strain the DNA and lower the kinetic barrier to base flipping.
The structures of early, unstable extrahelical intermediates involving G and 8-oxo-G have been investigated by trapping enzyme-DNA complexes using the disulfide crosslinking (DXL) method developed in the Verdine lab (20–22, 26, 27, 28). In the case of MutM, the beneficial effect of early DNA bending has been recently investigated in the context of a loop mutant that is deficient in stabilizing an extrahelical base (27). In this context, DNA containing G or 8-oxo-G was trapped using DXL, and in both cases the base was found to still reside within the DNA stack, even in the presence of nearly complete DNA bending. The salient structural features of the intrahelical complex with this loop mutant that may lead to facilitated base extrusion are (i) insertion of a destabilizing Phe residue into the base stack adjacent to the target base pair, and (ii) conformational distortions in the DNA phosphate backbone and sugar pucker that introduce ground state strain and thereby lower the barrier to flipping out of the minor groove, which differs from the major groove pathway of UNG and hOGG (7). It is not clear whether this structure should be interpreted as an extremely early intermediate during the flipping process, or simply the lowest energy conformation to which the system relaxes in the absence of loop interactions that stabilize extrahelical G and 8-oxo-G. What can be inferred is that bending alone is not sufficient to drive these bases into a stable extrahelical state. As with any crystal structure, it is never possible to discern the order of events that lead to an observed conformation. Thus, even in the case of DNA bending and destabilization by MutM, it is possible that intrinsic base pair dynamics initiates the process by allowing insertion of the destabilizing Phe residue, and damage discrimination would then arise from the increased dynamic behavior of 8-oxo-G:C base pairs as compared to G:C pairs (72).
Like UNG, both MutM and hOGG1 have taken a strategy of docking the extrahelical base in one or more transient binding sites along reaction coordinate, and some of these intermediates have been trapped using DXL strategies (Fig. 2). Kinetic studies of the 8-oxo-G flipping pathway establish that the interconversion of these intermediates is one or two orders of magnitude slower than the interconversion rates for late intermediates during UNG facilitated base flipping)(10, 23, 30). However, in all three cases a theme emerges where one or more discrete extrahelical intermediates form rapidly, and the last step before glycosidic bond cleavage involves a conformational change in each enzyme that results in the final docking interactions within the active site. This final step appears to involve the insertion of an amino acid plug into the DNA stack to fill the void left by the now fully extrahelical base. It is of interest to note that each intermediate that forms along the pathway to the active site can kinetically partition forward or backwards because of the similar activation barriers in both directions. In the case of hOGG1 and MutM, many of these steps are quite slow (~0.01 – 1 s−1), suggesting that normal bases never make it to these late intermediates, or else these enzymes would spend far too much time lingering on undamaged DNA. The slow rates for interconverting the late extrahelical intermediates for MutM and hOGG1 are appropriately tuned with the slow rate of glycosidic bond cleavage by these enzymes (~0.03 s−1 as compared to 110 s−1 for UNG): if these intermediates formed and decayed much quickly than the rate of chemistry, then the process would be extremely inefficient and unproductive.
DAMAGE REMOVAL IN THE CONTEXT OF CHROMATIN
A reasonable person can wonder what the study of enzymes interacting with short duplex DNAs in vitro has to do with DNA repair in the cell? Certainly, the work described above has relevance, but only so far as these enzymes execute repair on a naked DNA molecule. The vast majority of DNA in the human genome does not exist in this form on average, which begs the question of how efficiently these DNA glycosylases operate on DNA wrapped in nucleosomes or in the context of chromatin. Several investigations into the activity of UNG against uracil sites in the context of nucleosome bound DNA have been reported, and the findings are that uracil can be efficiently excised in a position dependent manner that is far from well-understood (73, 74). The observation of DNA glycosylase activity in the context of nucleosomes pushes the study of DNA repair into a new realm where enzymatic damage detection may be closely coupled with the local and global dynamics of histone-bound DNA. The largely 3D hopping strategies used by these enzymes is highly suited for bypassing nucleosome bound DNA when it is encountered, and for short range scanning of transient stretches of naked DNA. However, it is now clear that when nucleosomes are encountered, the remarkable DNA glycosylases have adapted mechanisms to detect and repair damage in the context of this very different substrate.
Figure 7.
Thermally induced flipping of a thymidine base results in an open state that closely resembles the conformation of thymine when bound in the exosite of UNG (see text). The figure is a view down the DNA helical axis, and depicts the conformation of intrahelical thymine in B DNA (blue), the minimum extrahelical state of thymine that is required for imino proton exchange (yellow), and the conformation of thymine when bound to the exosite of UNG (red). To facilitate visual comparisons, the bases have been projected onto a common plane, and the syn-conformation of thymine in the exosite structure is shown in the anti form for ease of comparison with the other two states. For reference, the circle depicts the circumference described by the DNA sugar phosphate backbone.
Abbreviations
- SC
search complex
- IC
interrogation complex
- EC
excision complex
Footnotes
Stivers, J.T. and Parker, J.B., unpublished measurements (2010).
This work was supported by NIH grant GM056834 (J.T.S).
References
- 1.Lindahl T. An N-Glycosidase from Escherichia Coli that Releases Free Uracil from DNA Containing Deaminated Cytosine Residues. Proc Natl Acad Sci U S A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stivers JT, Jiang YL. A Mechanistic Perspective on the Chemistry of DNA Repair Glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 3.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic Model of a Pyrimidine Dimer Excision Repair Enzyme Complexed with a DNA Substrate: Structural Basis for Damaged DNA Recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 4.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A Nucleotide-Flipping Mechanism from the Structure of Human Uracil-DNA Glycosylase Bound to DNA [See Comments] Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 5.Roberts RJ, Cheng X. Base Flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 6.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic Capture of an Extrahelical Thymine in the Search for Uracil in DNA. Nature. 2007;449:433–438. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA Glycosylase-DNA Substrate and Product Structures: Conformational Strain Promotes Catalytic Efficiency by Coupled Stereoelectronic Effects. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchet MA, Seiple LA, Jiang YL, Ichikawa Y, Amzel LM, Stivers JT. Electrostatic Guidance of Glycosyl Cation Migration Along the Reaction Coordinate of Uracil DNA Glycosylase. Biochemistry. 2003;42:12455–12460. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 9.Werner RM, Stivers JT. Kinetic Isotope Effect Studies of the Reaction Catalyzed by Uracil DNA Glycosylase: Evidence for an Oxocarbonium Ion-Uracil Anion Intermediate. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 10.Jiang YL, Song F, Stivers JT. Base Flipping Mutations of Uracil DNA Glycosylase: Substrate Rescue using a Pyrene Nucleotide Wedge. Biochemistry. 2002;41:11248–11254. doi: 10.1021/bi026227j. [DOI] [PubMed] [Google Scholar]
- 11.Krosky DJ, Song F, Stivers JT. The Origins of High-Affinity Enzyme Binding to an Extrahelical DNA Base. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 12.Bellamy SR, Krusong K, Baldwin GS. A Rapid Reaction Analysis of Uracil DNA Glycosylase Indicates an Active Mechanism of Base Flipping. Nucleic Acids Res. 2007;35:1478–1487. doi: 10.1093/nar/gkm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-State Analysis of a Single Catalytic Turnover by Escherichia Coli Uracil-DNA Glycosylase Reveals a “Pinch-Pull-Push” Mechanism. J Biol Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 14.Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia Coli and Rat Liver Mitochondrial Uracil-DNA Glycosylase is Affected by NaCl Concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 15.Porecha RH, Stivers JT. Uracil DNA Glycosylase Uses DNA Hopping and Short-Range Sliding to Trap Extrahelical Uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated Cleavage of Single- and Double-Stranded Substrates by Uracil-DNA Glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Higley M, Lloyd RS. Processivity of Uracil DNA Glycosylase. Mutat Res. 1993;294:109–116. doi: 10.1016/0921-8777(93)90019-d. [DOI] [PubMed] [Google Scholar]
- 18.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase SMUG1. Mol Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 19.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal Structure of Human Thymine DNA Glycosylase Bound to DNA Elucidates Sequence-Specific Mismatch Recognition. Proc Natl Acad Sci U S A. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A, Verdine GL. A Nucleobase Lesion Remodels the Interaction of its Normal Neighbor in a DNA Glycosylase Complex. Proc Natl Acad Sci U S A. 2006;103:15020–15025. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a Repair Enzyme Interrogating Undamaged DNA Elucidates Recognition of Damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 22.Bruner SD, Norman DP, Verdine GL. Structural Basis for Recognition and Repair of the Endogenous Mutagen 8-Oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 23.Kuznetsov NA, Koval VV, Nevinsky GA, Douglas KT, Zharkov DO, Fedorova OS. Kinetic Conformational Analysis of Human 8-Oxoguanine-DNA Glycosylase. J Biol Chem. 2007;282:1029–1038. doi: 10.1074/jbc.M605788200. [DOI] [PubMed] [Google Scholar]
- 24.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of Substrate Recognition and Cleavage by Human 8-Oxoguanine-DNA Glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidorenko VS, Zharkov DO. Correlated Cleavage of Damaged DNA by Bacterial and Human 8-Oxoguanine-DNA Glycosylases. Biochemistry. 2008;47:8970–8976. doi: 10.1021/bi800569e. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee A, Santos WL, Verdine GL. Structure of a DNA Glycosylase Searching for Lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 27.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and Extrusion of an Intrahelical Lesion by a DNA Repair Enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fromme JC, Verdine GL. DNA Lesion Recognition by the Bacterial Repair Enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 29.Koval VV, Kuznetsov NA, Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA, Fedorova OS. Pre-Steady-State Kinetics shows Differences in Processing of various DNA Lesions by Escherichia Coli Formamidopyrimidine-DNA Glycosylase. Nucleic Acids Res. 2004;32:926–935. doi: 10.1093/nar/gkh237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koval VV, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Real-Time Studies of Conformational Dynamics of the Repair Enzyme E. Coli Formamidopyrimidine-DNA Glycosylase and its DNA Complexes during Catalytic Cycle. Mutat Res. 2010;685:3–10. doi: 10.1016/j.mrfmmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural Basis for Removal of Adenine Mispaired with 8-Oxoguanine by MutY Adenine DNA Glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 32.McCann JA, Berti PJ. Transition-State Analysis of the DNA Repair Enzyme MutY. J Am Chem Soc. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 33.Wong I, Bernards AS, Miller JK, Wirz JA. A Dimeric Mechanism for Contextual Target Recognition by MutY Glycosylase. J Biol Chem. 2003;278:2411–248. doi: 10.1074/jbc.M209802200. [DOI] [PubMed] [Google Scholar]
- 34.Francis AW, David SS. Escherichia Coli MutY and Fpg Utilize a Processive Mechanism for Target Location. Biochemistry. 2003;42:801–810. doi: 10.1021/bi026375+. [DOI] [PubMed] [Google Scholar]
- 35.Hollis T, Ichikawa Y, Ellenberger T. DNA Bending and a Flip-Out Mechanism for Base Excision by the Helix- Hairpin-Helix DNA Glycosylase, Escherichia Coli AlkA. EMBO J. 2000;19:758–66. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular Basis for Discriminating between Normal and Damaged Bases by the Human Alkyladenine Glycosylase, AAG. Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal Structure of a Human Alkylbase-DNA Repair Enzyme Complexed to DNA: Mechanisms for Nucleotide Flipping and Base Excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 38.Hedglin M, O’Brien PJ. Human Alkyladenine DNA Glycosylase Employs a Processive Search for DNA Damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metz AH, Hollis T, Eichman BF. DNA Damage Recognition and Repair by 3-Methyladenine DNA Glycosylase I (TAG) EMBO J. 2007;26:2411–2420. doi: 10.1038/sj.emboj.7601649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fromme JC, Verdine GL. Structure of a Trapped Endonuclease III-DNA Covalent Intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindahl T. Instability and Decay of the Primary Structure of DNA [See Comments] Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 42.Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of Base Excision Repair are Not Solely Dependent on Levels of Initiating Enzymes. Carcinogenesis. 2001;22:387–393. doi: 10.1093/carcin/22.3.387. [DOI] [PubMed] [Google Scholar]
- 43.Halford SE, Marko JF. How do Site-Specific DNA-Binding Proteins Find their Targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmo-Fonseca M, Platani M, Swedlow JR. Macromolecular Mobility Inside the Cell Nucleus. Trends Cell Biol. 2002;12:491–495. doi: 10.1016/s0962-8924(02)02387-5. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd RS, Hanawalt PC, Dodson ML. Processive Action of T4 Endonuclease V on Ultraviolet-Irradiated DNA. Nucleic Acids Res. 1980;8:5113–527. doi: 10.1093/nar/8.21.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg HC. Random Walks in Biology. Princeton University Press; 1993. [Google Scholar]
- 47.Kampmann M. Facilitated Diffusion in Chromatin Lattices: Mechanistic Diversity and Regulatory Potential. Mol Microbiol. 2005;57:889–899. doi: 10.1111/j.1365-2958.2005.04707.x. [DOI] [PubMed] [Google Scholar]
- 48.Zharkov DO, Grollman AP. The DNA Trackwalkers: Principles of Lesion Search and Recognition by DNA Glycosylases. Mutat Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Givaty O, Levy Y. Protein Sliding Along DNA: Dynamics and Structural Characterization. J Mol Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 50.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically Bound Proteins Spin while Diffusing Along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gowers DM, Wilson GG, Halford SE. Measurement of the Contributions of 1D and 3D Pathways to the Translocation of a Protein Along DNA. Proc Natl Acad Sci U S A. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halford SE. An End to 40 Years of Mistakes in DNA-Protein Association Kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 53.Stivers JT. Extrahelical Damaged Base Recognition by DNA Glycosylase Enzymes. Chemistry. 2008;14:786–793. doi: 10.1002/chem.200701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammes-Schiffer S, Benkovic SJ. Relating Protein Motion to Catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 55.Priyakumar UD, MacKerell AD., Jr Computational Approaches for Investigating Base Flipping in Oligonucleotides. Chem Rev. 2006;106:489–505. doi: 10.1021/cr040475z. [DOI] [PubMed] [Google Scholar]
- 56.Zharkov DO, Mechetin GV, Nevinsky GA. Uracil-DNA Glycosylase: Structural, Thermodynamic and Kinetic Aspects of Lesion Search and Recognition. Mutat Res. 2010;685:11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David SS, O’Shea VL, Kundu S. Base-Excision Repair of Oxidative DNA Damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purmal AA, Lampman GW, Pourmal EI, Melamede RJ, Wallace SS, Kow YW. Uracil DNA N-Glycosylase Distributively Interacts with Duplex Polynucleotides Containing Repeating Units of either TGGCCAAGCU Or TGGCCAAGCTTGGCCAAGCU. J Biol Chem. 1994;269:22046–22053. [PubMed] [Google Scholar]
- 59.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA Binding Shapes the Dynamic Landscape for Enzymatic Recognition of DNA Damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Werner RM, Jiang YL, Gordley RG, Jagadeesh GJ, Ladner JE, Xiao G, Tordova M, Gilliland GL, Stivers JT. Stressing-Out DNA? The Contribution of Serine-Phosphodiester Interactions in Catalysis by Uracil DNA Glycosylase. Biochemistry. 2000;39:12585–1294. doi: 10.1021/bi001532v. [DOI] [PubMed] [Google Scholar]
- 61.Banavali NK, MacKerell AD. Re-Examination of the Intrinsic, Dynamic and Hydration Properties of Phosphoramidate DNA. Nucleic Acids Res. 2001;29:3219–3230. doi: 10.1093/nar/29.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gueron M, Charretier E, Hagerhorst J, Kochoyan M, Leroy JL, Moraillon A, Charretier E, Hagerhorst J, Kochoyan M, Leroy JL, Moraillon A. In: Struct Methods; Applications of Imino Proton Exchange to Nucleic Acid Kinetics and Structures. Gueron M, editor. Adenine press; 1990. [Google Scholar]
- 63.Cao C, Jiang YL, Stivers JT, Song F. Dynamic Opening of DNA during the Enzymatic Search for a Damaged Base. Nat Struct Mol Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 64.Jiang YL, Stivers JT. Mutational Analysis of the Base Flipping Mechanism of Uracil DNA Glycosylase. Biochemistry. 2002;41:11236–11247. doi: 10.1021/bi026226r. [DOI] [PubMed] [Google Scholar]
- 65.Wong AW, He S, Grubb JH, Sly WS, Withers SG. Identification of Glu-540 as the Catalytic Nucleophile of Human Beta- Glucuronidase using Electrospray Mass Spectrometry. J Biol Chem. 1998;273:34057–3462. doi: 10.1074/jbc.273.51.34057. [DOI] [PubMed] [Google Scholar]
- 66.Moe JG, Russu IM. Kinetics and Energetics of Base-Pair Opening in 5′-d(CGCGAATTCGCG)-3′ and a Substituted Dodecamer Containing G.T Mismatches. Biochemistry. 1992;31:8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- 67.Giudice E, Varnai P, Lavery R. Base Pair Opening within B-DNA: Free Energy Pathways for GC and AT Pairs from Umbrella Sampling Simulations. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao C, Jiang YL, Krosky DJ, Stivers JT. The Catalytic Power of Uracil DNA Glycosylase in the Opening of Thymine Base Pairs. J Am Chem Soc. 2006;128:13034–13035. doi: 10.1021/ja062978n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A Base-Excision DNA-Repair Protein Finds Intrahelical Lesion Bases by Fast Sliding in Contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative Dynamic DNA Scanning by Nucleotide Excision Repair Proteins Investigated by Single- Molecule Imaging of Quantum-Dot-Labeled Proteins. Mol Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L, Haushalter KA, Lieber CM, Verdine GL. Direct Visualization of a DNA Glycosylase Searching for Damage. Chem Biol. 2002;9:345–350. doi: 10.1016/s1074-5521(02)00120-5. [DOI] [PubMed] [Google Scholar]
- 72.Cheng X, Kelso C, Hornak V, de los Santos C, Grollman AP, Simmerling C. Dynamic Behavior of DNA Base Pairs Containing 8-Oxoguanine. J Am Chem Soc. 2005;127:13906–13918. doi: 10.1021/ja052542s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beard BC, Wilson SH, Smerdon MJ. Suppressed Catalytic Activity of Base Excision Repair Enzymes on Rotationally Positioned Uracil in Nucleosomes. Proc Natl Acad Sci U S A. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA Glycosylase Activity on Nucleosomal DNA Depends on Rotational Orientation of Targets. J Biol Chem. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lloyd RS, Hanawalt PC, Dodson ML. Processive action of T4 endonuclease V on ultraviolet-irradiated DNA. Nucleic Acids Res. 1980;8:5113–5127. doi: 10.1093/nar/8.21.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]