Summary
It is generally believed that cholesterol homoeostasis in the brain is both linked to and impacted by Alzheimer’s disease (AD). For example, elevated levels of cholesterol in neuronal plasma and endosome membranes appears to be a pro-amyloidogenic factor. The recent observation that the C-terminal transmembrane domain (C99, also known as the β-CTF) of the amyloid precursor protein (APP) specifically binds cholesterol helps to tie together previously loose ends in the web of our understanding of Alzheimer’s-cholesterol relationships. In particular, binding of cholesterol to C99 appears to favor the amyloidogenic pathway in cells by promoting localization of C99 in lipid rafts. In turn, the products of this pathway—amyloid-β and the intracellular domain of the APP (AICD)—may down-regulate ApoE-mediated cholesterol uptake and cholesterol biosynthesis. If confirmed, this negative-feedback loop for membrane cholesterol levels has implications for understanding the function of the APP and for devising anti-amyloidogenic preventive strategies in AD.
Keywords: Alzheimer’s disease, amyloid precursor protein, APP, cholesterol, NMR, lipid rafts, membranes, structure, trafficking
Introduction
An early-2010 PubMed search on “Cholesterol AND Alzheimer’s” yielded 1500 hits, exemplifying the large amount of attention devoted to unraveling this lipid-disease relationship. However, there remain many challenges to attaining a definitive understanding. For example, the results of experiments designed to elucidate linkages between the many proteins involved in cholesterol homeostasis and processes believed to be central to the etiology of Alzheimer’s disease (AD) are rarely unambiguous because of pleiotropy and because of difficulties in extrapolating results from experiments with cultured cells to physiological conditions. Another set of challenges relates to detecting and monitoring the AD-relevant pools of cholesterol in neurons and glial cells. While familial hypercholesterolemia and obesity are generally regarded as risk factors for AD(1), cholesterol metabolism in the brain is largely isolated by the blood-brain barrier from cholesterol metabolism in the rest of the body, with nearly all brain cholesterol being synthesized in situ(2–6). Measurement of total cholesterol levels in the CNS is not informative because 70–80% of brain cholesterol is dedicated to a structural role in myelin membranes(7). Modest quantities of cholesterol leave the brain primarily in the form of 24S-hydroxycholesterol, but enters in very small amounts only—in the form of 27-hydroxycholesterol(2;8). While cholesterol’s mean half-life in the brain has been estimated to be at least 5 years its turnover in metabolically-active neurons is believed to occur much more rapidly than in glia and in myelin (2;5;9). There is data that suggests cholesterol-lowering statins reduce the risk of AD(1;10), although this is not without controversy. It is clear that apolipoprotein E (ApoE) is responsible for transport of cholesterol between cells in the brain and that the ε4 isoform of the protein is a major risk factor for the common late-onset form of AD; however, the exact mechanisms by which different variants of ApoE and related lipoprotein receptors variously promote or suppress AD pathogenesis are not understood(11–13).
Despite such limitations, there is a large body of evidence suggesting that increased levels of plasma membrane cholesterol promote the amyloidogenic processing of the amyloid precursor protein (APP) and thereby contribute to the key series of molecular (proteolytic) events widely believed to underlay with the etiology of AD(1;14–22). A number of genes involved in cholesterol homeostasis have been identified as susceptibility loci for sporadic or late-onset Alzheimer’s disease (13;23–25). There is also evidence that cholesterol and other specific lipids may enhance the propensity of the amyloid-β (Aβ) polypeptide to form neurotoxic aggregates(26–32), although these latter studies are outside of the scope of this contribution. Amyloid plaques found in the brains of Alzheimer’s patients are highly enriched in cholesterol(33).
As referenced above there are a number of excellent recent reviews on various aspects of the cholesterol-AD relationship, such that a comprehensive survey is not here merited. Instead, this paper is focused on very recent results from our own lab that offer a mechanistic hypothesis that links together a number of previous observations regarding how cholesterol promotes amyloidogenesis and how processing of the amyloid precursor protein may in turn impact cholesterol homeostasis.
Structure of C99, the Transmembrane C-Terminal Domain of the Amyloid Precursor Protein (APP)
While the structures of the Aβ polypeptides in various states have been the subject of numerous biophysical studies, there have been few attempts to study the structure of the immediate precursor to Aβ: the 99 residue transmembrane C-terminal domain (C99) of APP. C99 is the product of β-secretase cleavage of APP and is the substrate of γ-secretase cleavage. We recently initiated structural studies of C99 using solution nuclear magnetic resonance (NMR) spectroscopy to examine this protein under conditions in which it is solubilized in membrane-mimetic lyso-myristoylphosphatidylglycerol micelles(34). While the use of detergent micelles in biophysical studies of membrane proteins is not without pitfalls, the lyso-phospholipids used in our study are structurally similar to true phospholipids and are generally thought to be “mild” in the sense that they are good at maintaining native-like membrane protein function and stability (see review in(35)). This study of full length C99 complements and extends results from studies of fragments of this protein (36–48), most of which did not include an intact transmembrane segment.
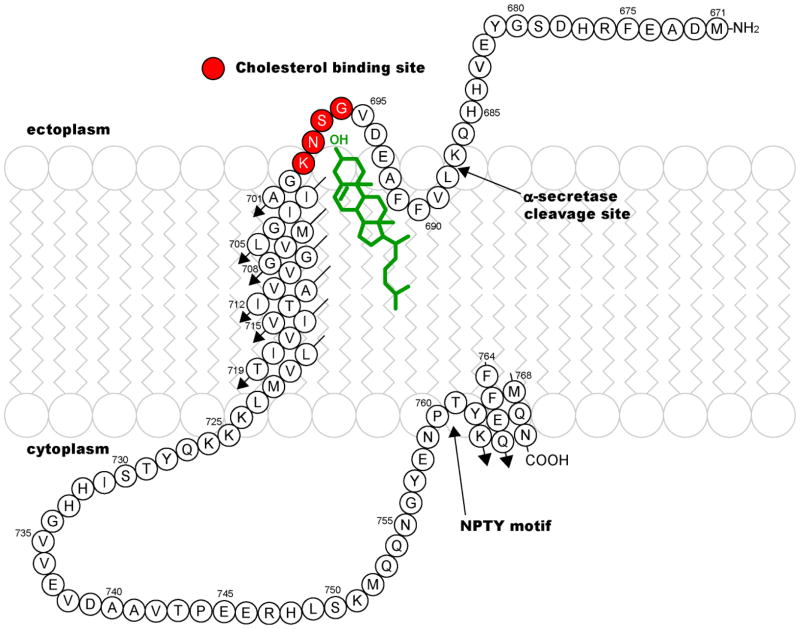
While the process of completing a high resolution structure for C99 is on-going, its secondary structure and membrane topology have been characterized as summarized in Figure 1. The transmembrane (TM) domain appears to be an unbroken helix(34;45). Calculations suggest that the stability of this helix in the vicinity of the γ-secretase cleavage sites may be lower than normal for standard TM helices(49), such that it may be relatively easy to disrupt this helix when it binds to the active site of γ-secretase, as required for cleavage. The cytosolic distal C-terminus of C99 is an amphipathic helix that is associated with the membrane surface(34). This domain starts right after the important NPTY Tyr-phosphorylation motif, which is thought to be critical to protein-protein interactions that play a role in regulating intracellular trafficking of APP and derived C99(38;42;50;51). A second surface-associated segment was found in the extracellular domain, where a short hydrophobic sequence (LVFFA) dips into the membrane, followed by a short connecting loop to the beginning the TM domain(34). The site for non-amyloidogenic α-secretase cleavage is located just before this membrane-interactive motif. It is likely that the affinities of both the extracellular hydrophobic segment and the distal C-terminus for the membrane surface are not very high, such that the association of these segments with the membrane surface in vivo may be modulated by the local membrane lipid composition and/or (for the C-terminus) phosphorylation of Tyr762. One can also speculate that α-secretase cleavage of C99 and/or C99 trafficking may be coupled to the state of membrane association of one or both of these segments.
Figure 1.
Secondary structure and membrane topology of C99 and location of the focal point for its recognition and binding of cholesterol (red-highlighted sites), as derived from NMR studies of this protein in LMPG micelles(34).
The NMR results were also consistent with a reported tendency for C99 to homodimerize(40;45;52–55), although the avidity of C99 dimers in the lyso-phospholipid micelles used in the NMR studies appears to be only modest(34), suggesting that the oligomeric state of C99 in cells may favor either monomer or dimer depending on C99 concentration, membrane composition, and state of interactions with other proteins.
Evidence that the C-terminal Transmembrane Domain of APP Binds Cholesterol
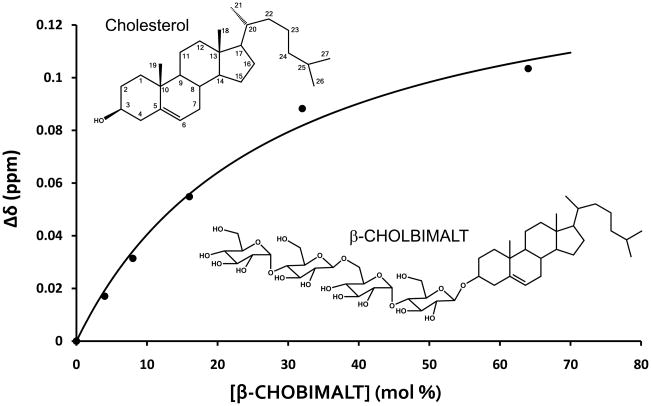
The binding of cholesterol and cholesterol analogs to C99 in lyso-phospholipid micelles was monitored using NMR(34). At concentrations that approached its (low) solubility limit cholesterol induced significant shifts in a sub-set of C99’s backbone amide NMR resonances. The residues associated with the peaks that underwent the largest shifts in response to cholesterol were localized almost exclusively to the extracellular loop connecting the surface-associated hydrophobic segment in the extracellular domain and the start of the TMD, suggestive of a possible cholesterol binding site centered at this position (see Figure 1). However, because the solubility limit of cholesterol was reached before saturation of binding could be attained this result alone did not exclude the possibility that the changes in the NMR spectrum reflect non-specific interactions of cholesterol with C99 rather than formation of a stoichiometric complex. For this reason, we turned to the use of a recently developed water soluble cholesterol analog, “CHOBIMALT”, in which the –OH cholesterol has been replaced by a tetrasaccharide (Figure 2). When C99 was titrated with CHOBIMALT, the peaks that underwent maximal shifts were seen to be the same as those that shifted the most in response to bona fide cholesterol (34). This indicates that CHOBIMALT’s mode of interaction with C99 faithfully mimics that of cholesterol and is not significantly perturbed by the analog’s tetrasaccharide head group. It was possible to complete titration of C99 with this compound. When the averaged chemical shift changes of the residues located in the extracellular connecting loop were plotted as function of the CHOBIMALT concentration, it was observed that the binding curve approaches a plateau, strongly suggestive of saturation of binding (Figure 2)—a hallmark for the formation of a stoichiometric complex. As shown in Figure 2 this data is well fit by a 1:1 binding model. Kd was determined to be a very modest 28 mol% ± 14 mol%. The mol% unit is used to express the concentrations of molecules in membranes and is defined as: 100 X moles of CHOBIMALT/(moles of CHOBIMALT + moles of micellar lyso-phospholipid).
Figure 2.
NMR titration data for binding of a cholesterol analog (CHOBIMALT) to C99. A 1:1 binding model has been fit to the data, which is taken from(34) and represents the averaged values of the changes of chemical shifts for the backbone amide resonances of the residues located at the focal point for cholesterol binding to the C99 (the residues highlighted in red in Figure 1).
It is probable that the binding of cholesterol to C99 in actual membranes would be somewhat more avid than the ca. 25 mol% Kd, both because cholesterol is likely to bind more tightly to C99 than to CHOBIMALT and because the membrane should provide a more favorable environment for lipid-protein binding than detergent micelles. An important priority for future work will be to examine this matter experimentally. In any case it is clear that C99’s avidity for cholesterol is modest, being weaker by orders of magnitude, for example, than the binding of a typical agonist to a typical G protein-coupled receptor(56). However, given that steady state cholesterol levels in the plasma membrane (PM) are often 40–50 mol% (57), C99’s affinity for cholesterol appears to be well-matched for this protein to sense membrane cholesterol levels. At PM concentrations of cholesterol that are probably at the lower end of the physiological range, most C99 would be expected to be uncomplexed with cholesterol, while at the upper end it would be expected to be ≫50% saturated. The possible functional relevance of this observation is discussed later in this review.
While the structural details of the cholesterol-C99 complex await full elucidation, the NMR titration results clearly indicate that the loop between the extracellular membrane surface-associated segment and the start of the TMD is the epicenter for C99-cholesterol interactions (see Figure 1). This loop includes the sequence VGSNK. With its interfacial location and several residues capable of both accepting and donating hydrogen bonds, it is probable that this loop forms hydrogen bonds with cholesterol’s –OH moiety. A cholesterol binding sequence found in a number of proteins has previously been described(58;59)— the CRAC motif: L/V-X1–5-Y-X1-5-R/K This motif is believed to drive cholesterol binding by specifically recognizing its –OH moiety. While C99 does not contain the canonical CRAC sequence, it has previously been noted that its VGSNK motif bears some resemblance(60), suggesting that C99 binds cholesterol through interactions akin to those previously documented for other cholesterol-binding proteins. Whether cholesterol binds with similar affinity to both C99 monomers and dimers is unclear, as is the question of whether a single cholesterol molecule makes contacts with 2 subunits in a dimer.
The fact that C99 binds cholesterol suggests that full length APP does so as well. However, it should be pointed out that while C99 is often observed to be found associated with the detergent-resistant membrane fractions of cell extracts, full length APP is often seen to fractionate with detergent-extractable membranes(61;62), suggesting either that these two molecules have very different affinities for cholesterol or that there are additional factors that differentially contribute to determining their membrane localization.
The sites in C99 believed to be central to binding of cholesterol are also present in the Aβ polypeptides. There have been a number of studies that indicate that Aβ interacts with cholesterol, typically in a manner that is believed to promote oligomerization, fibrillization, and/or cholesterol oxidation(26–32;63–66). It is possible that the cholesterol binding site we have documented in C99 may play an important role in cholesterol-Aβ interactions even though Aβ has significant aqueous solubility and interacts with membranes in ways that are generally different from C99.
Why Binding of Cholesterol to C99 is Expected to Promote Amyloidogenesis
The propensity of C99 and, possibly, full length APP to form stoichiometric complexes with cholesterol may represent a factor that favors the amyloidogenic pathway by promoting localization of APP/C99 to cholesterol-rich membrane domains and organelles where γ-secretase and possibly β-secretase reside. There is now a large body of data showing that significant amounts of both β-secretase and γ-secretase, but not α-secretase, reside within cholesterol-rich membrane microdomains, often referred to as “lipid rafts”—a term we will use herein for the sake of simplicity. Lipid rafts are found primarily in the trans-Golgi, plasma, and endosomal membranes(57;67;68). β-secretase cleavage of APP has been proposed to occur primarily in membrane rafts of the PM and endosomes (69–75), although this is not without controversy(61;76) and significant quantities of β-secretase is sometimes found in the bulk membrane(61;62). γ-secretase cleavage of C99 appears to occur primarily in rafts located in the endosomes (16;22;75;77–81). On the other hand, non-amyloidogenic α-secretase is believed to reside primarily in the bulk (non-raft) phase of the plasma membrane(71;82–84) and to be inactivated when forced to associate with rafts(85).
Membrane cholesterol within the plasma or endosomal membranes is distributed between two pools: free cholesterol in the bulk membrane phase and cholesterol that is associated with lipid rafts or related ordered microdomains. Based on studies with model membranes of defined composition it is thought that for a given membrane lipid composition, the partitioning ratio between free cholesterol and raft-associated cholesterol will be determined partly by the total mol% of cholesterol in the membrane, with higher cholesterol concentrations resulting in higher ratios in raft domains(57;86). When considered in this conceptual framework, three populations of C99 or APP are predicted. First is C99/APP that is uncomplexed with cholesterol. This population of protein would be prominent in membranes that have a mol% cholesterol below the Kd for binding to C99/APP, which appears to be roughly 25 mol% based on our NMR studies(34). This population would be subject to proteolytic processing primarily via α-secretase. Second is C99/APP that is complexed with cholesterol but remains in the bulk membrane phase. This population of protein would predominate only if the mol% of cholesterol in the membrane is high enough to exceed Kd and yet is low enough so that the amount of free cholesterol relative to the population of raft-associated cholesterol is low. If indeed the ca. 25 mol% Kd measured for cholesterol/C99 interaction under micellar conditions approximates the Kd in real membranes we wonder if non-raft associated cholesterol-C99/APP complexes would ever dominate since this level of cholesterol is well above the cholesterol concentration at which rafts will begin to form, at least in typical model membranes. A third population of C99/APP would be complexed with cholesterol in rafts. This is the population of the protein that is widely thought to be subject to amyloidogenic processing by β- and γ-secretases. This population is expected to predominate when the total cholesterol concentrations are on the order of Kd or higher.
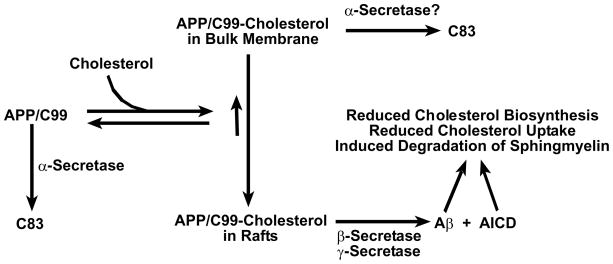
To summarize this model, at low plasma membrane/endosomal cholesterol concentrations, free C99 and APP will predominate in the plasma membrane and the protein will primarily be subject to α-secretase-initiated processing. However, as cholesterol levels in the membrane rise to the point where a majority of this lipid is found in rafts, most C99 and possibly APP will complex with cholesterol, most of which will be located in lipid rafts. Localization of a majority of APP to rafts is expected to result in a tip in the balance between α-secretase- and β-secretase-initiated processing towards β-secretase, with subsequent γ-secretase cleavage of C99 also being promoted by cholesterol complexation and raft localization (Figure 3). We regard the latter (γ-secretase/C99) part of this model as being more uniformly supported by the existing data, with the β-secretase/APP part of this model being more controversial.
Figure 3.
Working model for how cholesterol binding to C99/APP is related to the amyloidogenic and non-amyloidogenic pathways for processing APP and for how the amyloidogenic pathway may be linked to cellular cholesterol reduction. Given that the transmembrane product of α-secretase cleavage of APP (C83) contains the sequence motif believed to be central to cholesterol binding by C99/APP (see Figure 1), C83 probably also binds cholesterol. In this case, elevated cholesterol levels would also result in raft localization of C83, subsequent γ-secretease cleavage of which would then release AICD and an apparently harmless peptide known as p3. Whether C99/APP-cholesterol complexes in the bulk (non-raft) membranes can be cleaved by α-secretase is unclear. It is known that both full length APP and C99 can serve as substrates for α-secretase cleavage in the bulk membrane(138).
The above model assumes both that (i) both C99 and full length APP have a similar affinity for cholesterol, (ii) that C99/APP has a similar affinity for cholesterol in both rafts and in the bulk membrane and (ii) that complexation of C99/APP with cholesterol does not grossly perturb cholesterol’s partitioning between bulk membrane and rafts. These assumptions have yet to be tested. Moreover, this model should be regarded as a nested model in the sense that cholesterol binding to APP/C99 is likely to prove to be only one of a number of factors that collude to induce intracellular trafficking and membrane/raft localization of APP/C99 (c.f., (22;51;87;88)). This may help to explain, for example, why the distribution of C99 vs. full length APP between detergent-resistant membranes and the bulk membrane is not always seen to be the same (c.f.,(62)). It should also be recognized that other factors besides cholesterol levels—such as sphingolipid content—may also help determine lipid raft content in vivo. Despite these caveats, there appears to be good reasons to suppose that the propensity of C99 and possibly APP to bind cholesterol is an important factor that promotes the amyloidogenic pathway.
A second possible mechanism by which cholesterol complexation with APP/C99 could promote amyloidogenesis is by directly modulating substrate binding, catalysis, or product dissociation by β- or γ-secretase. These enzymes have been shown to exhibit much higher activities in the presence of cholesterol than in the absence(89;90), although there is contrary data in the case of γ-secretase(91). Whether cholesterol’s promotion of the activities of these secretases is due to alterations in the membrane microenvironment surrounding these enzymes or could involve a more specific allosteric regulation, possibly involving direct substrate-cholesterol interactions is not yet clear. It is interesting to note that a number of NSAID-class drugs are known to be γ-secretase modulators (GSMs), which inhibit and/or perturb γ-secretase cleavage of C99 to alter the Aβ42/Aβ40 production ratio (92–94). However, NSAIDs are not regarded as cholesterol mimics, although they may promote ordering of lipid bilayers in a way that resembles cholesterol(95).
Therapeutic Implications of Complex Formation Between C99/APP and Cholesterol
A serious concern regarding general inhibitors of β- or γ-secretase as potential preventative or therapeutic agents for Alzheimer’s disease is that these proteases cleave other proteins besides C99/APP in ways that are essential for normal health(49;96–98), leading to concerns about toxicity(99;100). While such inhibitors are, nevertheless, under clinical development(101–103), this concern has led to a search for strategies for specifically inhibiting or favorably modulating cleavage of C99/APP relative to other substrates. For example, small molecule “γ-secretase modulators” (GSMs) of C99 cleavage have been described (92–94) and have been proposed to act by binding directly to C99 to alter its substrate recognition by γ-secretase(93). However, that these compounds directly complex C99 has been disputed(104).
It has previously been proposed that specifically altering the trafficking of C99/APP away from lipid rafts is expected to be beneficial (c.f., (105;106)). That C99 forms a complex with cholesterol, which likely leads to its raft localization, suggests that drug-like molecules that can inhibit complex formation between C99/APP and cholesterol might provide a way of avoiding Aβ production. There is evidence that drug-like molecules can be developed that act by specifically-recognizing and binding to single-span membrane proteins(107;108), suggesting that this approach is feasible.
Possible Teleology for Cholesterol Binding to C99/APP
Though subject to much continuing inquiry, the functions of the APP remain only partially elucidated (109–113). While for the most part there are not obvious direct relationships between APP’s proposed functions and cholesterol, there is evidence that the products of the amyloidogenic pathway—both Aβ and the intracellular domain of the APP (AICD)—can play a direct role in regulating cholesterol homeostasis, acting to lower cellular cholesterol levels(114–116).
It has been shown that Aβ stimulates the release of cholesterol and some other lipids from cells in the form of lipoproteins(117) and also that fibrillar Aβ down-regulates cholesterol biosynthesis(118). Another study suggested that Aβ reduces biosynthesis of cholesterol and other lipids under conditions of ischemia(115;119). It has also been proposed that cytosolic Aβ acts as a (possibly indirect) inhibitor of HMG-CoA reductase (HMGR), the rate-limiting enzyme in the biosynthetic pathway for cholesterol(120). The same study suggested that Aβ production also stimulates sphingomyelinase activity, which would coordinate with cholesterol reduction to decrease lipid raft content in the cell membrane. These observations suggest that Aβ production can down-regulate cellular cholesterol and raft content. It should be noted that while one normally thinks of Aβ as being released primarily into the extracellular milieu, an appreciable fraction of γ-secretase cleavage of C99 appears to occur in endosomes, where Aβ is released into the lumen (reviews in (22;80;81;121–123)). While some C99 in this luminal pool would be expected to be exocytosed from the cell when endosome-derived vesicles recycle to the plasma membrane there is also evidence that significant amounts of Aβ traffic to the secretory pathway, to mitochondria, and even to the cytosol(122;124–127). Some Aβ that is secreted into the ectoplasm may also be taken up again by cells(122). Thus, the impact of Aβ on cholesterol homeostasis could derive from both intra- and extracellular peptide pools.
The AICD that is released upon γ-secretase cleavage of C99 translocates to the cell nucleus in concert with Fe65 and Tip60, where it then acts as a transcriptional suppressor to the gene that encodes the LRP1 protein, a major apolipoprotein E receptor in the brain that mediates cellular cholesterol uptake via endocytosis. This suggests that AICD production down-regulates cellular cholesterol uptake(128), although this has been questioned(129).
Given the above considerations, a possible function of the amyloidogenic pathway may be to reduce total cellular cholesterol levels, as previously proposed (114–116) and as illustrated in Figure 3. The fact that APP now appears to be a cholesterol binding protein suggests that one of its functions may be to act as a cellular cholesterol sensor/receptor. When membrane cholesterol levels are elevated APP forms a complex with cholesterol, which promotes the amyloidogenic pathway. The peptide products of this pathway ultimately reduce both cholesterol uptake and biosynthesis, completing a negative feedback loop.
If indeed APP does turn out to be involved in regulating cellular cholesterol levels, why is this only now being uncovered? One possibility is that APP’s role as a cellular cholesterol sensor is as a minor niche or backup player relative to the normally preeminent primary cellular cholesterol sensors(130;131). It is well known that that most genes do not appear to be essential to the organisms they support, in part because of functional redundancy(132–135)—for any given protein and its associated function, there is usually at least one other protein in the cell that can fulfill that function if the first protein is knocked out. If this is the case for APP, then it is easy to imagine how a role as a cellular cholesterol sensor would not be easily detected.
Many Unanswered Questions
As described in this review, the recent documentation that C99 and cholesterol form a complex under micellar conditions offers new insight into the nature of the relationships between cholesterol, APP processing, and Alzheimer’s disease. This raises many new questions, which include: Does APP/C99 bind to other cholesterol-related compounds such as 24S-hydroxycholesterol and the products of oxidative damage of cholesterol? Do any of the mutations in the APP that lead to familial early-onset AD impact C99/APP-cholesterol interactions? What is the relationship of C99/APP homodimerization to cholesterol binding and how does this relate to protein trafficking and interactions with the secretases? Does binding of cholesterol to C99/APP directly alter substrate association, cleavage, and/or product release by the β- and γ-secretases? Finally, to what degree is cholesterol binding to APP a decisive event that commits APP to the amyloidogenic pathway, as opposed to merely being a factor in a network of APP-protein or APP-lipid interactions that collectively determine this fate of APP in the cell? It is hoped that this review will spur interest in searching for answers to these questions.
Finally, it should be pointed out that for cholesterol to serve as a regulatory mechanism for proteolytic processing of APP/C99 the physiological concentrations of cholesterol in the plasma and endosomal membranes must vary enough throughout the lifetime of a cell to generate different outcomes at different points in time with regard to the rate of amyloidogenic processing and to the partitioning of APP/C99 between the amyloidogenic and non-amyloidogenic processing pathways. For model cell lines it has been shown that the overall cholesterol content of a cell can change by factors in the range of 2–4 at different phases of cell growth(136;137). However, even these relatively simple measurements do not appear to have been made for neurons in culture, much less under physiological conditions. Moreover, even for model cell lines almost nothing is known about the dynamic variability of cholesterol and lipid raft content in plasma or organellar membranes as a function of cellular physiological state. Quantitation of the dynamics of organelle-specific membrane cholesterol content in the neurons of living (and aging) organisms is a daunting goal, but one that may ultimately need to be surmounted as a key step toward elucidating the complex relationship between cholesterol and Alzheimer’s disease.
Acknowledgments
This work was supported by grants from NIH (P01 GM080513) and from the Alzheimer’s Association (IIRG-07-59379). We thank the editor and the anonymous reviewers for a their helpful comments on an earlier version of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fonseca AC, Resende R, Oliveira CR, Pereira CM. Cholesterol and statins in Alzheimer’s disease: Current controversies. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 3.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Dietschy JM. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biol Chem. 2009;390:287–293. doi: 10.1515/BC.2009.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vance JE, Hayashi H, Karten B. Cholesterol homeostasis in neurons and glial cells. Semin Cell Dev Biol. 2005;16:193–212. doi: 10.1016/j.semcdb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Kandiah N, Feldman HH. Therapeutic potential of statins in Alzheimer’s disease. J Neurol Sci. 2009;283:230–234. doi: 10.1016/j.jns.2009.02.352. [DOI] [PubMed] [Google Scholar]
- 11.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J, Donkin J, Wellington C. Greasing the wheels of Abeta clearance in Alzheimer’s disease: the role of lipids and apolipoprotein E. Biofactors. 2009;35:239–248. doi: 10.1002/biof.37. [DOI] [PubMed] [Google Scholar]
- 13.Martins IJ, Berger T, Sharman MJ, Verdile G, Fuller SJ, Martins RN. Cholesterol metabolism and transport in the pathogenesis of Alzheimer’s disease. J Neurochem. 2009;111:1275–1308. doi: 10.1111/j.1471-4159.2009.06408.x. [DOI] [PubMed] [Google Scholar]
- 14.Cordy JM, Hooper NM, Turner AJ. The involvement of lipid rafts in Alzheimer’s disease. Mol Membr Biol. 2006;23:111–122. doi: 10.1080/09687860500496417. [DOI] [PubMed] [Google Scholar]
- 15.Grimm MO, Grimm HS, Tomic I, Beyreuther K, Hartmann T, Bergmann C. Independent inhibition of Alzheimer disease beta- and gamma-secretase cleavage by lowered cholesterol levels. J Biol Chem. 2008;283:11302–11311. doi: 10.1074/jbc.M801520200. [DOI] [PubMed] [Google Scholar]
- 16.Guardia-Laguarta C, Coma M, Pera M, Clarimon J, Sereno L, Agullo JM, Molina-Porcel L, Gallardo E, Deng A, Berezovska O, Hyman BT, Blesa R, Gomez-Isla T, Lleo A. Mild cholesterol depletion reduces amyloid-beta production by impairing APP trafficking to the cell surface. J Neurochem. 2009;110:220–230. doi: 10.1111/j.1471-4159.2009.06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halford RW, Russell DW. Reduction of cholesterol synthesis in the mouse brain does not affect amyloid formation in Alzheimer’ disease, but does extend lifespan. Proc Natl Acad Sci U S A. 2009;106:3502–3506. doi: 10.1073/pnas.0813349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch-Reinshagen V, Burgess BL, Wellington CL. Why lipids are important for Alzheimer disease? Mol Cell Biochem. 2009;326:121–129. doi: 10.1007/s11010-008-0012-2. [DOI] [PubMed] [Google Scholar]
- 19.Sakurai T, Kaneko K, Okuno M, Wada K, Kashiyama T, Shimizu H, Akagi T, Hashikawa T, Nukina N. Membrane microdomain switching: a regulatory mechanism of amyloid precursor protein processing. J Cell Biol. 2008;183:339–352. doi: 10.1083/jcb.200804075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sjogren M, Mielke M, Gustafson D, Zandi P, Skoog I. Cholesterol and Alzheimer’s disease--is there a relation? Mech Ageing Dev. 2006;127:138–147. doi: 10.1016/j.mad.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Xiong H, Callaghan D, Jones A, Walker DG, Lue LF, Beach TG, Sue LI, Woulfe J, Xu H, Stanimirovic DB, Zhang W. Cholesterol retention in Alzheimer’s brain is responsible for high beta- and gamma-secretase activities and Abeta production. Neurobiol Dis. 2008;29:422–437. doi: 10.1016/j.nbd.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzolo MP, Bu G. Lipoprotein receptors and cholesterol in APP trafficking and proteolytic processing, implications for Alzheimer’ disease. Semin Cell Dev Biol. 2009;20:191–200. doi: 10.1016/j.semcdb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter CJ. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Int. 2007;50:12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Garcia AN, Muniz MT, Souza e Silva HR, da Silva HA, thayde-Junior L. Cyp46 polymorphisms in Alzheimer’s disease: a review. J Mol Neurosci. 2009;39:342–345. doi: 10.1007/s12031-009-9227-2. [DOI] [PubMed] [Google Scholar]
- 25.Sanders AE, Wang C, Katz M, Derby CA, Barzilai N, Ozelius L, Lipton RB. Association of a functional polymorphism in the cholesteryl ester transfer protein (CETP) gene with memory decline and incidence of dementia. JAMA. 2010;303:150–158. doi: 10.1001/jama.2009.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SI, Yi JS, Ko YG. Amyloid beta oligomerization is induced by brain lipid rafts. J Cell Biochem. 2006;99:878–889. doi: 10.1002/jcb.20978. [DOI] [PubMed] [Google Scholar]
- 27.Lau TL, Gehman JD, Wade JD, Masters CL, Barnham KJ, Separovic F. Cholesterol and Clioquinol modulation of A beta(1–42) interaction with phospholipid bilayers and metals. Biochim Biophys Acta. 2007;1768:3135–3144. doi: 10.1016/j.bbamem.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson AM, Ferreira A. Increased membrane cholesterol might render mature hippocampal neurons more susceptible to beta-amyloid-induced calpain activation and tau toxicity. J Neurosci. 2009;29:4640–4651. doi: 10.1523/JNEUROSCI.0862-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu L, Lewis A, Como J, Vaughn MW, Huang J, Somerharju P, Virtanen J, Cheng KH. Cholesterol modulates the interaction of beta-amyloid peptide with lipid bilayers. Biophys J. 2009;96:4299–4307. doi: 10.1016/j.bpj.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Usui K, Hulleman JD, Paulsson JF, Siegel SJ, Powers ET, Kelly JW. Site-specific modification of Alzheimer’s peptides by cholesterol oxidation products enhances aggregation energetics and neurotoxicity. Proc Natl Acad Sci U S A. 2009;106:18563–18568. doi: 10.1073/pnas.0804758106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagisawa K. Cholesterol and amyloid beta fibrillogenesis. Subcell Biochem. 2005;38:179–202. doi: 10.1007/0-387-23226-5_9. [DOI] [PubMed] [Google Scholar]
- 32.Yip CM, Elton EA, Darabie AA, Morrison MR, McLaurin J. Cholesterol, a modulator of membrane-associated Abeta-fibrillogenesis and neurotoxicity. J Mol Biol. 2001;311:723–734. doi: 10.1006/jmbi.2001.4881. [DOI] [PubMed] [Google Scholar]
- 33.Panchal M, Loeper J, Cossec JC, Perruchini C, Lazar A, Pompon D, Duyckaerts C. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J Lipid Res. 2010;51:598–605. doi: 10.1194/jlr.M001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 36.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 37.Kroenke CD, Ziemnicka-Kotula D, Xu J, Kotula L, Palmer AG., III Solution conformations of a peptide containing the cytoplasmic domain sequence of the beta amyloid precursor protein. Biochemistry. 1997;36:8145–8152. doi: 10.1021/bi9705669. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Koshiba S, Hayashi F, Tochio N, Tomizawa T, Kasai T, Yabuki T, Motoda Y, Harada T, Watanabe S, Inoue M, Hayashizaki Y, Tanaka A, Kigawa T, Yokoyama S. Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode. J Biol Chem. 2008;283:27165–27178. doi: 10.1074/jbc.M803892200. [DOI] [PubMed] [Google Scholar]
- 39.Marenchino M, Williamson PT, Murri S, Zandomeneghi G, Wunderli-Allenspach H, Meier BH, Kramer SD. Dynamics and Cleavability at the alpha-cleavage site of APP(684–726) in different lipid environments. Biophys J. 2008;95:1460–1473. doi: 10.1529/biophysj.108.129726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyashita N, Straub JE, Thirumalai D, Sugita Y. Transmembrane structures of amyloid precursor protein dimer predicted by replica-exchange molecular dynamics simulations. J Am Chem Soc. 2009;131:3438–3439. doi: 10.1021/ja809227c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson LK, Lu KP. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 42.Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008;9:1134–1140. doi: 10.1038/embor.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramelot TA, Gentile LN, Nicholson LK. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- 44.Ramelot TA, Nicholson LK. Phosphorylation-induced structural changes in the amyloid precursor protein cytoplasmic tail detected by NMR. J Mol Biol. 2001;307:871–884. doi: 10.1006/jmbi.2001.4535. [DOI] [PubMed] [Google Scholar]
- 45.Sato T, Tang TC, Reubins G, Fei JZ, Fujimoto T, Kienlen-Campard P, Constantinescu SN, Octave JN, Aimoto S, Smith SO. A helix-to-coil transition at the epsilon-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc Natl Acad Sci U S A. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao H, Jao S, Ma K, Zagorski MG. Solution structures of micelle-bound amyloid beta-(1–40) and beta-(1–42) peptides of Alzheimer’s disease. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 47.Wahlstrom A, Hugonin L, Peralvarez-Marin A, Jarvet J, Graslund A. Secondary structure conversions of Alzheimer’s Abeta(1–40) peptide induced by membrane-mimicking detergents. FEBS J. 2008;275:5117–5128. doi: 10.1111/j.1742-4658.2008.06643.x. [DOI] [PubMed] [Google Scholar]
- 48.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’rsi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 49.Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muresan Z, Muresan V. The amyloid-beta precursor protein is phosphorylated via distinct pathways during differentiation, mitosis, stress, and degeneration. Mol Biol Cell. 2007;18:3835–3844. doi: 10.1091/mbc.E06-07-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamayev R, Zhou D, D’Adamio L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Multhaup G. Amyloid precursor protein and BACE function as oligomers. Neurodegener Dis. 2006;3:270–274. doi: 10.1159/000095266. [DOI] [PubMed] [Google Scholar]
- 53.Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorman PM, Kim S, Guo M, Melnyk RA, McLaurin J, Fraser PE, Bowie JU, Chakrabartty A. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer’s disease mutants. BMC Neurosci. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kienlen-Campard P, Tasiaux B, Van HJ, Li M, Huysseune S, Sato T, Fei JZ, Aimoto S, Courtoy PJ, Smith SO, Constantinescu SN, Octave JN. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J Biol Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall A. The G-Protein Linked Receptor Facts-Book - Watson, S, Arkinstall, S. Nature. 1994;369:30. [Google Scholar]
- 57.van MG, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Epand RM. Proteins and cholesterol-rich domains. Biochim Biophys Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Yao Z, Degenhardt B, Teper G, Papadopoulos V. Cholesterol binding at the cholesterol recognition/interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc Natl Acad Sci U S A. 2001;98:1267–1272. doi: 10.1073/pnas.031461598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abad C, Martinez-Gil L, Tamborero S, Mingarro I. Membrane topology of gp41 and amyloid precursor protein: interfering transmembrane interactions as potential targets for HIV and Alzheimer treatment. Biochim Biophys Acta. 2009;1788:2132–2141. doi: 10.1016/j.bbamem.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abad-Rodriguez J, Ledesma MD, Craessaerts K, Perga S, Medina M, Delacourte A, Dingwall C, De SB, Dotti CG. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vetrivel KS, Cheng H, Kim SH, Chen Y, Barnes NY, Parent AT, Sisodia SS, Thinakaran G. Spatial segregation of gamma-secretase and substrates in distinct membrane domains. J Biol Chem. 2005;280:25892–25900. doi: 10.1074/jbc.M503570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson TJ, Alkon DL. Oxidation of cholesterol by amyloid precursor protein and beta-amyloid peptide. J Biol Chem. 2005;280:7377–7387. doi: 10.1074/jbc.M409071200. [DOI] [PubMed] [Google Scholar]
- 64.Opazo C, Huang X, Cherny RA, Moir RD, Roher AE, White AR, Cappai R, Masters CL, Tanzi RE, Inestrosa NC, Bush AI. Metalloenzyme-like activity of Alzheimer’s disease beta-amyloid. Cu-dependent catalytic conversion of dopamine, cholesterol, and biological reducing agents to neurotoxic H(2)O(2) J Biol Chem. 2002;277:40302–40308. doi: 10.1074/jbc.M206428200. [DOI] [PubMed] [Google Scholar]
- 65.Puglielli L, Friedlich AL, Setchell KD, Nagano S, Opazo C, Cherny RA, Barnham KJ, Wade JD, Melov S, Kovacs DM, Bush AI. Alzheimer disease beta-amyloid activity mimics cholesterol oxidase. J Clin Invest. 2005;115:2556–2563. doi: 10.1172/JCI23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ashley RH, Harroun TA, Hauss T, Breen KC, Bradshaw JP. Autoinsertion of soluble oligomers of Alzheimer’s Abeta(1–42) peptide into cholesterol-containing membranes is accompanied by relocation of the sterol towards the bilayer surface. BMC Struct Biol. 2006;6:21. doi: 10.1186/1472-6807-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mesmin B, Maxfield FR. Intracellular sterol dynamics. Biochim Biophys Acta. 2009;1791:636–645. doi: 10.1016/j.bbalip.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mondal M, Mesmin B, Mukherjee S, Maxfield FR. Sterols are mainly in the cytoplasmic leaflet of the plasma membrane and the endocytic recycling compartment in CHO cells. Mol Biol Cell. 2009;20:581–588. doi: 10.1091/mbc.E08-07-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang BL. Neuronal protein trafficking associated with Alzheimer disease: From APP and BACE1 to glutamate receptors. Cell Adh Migr. 2009;3 doi: 10.4161/cam.3.1.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cordy JM, Hussain I, Dingwall C, Hooper NM, Turner AJ. Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2003;100:11735–11740. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, Schwille P, Schulz JB, Schroeder C, Simons M, Jennings G, Knolker HJ, Simons K. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 73.Riddell DR, Christie G, Hussain I, Dingwall C. Compartmentalization of beta-secretase (Asp2) into low-buoyant density, noncaveolar lipid rafts. Curr Biol. 2001;11:1288–1293. doi: 10.1016/s0960-9822(01)00394-3. [DOI] [PubMed] [Google Scholar]
- 74.Sakurai T, Kaneko K, Okuno M, Wada K, Kashiyama T, Shimizu H, Akagi T, Hashikawa T, Nukina N. Membrane microdomain switching: a regulatory mechanism of amyloid precursor protein processing. J Cell Biol. 2008;183:339–352. doi: 10.1083/jcb.200804075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiong H, Callaghan D, Jones A, Walker DG, Lue LF, Beach TG, Sue LI, Woulfe J, Xu H, Stanimirovic DB, Zhang W. Cholesterol retention in Alzheimer’s brain is responsible for high beta- and gamma-secretase activities and Abeta production. Neurobiol Dis. 2008;29:422–437. doi: 10.1016/j.nbd.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vetrivel KS, Meckler X, Chen Y, Nguyen PD, Seidah NG, Vassar R, Wong PC, Fukata M, Kounnas MZ, Thinakaran G. Alzheimer disease Abeta production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J Biol Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hur JY, Welander H, Behbahani H, Aoki M, Franberg J, Winblad B, Frykman S, Tjernberg LO. Active gamma-secretase is localized to detergent-resistant membranes in human brain. FEBS J. 2008;275:1174–1187. doi: 10.1111/j.1742-4658.2008.06278.x. [DOI] [PubMed] [Google Scholar]
- 78.Vetrivel KS, Cheng H, Lin W, Sakurai T, Li T, Nukina N, Wong PC, Xu H, Thinakaran G. Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes. J Biol Chem. 2004;279:44945–44954. doi: 10.1074/jbc.M407986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, Younkin LH, Younkin SG, Golde TE. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9:11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- 80.Wu F, Yao PJ. Clathrin-mediated endocytosis and Alzheimer’s disease: an update. Ageing Res Rev. 2009;8:147–149. doi: 10.1016/j.arr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bodovitz S, Klein WL. Cholesterol modulates alpha-secretase cleavage of amyloid precursor protein. J Biol Chem. 1996;271:4436–4440. doi: 10.1074/jbc.271.8.4436. [DOI] [PubMed] [Google Scholar]
- 83.Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci U S A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parvathy S, Hussain I, Karran EH, Turner AJ, Hooper NM. Cleavage of Alzheimer’s amyloid precursor protein by alpha-secretase occurs at the surface of neuronal cells. Biochemistry. 1999;38:9728–9734. doi: 10.1021/bi9906827. [DOI] [PubMed] [Google Scholar]
- 85.Harris B, Pereira I, Parkin E. Targeting ADAM10 to lipid rafts in neuroblastoma SH-SY5Y cells impairs amyloidogenic processing of the amyloid precursor protein. Brain Res. 2009;1296:203–215. doi: 10.1016/j.brainres.2009.07.105. [DOI] [PubMed] [Google Scholar]
- 86.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 87.Andersen OM, Willnow TE. Lipoprotein receptors in Alzheimer’s disease. Trends Neurosci. 2006;29:687–694. doi: 10.1016/j.tins.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Hoe HS, Rebeck GW. Functional interactions of APP with the apoE receptor family. J Neurochem. 2008;106:2263–2271. doi: 10.1111/j.1471-4159.2008.05517.x. [DOI] [PubMed] [Google Scholar]
- 89.Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J Biol Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 90.Osenkowski P, Ye W, Wang R, Wolfe MS, Selkoe DJ. Direct and potent regulation of gamma-secretase by its lipid microenvironment. J Biol Chem. 2008;283:22529–22540. doi: 10.1074/jbc.M801925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wada S, Morishima-Kawashima M, Qi Y, Misono H, Shimada Y, Ohno-Iwashita Y, Ihara Y. Gamma-secretase activity is present in rafts but is not cholesterol-dependent. Biochemistry. 2003;42:13977–13986. doi: 10.1021/bi034904j. [DOI] [PubMed] [Google Scholar]
- 92.Hirohata M, Ono K, Yamada M. Non-steroidal anti-inflammatory drugs as anti-amyloidogenic compounds. Curr Pharm Des. 2008;14:3280–3294. doi: 10.2174/138161208786404173. [DOI] [PubMed] [Google Scholar]
- 93.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Substrate-targeting gamma-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolfe MS. Inhibition and modulation of gamma-secretase for Alzheimer’s disease. Neurotherapeutics. 2008;5:391–398. doi: 10.1016/j.nurt.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gamerdinger M, Clement AB, Behl C. Cholesterol-like effects of selective cyclooxygenase inhibitors and fibrates on cellular membranes and amyloid-beta production. Mol Pharmacol. 2007;72:141–151. doi: 10.1124/mol.107.034009. [DOI] [PubMed] [Google Scholar]
- 96.Hoffmeister A, Dietz G, Zeitschel U, Mossner J, Rossner S, Stahl T. BACE1 is a newly discovered protein secreted by the pancreas which cleaves enteropeptidase in vitro. JOP. 2009;10:501–506. [PubMed] [Google Scholar]
- 97.Lleo A. Activity of gamma-secretase on substrates other than APP. Curr Top Med Chem. 2008;8:9–16. doi: 10.2174/156802608783334060. [DOI] [PubMed] [Google Scholar]
- 98.Willem M, Lammich S, Haass C. Function, regulation and therapeutic properties of beta-secretase (BACE1) Semin Cell Dev Biol. 2009;20:175–182. doi: 10.1016/j.semcdb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 99.Cole SL, Vassar R. The role of amyloid precursor protein processing by BACE1, the beta-secretase, in Alzheimer disease pathophysiology. J Biol Chem. 2008;283:29621–29625. doi: 10.1074/jbc.R800015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolfe MS. gamma-Secretase in biology and medicine. Semin Cell Dev Biol. 2009;20:219–224. doi: 10.1016/j.semcdb.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 101.Bergmans BA, De SB. gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 2010;9:215–226. doi: 10.1016/S1474-4422(09)70332-1. [DOI] [PubMed] [Google Scholar]
- 102.Guardia-Laguarta C, Pera M, Lleo A. gamma-Secretase as a Therapeutic Target in Alzheimer’s Disease. Curr Drug Targets. 2009 doi: 10.2174/138945010790980349. [DOI] [PubMed] [Google Scholar]
- 103.Panza F, Solfrizzi V, Frisardi V, Capurso C, D’Introno A, Colacicco AM, Vendemiale G, Capurso A, Imbimbo BP. Disease-modifying approach to the treatment of Alzheimer’s disease: from alpha-secretase activators to gamma-secretase inhibitors and modulators. Drugs Aging. 2009;26:537–555. doi: 10.2165/11315770-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 104.Beel AJ, Barrett P, Schnier PD, Hitchcock SA, Bagal D, Sanders CR, Jordan JB. Nonspecificity of binding of gamma-secretase modulators to the amyloid precursor protein. Biochemistry. 2009;48:11837–11839. doi: 10.1021/bi901839d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng H, Vetrivel KS, Gong P, Meckler X, Parent A, Thinakaran G. Mechanisms of disease: new therapeutic strategies for Alzheimer’s disease--targeting APP processing in lipid rafts. Nat Clin Pract Neurol. 2007;3:374–382. doi: 10.1038/ncpneuro0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fahrenholz F, Postina R. Alpha-secretase activation--an approach to Alzheimer’s disease therapy. Neurodegener Dis. 2006;3:255–261. doi: 10.1159/000095264. [DOI] [PubMed] [Google Scholar]
- 107.Caputo GA, Litvinov RI, Li W, Bennett JS, DeGrado WF, Yin H. Computationally designed peptide inhibitors of protein-protein interactions in membranes. Biochemistry. 2008;47:8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yin H. Exogenous agents that target transmembrane domains of proteins. Angew Chem Int Ed Engl. 2008;47:2744–2752. doi: 10.1002/anie.200704780. [DOI] [PubMed] [Google Scholar]
- 109.Annaert W, De SB. A cell biological perspective on Alzheimer’s disease. Annu Rev Cell Dev Biol. 2002;18:25–51. doi: 10.1146/annurev.cellbio.18.020402.142302. [DOI] [PubMed] [Google Scholar]
- 110.Jacobsen KT, Iverfeldt K. Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors. Cell Mol Life Sci. 2009;66:2299–2318. doi: 10.1007/s00018-009-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Russo C, Venezia V, Repetto E, Nizzari M, Violani E, Carlo P, Schettini G. The amyloid precursor protein and its network of interacting proteins: physiological and pathological implications. Brain Res Brain Res Rev. 2005;48:257–264. doi: 10.1016/j.brainresrev.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 112.Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 113.Wolfe MS, Guenette SY. APP at a glance. J Cell Sci. 2007;120:3157–3161. doi: 10.1242/jcs.03481. [DOI] [PubMed] [Google Scholar]
- 114.Grimm MO, Grimm HS, Hartmann T. Amyloid beta as a regulator of lipid homeostasis. Trends Mol Med. 2007;13:337–344. doi: 10.1016/j.molmed.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 115.Koudinov AR, Koudinova NV. Cholesterol homeostasis failure as a unifying cause of synaptic degeneration. J Neurol Sci. 2005;229–230:233–240. doi: 10.1016/j.jns.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 116.Michikawa M. The role of cholesterol in pathogenesis of Alzheimer’s disease: dual metabolic interaction between amyloid beta-protein and cholesterol. Mol Neurobiol. 2003;27:1–12. doi: 10.1385/MN:27:1:1. [DOI] [PubMed] [Google Scholar]
- 117.Michikawa M, Gong JS, Fan QW, Sawamura N, Yanagisawa K. A novel action of alzheimer’s amyloid beta-protein (Abeta): oligomeric Abeta promotes lipid release. J Neurosci. 2001;21:7226–7235. doi: 10.1523/JNEUROSCI.21-18-07226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gong JS, Sawamura N, Zou K, Sakai J, Yanagisawa K, Michikawa M. Amyloid beta-protein affects cholesterol metabolism in cultured neurons: implications for pivotal role of cholesterol in the amyloid cascade. J Neurosci Res. 2002;70:438–446. doi: 10.1002/jnr.10347. [DOI] [PubMed] [Google Scholar]
- 119.Koudinova NV, Koudinov AR, Yavin E. Alzheimer’s Abeta1–40 peptide modulates lipid synthesis in neuronal cultures and intact rat fetal brain under normoxic and oxidative stress conditions. Neurochem Res. 2000;25:653–660. doi: 10.1023/a:1007511120099. [DOI] [PubMed] [Google Scholar]
- 120.Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De SB, Muller U, Shen J, Hartmann T. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- 121.Gouras GK, Almeida CG, Takahashi RH. Intraneuronal Abeta accumulation and origin of plaques in Alzheimer’s disease. Neurobiol Aging. 2005;26:1235–1244. doi: 10.1016/j.neurobiolaging.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 122.LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 123.Li M, Chen L, Lee DH, Yu LC, Zhang Y. The role of intracellular amyloid beta in Alzheimer’s disease. Prog Neurobiol. 2007;83:131–139. doi: 10.1016/j.pneurobio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 124.Greenfield JP, Tsai J, Gouras GK, Hai B, Thinakaran G, Checler F, Sisodia SS, Greengard P, Xu H. Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer beta-amyloid peptides. Proc Natl Acad Sci U S A. 1999;96:742–747. doi: 10.1073/pnas.96.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nishitsuji K, Tomiyama T, Ishibashi K, Ito K, Teraoka R, Lambert MP, Klein WL, Mori H. The E693Delta mutation in amyloid precursor protein increases intracellular accumulation of amyloid beta oligomers and causes endoplasmic reticulum stress-induced apoptosis in cultured cells. Am J Pathol. 2009;174:957–969. doi: 10.2353/ajpath.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reddy PH. Amyloid beta, mitochondrial structural and functional dynamics in Alzheimer’s disease. Exp Neurol. 2009;218:286–292. doi: 10.1016/j.expneurol.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schmitz A, Herzog V. Endoplasmic reticulum-associated degradation: exceptions to the rule. Eur J Cell Biol. 2004;83:501–509. doi: 10.1078/0171-9335-00412. [DOI] [PubMed] [Google Scholar]
- 128.Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Muller T, Meyer HE, Egensperger R, Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer’s disease. Prog Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 130.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50(Suppl):S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 132.Hentges KE, Pollock DD, Liu B, Justice MJ. Regional variation in the density of essential genes in mice. PLoS Genet. 2007;3:e72. doi: 10.1371/journal.pgen.0030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mahadevan R, Lovley DR. The degree of redundancy in metabolic genes is linked to mode of metabolism. Biophys J. 2008;94:1216–1220. doi: 10.1529/biophysj.107.118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shastry BS. More to learn from gene knockouts. Mol Cell Biochem. 1994;136:171–182. doi: 10.1007/BF00926078. [DOI] [PubMed] [Google Scholar]
- 135.Thomas JH. Thinking about genetic redundancy. Trends Genet. 1993;9:395–399. doi: 10.1016/0168-9525(93)90140-d. [DOI] [PubMed] [Google Scholar]
- 136.Cansell M, Gouygou JP, Jozefonvicz J, Letourneur D. Lipid composition of cultured endothelial cells in relation to their growth. Lipids. 1997;32:39–44. doi: 10.1007/s11745-997-0006-3. [DOI] [PubMed] [Google Scholar]
- 137.Takahashi M, Murate M, Fukuda M, Sato SB, Ohta A, Kobayashi T. Cholesterol controls lipid endocytosis through Rab11. Mol Biol Cell. 2007;18:2667–2677. doi: 10.1091/mbc.E06-10-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jager S, Leuchtenberger S, Martin A, Czirr E, Wesselowski J, Dieckmann M, Waldron E, Korth C, Koo EH, Heneka M, Weggen S, Pietrzik CU. alpha-secretase mediated conversion of the amyloid precursor protein derived membrane stub C99 to C83 limits Abeta generation. J Neurochem. 2009;111:1369–1382. doi: 10.1111/j.1471-4159.2009.06420.x. [DOI] [PubMed] [Google Scholar]