Abstract
The membrane integration of tail-anchored proteins at the endoplasmic reticulum (ER) is post-translational, with different tail-anchored proteins exploiting distinct cytosolic factors. For example, mammalian TRC40 has a well-defined role during delivery of tail-anchored proteins to the ER. Although its Saccharomyces cerevisiae equivalent, Get3, is known to function in concert with at least four other components, Get1, Get2, Get4 and Get5 (Mdy2), the role of additional mammalian proteins during tail-anchored protein biogenesis is unclear. To this end, we analysed the cytosolic binding partners of Sec61β, a well-defined substrate of TRC40, and identified Bat3 as a previously unknown interacting partner. Depletion of Bat3 inhibits the membrane integration of Sec61β, but not of a second, TRC40-independent, tail-anchored protein, cytochrome b5. Thus, Bat3 influences the in vitro membrane integration of tail-anchored proteins using the TRC40 pathway. When expressed in Saccharomyces cerevisiae lacking a functional GET pathway for tail-anchored protein biogenesis, Bat3 associates with the resulting cytosolic pool of non-targeted chains and diverts it to the nucleus. This Bat3-mediated mislocalisation is not dependent upon Sgt2, a recently identified component of the yeast GET pathway, and we propose that Bat3 either modulates the TRC40 pathway in higher eukaryotes or provides an alternative fate for newly synthesised tail-anchored proteins.
Keywords: Asna-1, SGTA, Get3, Sec61β, TRC40
Introduction
Tail-anchored proteins are a distinct class of integral membrane proteins, distinguished by the presence of a single C-terminal transmembrane region that targets the polypeptide for membrane integration and anchors the protein in the lipid bilayer (Borgese et al., 2007; Kutay et al., 1993; Rabu et al., 2009). Tail-anchored proteins destined for locations within the eukaryotic secretory pathway are all synthesised at the endoplasmic reticulum (ER) and can then be retained or sorted to various subcellular compartments (Behrens et al., 1996; Borgese et al., 2007; Kutay et al., 1995; Linstedt et al., 1995). The biogenesis of tail-anchored (TA) proteins at the ER has been of particular interest because the process is post-translational, and hence quite distinct from the classical signal recognition particle dependent, co-translational, pathway associated with protein synthesised at this location (Cross et al., 2009). In vitro systems have revealed several different pathways that can deliver TA proteins to the mammalian ER, with different TA protein substrates preferentially using distinct cytosolic factors to assist their biogenesis (for reviews, see Borgese et al., 2007; Rabu et al., 2009). Pathway selection is influenced by the relative hydrophobicity of the tail-anchor region, presumably by mediating the recruitment of specific cytosolic factors (Rabu et al., 2008; Rabu et al., 2009). Several recent studies have focussed on the role of mammalian TRC40 (Asna-1), and its Saccharomyces cerevisiae equivalent, Get3, during the post-translational targeting of TA proteins to the ER (Favaloro et al., 2008; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). TRC40 was shown to promote the membrane integration of a number of model TA proteins with comparatively hydrophobic tail-anchor regions, including Sec61β and RAMP4 (Favaloro et al., 2008; Stefanovic and Hegde, 2007). By contrast, perturbation of the TRC40 pathway appears to have little or no effect on cytochrome b5 (Cytb5) integration at the ER membrane, as judged by in vitro assays (Colombo et al., 2009; Stefanovic and Hegde, 2007). This correlates with data suggesting that the cytosolic molecular chaperones Hsc70 and Hsp40 can facilitate the ER integration of proteins with moderately hydrophobic tail-anchor regions, including Cytb5 (Rabu et al., 2008). Alternatively, the lack of TRC40 dependency for Cytb5 integration might reflect a role for new cytosolic components, or even an unassisted mechanism (Colombo et al., 2009).
TRC40 and Get3 are conserved ATPases, which mediate ATP-dependent TA-protein integration at the ER membrane (Favaloro et al., 2008; Favaloro et al., 2010; Rabu et al., 2009; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). Furthermore, several recent studies of Get3 provide structural insights into the mechanisms that underlie its substrate binding and release, and provide models for how ATP binding and hydrolysis might influence these steps (Bozkurt et al., 2009; Hu et al., 2009; Mateja et al., 2009; Suloway et al., 2009). Studies of S. cerevisiae have shown that several components function in concert with Get3: Get1 and Get2 are ER-localised membrane receptors for the GET pathway of TA-protein delivery (Schuldiner et al., 2008), whereas Get4 and Get5 are cytosolic components that appear to act in concert with Get3 before membrane delivery (Jonikas et al., 2009; Rabu et al., 2009). It is assumed that higher eukaryotes possess functional equivalents of these additional components (Rabu et al., 2009), and indeed TRC40 appears to be part of a larger cytosolic complex (Stefanovic and Hegde, 2007). To address the identity of other components that might contribute to this pathway, we analysed the cytosolic binding partners of Sec61β – a well-defined TRC40 substrate (Stefanovic and Hegde, 2007).
We identified Bat3 (Kabbage and Dickman, 2008) as a new interacting component that binds to both Sec61β and RAMP4, but not to a version of Sec61β that lacks the hydrophobic TA region. Strikingly, Bat3 depletion from reticulocyte lysate inhibited the membrane integration of recombinant Sec61β but did not affect Cytb5 insertion, specifically implicating Bat3 in the TRC40 pathway. When biosynthetic intermediates were analysed, the Sec61β chains that co-fractionated with Bat3 appeared distinct from the integration-competent population associated with TRC40. When expressed in S. cerevisiae lacking a functional GET pathway, mammalian Bat3 associated with the resulting cytosolic pool of non-targeted TA proteins and diverted it to the nucleus. Sgt2 has recently been identified, both biochemically and genetically, as an additional component of the yeast GET pathway (Chang et al., 2010; Costanzo et al., 2010). Its mammalian equivalent, SGTA is known to associate with Bat3 (Winnefeld et al., 2006), suggesting that these components function in concert during TA-protein biogenesis. We found that SGTA was preferentially associated with tail-anchor regions in a similar fashion to Bat3. However, the Bat3-dependent relocalisation of TA proteins occurs in the absence of Sgt2, confirming a role for Sgt2 in the GET pathway but ruling out any requirement for this component to enable Bat3 to redirect non-targeted TA proteins to the nucleus in yeast. S. cerevisiae lack an obvious Bat3 equivalent, and we propose that Bat3 either modulates the TRC40 pathway in higher eukaryotes, or provides an alternative fate for newly synthesised TA proteins that complements the role of the TRC40 complex.
Results
Recombinant Sec61β requires cytosolic factors and ATP for membrane integration
Previous studies of TA-protein biogenesis have successfully analysed the behaviour and binding partners of in vitro synthesised polypeptides to identify key components and to understand the pathways that mediate TA-protein delivery to the ER membrane (for a review, see Rabu et al., 2009). As an alternative strategy, we have now exploited recombinant polypeptides expressed in Escherichia coli to identify novel cytosolic factors that bind the TA region of Sec61β and contribute to its membrane integration. To this end, we expressed human Sec61β, with a short C-terminal extension bearing an N-glycosylation site (Abell et al., 2007; Colombo et al., 2009; Kutay et al., 1995; Rabu et al., 2008), as a polyhistidine-tagged fusion protein in E. coli (supplementary material Fig. S1A). The recombinant polypeptide was purified by nickel affinity chromatography and released from the affinity tag by proteolysis (Colombo et al., 2009), generating full-length recombinant Sec61β and a small amount of truncated material, which probably lacks a few residues at the N-terminus (supplementary material Fig. S1B).
To ensure that our recombinant Sec61β was capable of binding relevant cytosolic factors, we confirmed that the protein was efficiently integrated into ER-derived microsomes as judged by a well-established N-glycosylation assay (Abell et al., 2007; Kutay et al., 1995; Rabu et al., 2008) (supplementary material Fig. S1C). Efficient membrane integration of recombinant Sec61β requires the addition of cytosol (Fig. 1A), and membrane insertion is enhanced when the proportion of lysate is increased (Fig. 1B). As expected from previous studies (Abell et al., 2007; Favaloro et al., 2008; Stefanovic and Hegde, 2007), the membrane integration of recombinant Sec61β was also exquisitely sensitive to the presence of nucleotide triphosphate (Fig. 1C). Hence, the requirements for the membrane integration of recombinant Sec61β appear to be identical to those previously defined using in vitro synthesised polypeptides. Thus, the recombinant protein provides a viable tool for the biochemical analysis of the cytosolic factors that promote membrane integration (Colombo et al., 2009).
Fig. 1.
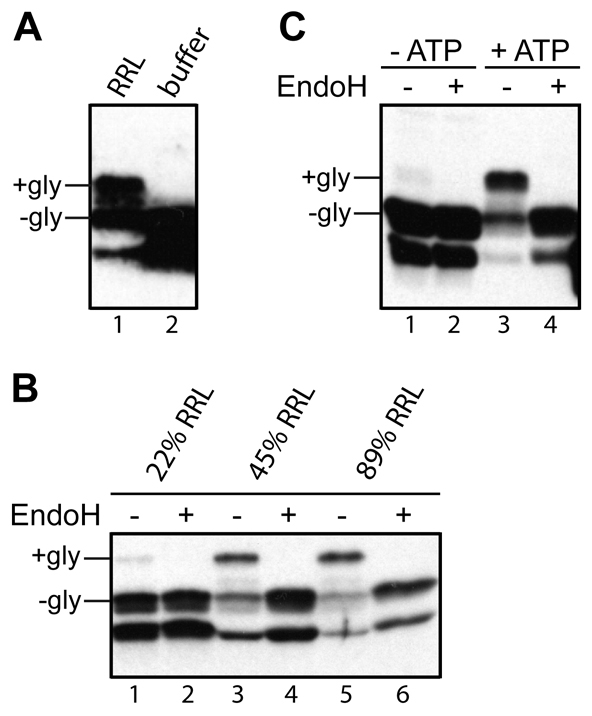
Sec61β-OPG membrane integration requires cytosol and nucleotide triphosphates. (A) A membrane-integration reaction of recombinant Sec61βOPG was performed in the presence of sheep microsomes and either rabbit reticulocyte lysate (RRL) or a buffer control. The membrane fraction was isolated and analysed by immunoblotting with a monoclonal antibody recognising the OPG tag. N-glycosylated (+gly) and non-glycosylated (−gly) forms of Sec61βOPG are labelled. The lower product is a truncated version of Sec61βOPG (supplementary material Fig. S1B,C). (B) Three 50 μl membrane-integration reactions containing ~1.1 μM Sec61β-OPG and ~2.1 OD280/ml sheep pancreatic microsomes supplemented with increasing amounts of rabbit reticulocyte lysate (% of total reaction volume shown) were performed and analysed by EndoH treatment and immunoblotting. (C) A standard membrane-integration reaction carried out with untreated rabbit reticulocyte lysate in the presence or absence of an energy regenerating system, as indicated.
The tail-anchor of Sec61β recruits several cytosolic factors
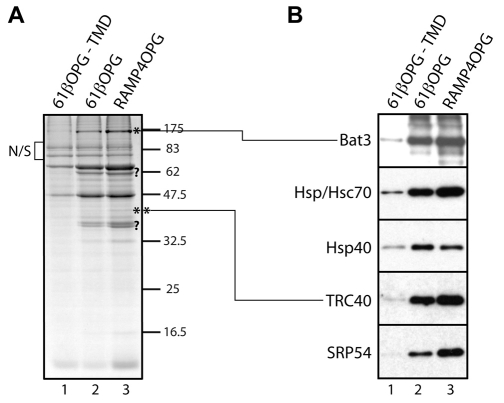
To identify novel cytosolic components that might contribute to the biogenesis of TA proteins, we looked for proteins that bound preferentially to recombinant polypeptides with an intact tail-anchor region (supplementary material Fig. S1A). To this end, recombinant opsin-epitope-tagged versions of Sec61β (Sec61β-OPG) with and without the tail-anchor region were coupled to an Ultralink resin and then used as bait for binding to components present in reticulocyte lysate. Upon analysis of the bound material, we noted that the binding of several proteins was substantially enhanced by the presence of the tail anchor (Fig. 2A). Strikingly, when a second recombinant protein with an intact tail-anchor region, RAMP4-OPG, was immobilised, an almost identical pattern of binding partners was detected (Fig. 2A). The behaviour of these components was consistent with a role in TA-protein biogenesis (Favaloro et al., 2008; Stefanovic and Hegde, 2007) and we investigated their identity using a combination of mass spectrometry and immunoblotting. In the case of cytosolic factors previously implicated in TA-protein biogenesis, we found that the recovery of SRP54, TRC40, Hsp/Hsc70 and Hsp40 were all enhanced by the presence of the hydrophobic tail-anchor regions of Sec61β and RAMP4 (Fig. 2B). The binding of an additional component of ~175 kDa was only apparent when the tail anchor was present (Fig. 2A, lanes 2 and 3, see asterisk). We were able to identify this protein as Bat3 by mass spectrometry, and confirmed its preferential binding to recombinant proteins with an intact tail-anchor region by immunoblotting (Fig. 2B). Bat3 (HLA-B associated transcript 3, also known as Scythe and Bag6) has no obvious homology with TRC40 and is involved in a variety of biological processes (see Discussion).
Fig. 2.
Identification of tail-anchor-specific cytosolic factors. (A) Comparable amounts of opsin epitope-tagged versions (OPG) of a Sec61β variant lacking the transmembrane domain (−TMD), full-length Sec61β and RAMP4 (supplementary material Fig. S1A) were immobilised on UltraLink Biosupport, incubated with rabbit reticulocyte lysate and binding partners eluted with 0.1% (v/v) Triton X-100 after extensive washing. Following SDS-PAGE and Coomassie Blue staining, components strongly enriched in lanes 2 and 3 were further characterised. Relevant proteins successfully identified by mass spectrometry are shown (*, **), other candidates (?) remain uncharacterised. (B) Eluted material was analysed by immunoblotting for specific components, as indicated.
Depletion of Bat3 specifically inhibits membrane integration of Sec61β
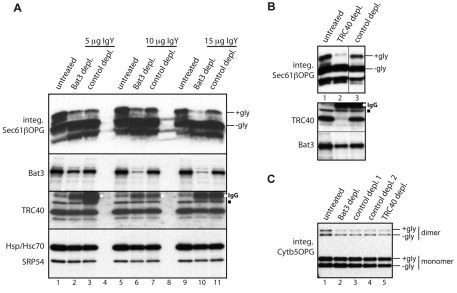
Since the membrane integration of recombinant Sec61β is entirely dependent upon the addition of cytosol in the form of reticulocyte lysate (Fig. 1A,B), we reasoned that if Bat3 contributed to Sec61β integration, its removal would inhibit this process. We therefore used an immunodepletion approach previously exploited to establish the dependence of TA-protein integration upon TRC40 (Colombo et al., 2009). The efficiency of Bat3 immunodepletion from reticulocyte lysate reflected the amount of primary antibody used, whereas significant levels remained in the paired controls (Fig. 3A). When the resulting lysates were analysed for their ability to stimulate Sec61β membrane insertion, integration was substantially reduced when Bat3 levels had been efficiently depleted (Fig. 3A, integ. Sec61β-OPG). To address whether Bat3 depletion simply removed other known components, we analysed various cytosolic factors implicated in Sec61β integration. No obvious differences in the levels of TRC40, Hsp/Hsc70 or SRP54 were apparent (Fig. 3A, TRC40 and Hsp/Hsc70 plus SRP54 panels). Strikingly, the reduction in Sec61β integration observed upon our most efficient Bat3 immunodepletion (Fig. 3A, integ. Sec61β-OPG) was comparable with that seen upon TRC40 depletion (Fig. 3B, integ. Sec61β-OPG). In this case, although depletion of TRC40 was effective, any reduction in Bat3 levels was extremely modest (Fig. 3B) and inconsistent with any defect in Sec61β-OPG membrane integration (Fig. 3A).
Fig. 3.
Bat3 is required for Sec61β-OPG membrane integration. Samples of rabbit reticulocyte lysate were immunodepleted of Bat3 or TRC40, subjected to a parallel control immunodepletion using appropriate chicken or rabbit antibodies, or left completely untreated, as indicated. (A) Upper panel, the ability of different reticulocyte lysate preparations to support the membrane integration of Sec61β-OPG was assessed via N-glycosylation (+gly and −gly) following immunoblotting with the anti-opsin monoclonal antibody. Lower panels, reticulocyte lysate was left untreated or treated with increasing amounts of a chicken anti-Bat3 IgY or a control chicken IgY as indicated, and samples immunodepleted using a secondary antibody. The resulting levels of Bat3, TRC40, Hsp/Hsc70 and SRP54 were determined by immunoblotting as indicated. Individual boxes show scans from one exposure of a single piece of film. (B) Upper panel, the ability of different lysate preparations to support the membrane integration of Sec61β-OPG was determined as described in A. This panel shows the scan from a single piece of film with two irrelevant lanes separating the TRC40 depletion (lane 2) and the parallel control sample (lane 3) removed for simplicity, as indicated by the vertical line. Lower panel, the levels of TRC40 and Bat3 were determined as for A. (C) The ability of different lysate preparations to support the membrane integration of Cytb5-OPG was determined as for A. A small amount of SDS-resistant Cytb5-OPG dimer was observed. In some cases, samples subjected to immunodepletion contained residual immunoglobulin heavy chain (labelled IgG) that crossreacted with the secondary antibody (see A and B, TRC40 immunoblot, white dots). A minor species observed with the TRC40 serum is also indicated (B and C, TRC40 panel, filled square).
The biogenesis of the TA-protein cytochrome b5 (Cytb5) is not dependent upon the canonical TRC40-mediated pathway (Favaloro et al., 2008; Favaloro et al., 2010; Rabu et al., 2008; Stefanovic and Hegde, 2007), and its membrane insertion is unaffected by the immunodepletion of this component (Colombo et al., 2009). In contrast to the clear reduction in Sec61β insertion, depletion of Bat3 had no effect upon the membrane integration of Cytb5 (Fig. 3C). Likewise, TRC40 immunodepletion had no effect on Cytb5 integration (Fig. 3C). This assay confirms that Bat3 depletion does not affect N-glycosylation per se. More importantly, the substrate specificity of Bat3 depletion for inhibiting the integration of Sec61β, but not Cytb5 (Fig. 3), combined with the tail-anchor-dependent association of Bat3 with Sec61β and RAMP4 (Fig. 2), suggests that Bat3 influences the TRC40 mediated route for TA-protein integration (Rabu et al., 2009).
A Bat3-enriched fraction rescues Sec61β membrane integration
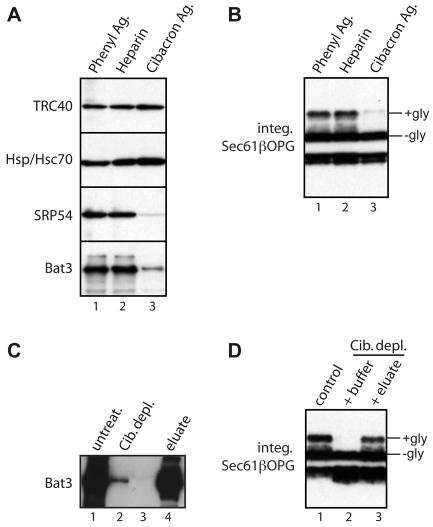
As an alternative approach for identifying cytosolic factors that facilitate TA-protein biogenesis, we developed a strategy to selectively deplete candidate components on the basis of their differential binding to various resins (Gorlich et al., 1994). After an empirical screening of a range of matrices, Cibacron Blue agarose was identified as a resin that substantially reduces the levels of Bat3 and SRP54 present in reticulocyte lysate, whilst leaving both TRC40 and Hsp/Hsc70 unaffected (Fig. 4A). When Cibacron-treated lysate was tested in a membrane-integration assay, its ability to promote the membrane integration of Sec61β was substantially impaired, although comparable treatments with other resins did not perturb this process (Fig. 4B). Thus, we found a correlation between Bat3 levels and the ability of reticulocyte lysate to promote Sec61β integration. We detected a substantial amount of Bat3 in the mixture of proteins that could be eluted from the Cibacron resin using a high-salt wash (Fig. 4C; supplementary material Fig. S2). Furthermore, when this eluted material was added back to the previously depleted reticulocyte lysate, Sec61β membrane integration was restored (Fig. 4D). Additional fractionation using nickel-NTA agarose reiterated a clear correlation between the presence of Bat3 and the ability of an eluted fraction to promote membrane integration (supplementary material Fig. S2). In summary, although TRC40 remains in reticulocyte lysate following either Bat3 immunodepletion (Fig. 3A) or Cibacron Blue agarose treatment (Fig. 4A), efficient Sec61β integration is only observed when Bat3 and TRC40 are both present in the lysate (Fig. 3A,B; Fig. 4).
Fig. 4.
A Bat3-containing fraction restores Sec61β-OPG membrane integration. (A) Reticulocyte lysate was incubated with different resins and the depletion of various cytosolic factors determined by immunoblotting, as indicated. (B) The resin-depleted lysate preparations were used to stimulate the membrane integration of recombinant Sec61βOPG. (C) Material bound to Cibacron Blue agarose was recovered and analysed for Bat3 content by immunoblotting and comparison with equivalent amounts of untreated lysate and Cibacron-depleted lysate as indicated. Lane 3 is empty. (D) The membrane integration of recombinant Sec61β-OPG was determined in the presence of untreated lysate (lane 1), Cibacron-depleted lysate supplemented with buffer (lane 2), or Cibacron-depleted lysate supplemented with the eluate from the Cibacron resin (lane 3). Individual boxes all represent a single exposure, as described for Fig. 3.
Bat3 and TRC40 associate with distinct populations of Sec61β polypeptides
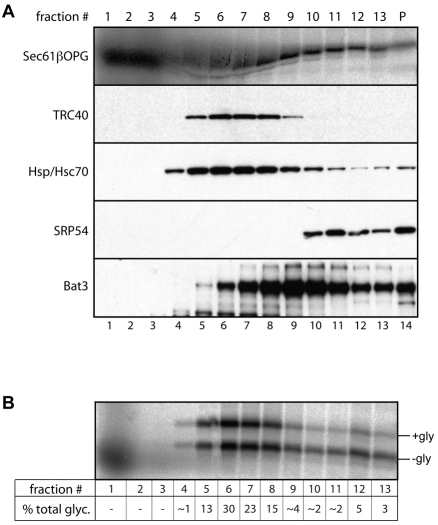
TRC40/Get3 forms a stable association with newly synthesised TA proteins (Favaloro et al., 2008; Stefanovic and Hegde, 2007), and functions at a late stage of the ER-delivery process (Bozkurt et al., 2009). We investigated the relationship of Bat3 with newly synthesised Sec61β-OPG chains generated by cell-free translation. Radiolabelled Sec61β chains were generated using a reticulocyte-lysate translation system and then fractionated by centrifugation through a sucrose gradient (Favaloro et al., 2008; Stefanovic and Hegde, 2007). Each fraction was analysed for the presence of both radiolabelled Sec61β polypeptides and a variety of cytosolic factors including Bat3 and TRC40 (Fig. 5A). In parallel, equivalent samples were analysed for their capacity to support the membrane integration of the radiolabelled Sec61βOPG chains present in the fraction, using N-glycosylation as the readout (Fig. 5B). These experiments show that the bulk of TRC40 co-migrates with the peak of integration competent Sec61β chains (~80% of total N-glycosylation) found in fractions 5 to 8 of the gradient (Fig. 5A,B).
Fig. 5.
Bat3 and TRC40 are associated with distinct populations of Sec61β-OPG polypeptides. (A) To recapitulate biosynthetic associations, Sec61β-OPG was translated in rabbit reticulocyte lysate, the material subjected to centrifugation through a 5-25% sucrose gradient, and 13 fractions plus the pellet recovered (fraction 1=top, p=pellet). Following SDS-PAGE, the location of Sec61βOPG chains was determined by phosphorimaging of the radiolabelled chains (upper panel), whereas the migration of various cytosolic factors was established by immunoblotting (as labelled). The distortion of the radiolabelled Sec61βOPG samples seen in fractions 4-8 results from the presence of large quantities of unlabelled globin chains present in the lysate used for in vitro translation. (B) A portion of each sucrose gradient fraction was incubated with canine pancreatic microsomes, the membranes recovered and the glycosylation status of Sec61β-OPG determined by phosphorimaging. The proportion of the total N-glycosylated material resulting from each individual fraction (% total glyc.) was estimated to provide a comparative measure of membrane integration.
The distribution of Bat3 is clearly distinct from that of TRC40, showing a partial overlap but peaking at a lower part of the gradient from fractions 7 to 11 (Fig. 5A). In particular, we noted that although fraction 9 contains the strongest Bat3 signal, it supports rather low levels of Sec61β membrane integration (Fig. 5A,B; ~4% of total N-glycosylated material). Hence, although fraction 9 contains levels of Sec61β chains comparable with fractions 7 and 8 (Fig. 5A), the population of TA proteins found in the same fraction as the peak of Bat3 appears far less competent for membrane integration (Fig. 5B, compare fraction 7 with fraction 9).
Bat3 can influence the fate of a tail-anchored protein in S. cerevisiae
We found no evidence to suggest that a stable complex is formed between the bulk of Bat3 and TRC40 present in reticulocyte lysate (Figs 3, 4 and 5); therefore, to gain further insight into the role of Bat3 during TA-protein biogenesis, we exploited an experimental system originally designed to characterise components of the equivalent GET pathway in S. cerevisiae. This assay relies on the observation that the deletion of components of the GET pathway inhibits the ER integration of a model TA protein, GFP-Sed5 (Weinberger et al., 2005), reducing its trafficking to the Golgi and concomitantly increasing the level of cytosolic GFP-Sed5 that has failed to become membrane integrated (Jonikas et al., 2009; Schuldiner et al., 2008).
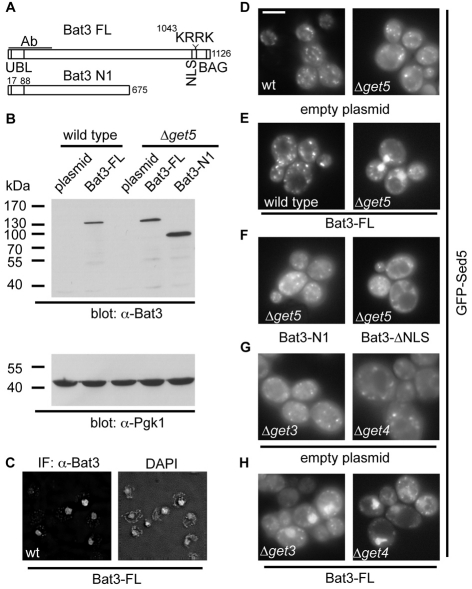
Although quite clearly distinct, mammalian Bat3 and yeast Mdy2 (Get5) both contain a ubiquitin-like domain (Hu et al., 2006; Kabbage and Dickman, 2008) and we tested the hypothesis that Bat3 is a functional equivalent of one of the two novel cytosolic components, Get4 and Mdy2 (Get5), which have been recently identified in S. cerevisiae (Jonikas et al., 2009) and are presumed to have mammalian homologues (Rabu et al., 2009). Full-length human Bat3 could be expressed in both wild-type and Δmdy2 (Δget5) deletion strains (Fig. 6A,B), and immunofluorescence microscopy showed that Bat3 was readily detected in the nucleus of both strains (Fig. 6C and supplementary material Fig. S3A). This is consistent with studies in cultured mammalian cells where populations of Bat3 in both the nucleus and the cytosol can be observed (Desmots et al., 2008). Bat3 expression had no effect on the location of GFP-Sed5 in wild-type cells, where numerous punctae were seen, consistent with efficient membrane integration and Golgi localization (Fig. 6D,E and supplementary material Fig. S3C, wild-type panels) (Jonikas et al., 2009; Schuldiner et al., 2008; Weinberger et al., 2005). By contrast, the cytosolic accumulation of GFP-Sed5 that was apparent upon the loss of Mdy2 (Get5) (Jonikas et al., 2009) was reduced by expression of full-length Bat3, with GFP-Sed5 being redirected to the nucleus (Fig. 6D,E and supplementary material Fig. S3C, Δget5 panels). This reduction in cytosolic fluorescence and nuclear localisation of GFP-Sed5 required the full-length Bat3 protein with an intact nuclear localisation signal (NLS). Hence, although an N-terminal fragment of Bat3 and a full-length version with the NLS rendered non-functional were both efficiently expressed (Fig. 6A,B and supplementary material Fig. S3E), neither Bat3 variant supported the nuclear relocalisation of GFP-Sed5 (Fig. 6F). Immunofluorescence microscopy confirmed the cytosolic localisation of the two Bat3 mutants (supplementary material Fig. S3B,D), and we conclude that full-length Bat3 associates with GFP-Sed5 and redirects this TA protein to the nucleus by virtue of its previously defined NLS (Manchen and Hubberstey, 2001).
Fig. 6.
Bat3 relocalises GFP-Sed5 in S. cerevisiae GET mutants. (A) Outline of full-length Bat3 (isoform 2), and the N-terminal fragment used in this study. The locations of the ubiquitin-like domain, NLS and BAG domains and the antibody-binding region are indicated. In the ΔNLS mutant, the KRRK motif shown is altered to KRSL to disrupt nuclear targeting of Bat3 (Manchen and Hubberstey, 2001). (B) Immunoblot showing Bat3 and phosphoglycerate kinase 1 (Pgk1) levels in wild-type or Δmdy2 (Δget5)-transformed S. cerevisiae cells. (C) Subcellular localisation of full-length Bat3 expressed in wild-type S. cerevisiae and DAPI staining of nuclei visualised by immunofluorescence microscopy. (D-H) The effect of Bat3 expression upon the subcellular localisation of GFP-Sed5 was determined by live-cell imaging in wild-type, Δget3, Δget4 or Δmdy2 (Δget5) cells, as indicated. Full-length Bat3, an N-terminal fragment or the ΔNLS mutant, were used as indicated. See also supplementary material Fig. S2C for fixed and immunostained cells of the same genotype demonstrating co-localisation of Bat3 and GFP-Sed5 immunoreactivity with DAPI staining of the nucleus in Δmdy2 (Δget5) cells. Scale bar: 5 μm.
Although the expression of full-length Bat3 did not restore a wild-type phenotype for GFP-Sed5 localisation in the mdy2-deletion strains, the protein had a clear effect on the fate of the TA substrate. To establish whether this effect simply resulted from some perturbation of the remaining cytosolic GET complex, we looked at the outcome of Bat3 expression in yeast strains lacking other components of the GET pathway. A strikingly similar effect, namely a reduction in cytosolic labelling and the appearance of a nuclear GFP-Sed5 signal, was also observed in strains lacking either Get3 or Get4 (Fig. 6G,H). Clearly Bat3 is not compensating for the loss of a specific component of the GET pathway in yeast, but rather providing a non-physiological alternative under conditions where the GET pathway is perturbed. This conclusion was further supported by the lack of any apparent perturbation of GFP-Sed5 localisation following Bat3 expression in wild-type cells (Fig. 6E). Since Bat3 is able to redirect GFP-Sed5 to the nucleus in all three of the GET-pathway mutants tested, the association of Bat3 with a TA protein does not require Get3, Get4 or Mdy2 (Get5).
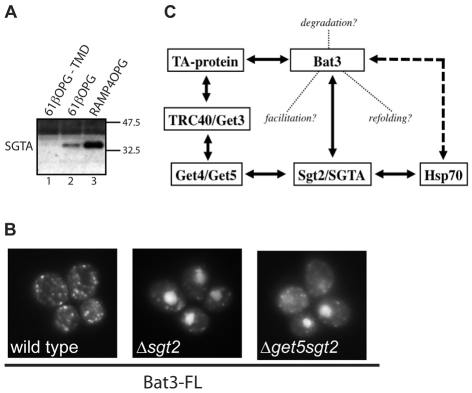
Yeast Sgt2 has recently been implicated in the GET pathway for TA-protein biogenesis (Chang et al., 2010; Costanzo et al., 2010) and its mammalian equivalent, SGTA (SGT), is a known interacting partner of Bat3 and Hsp/Hsc70 in mammalian cells (Winnefeld et al., 2006). We therefore addressed the possibility that any role for Bat3 during TA-protein biogenesis might also involve SGTA and/or its yeast equivalent. When the mammalian cytosolic components that bind tail-anchor regions were re-analysed by immunoblotting, we found that SGTA was enriched in fractions eluted from full-length TA proteins (Fig. 7A), consistent with a potential role in TA-protein biogenesis (Fig. 2). However, the ability of Bat3 to relocalise GFP-Sed5 to the nucleus of a Δget5 strain was not dependent upon the presence of Sgt2 (Fig. 7B). Furthermore, the loss of Sgt2 alone proved sufficient to enable Bat3-mediated relocalisation (Fig. 7B). Thus, Bat3 can associate with TA proteins in the absence of any of the known soluble proteins of the yeast GET pathway, including Sgt2.
Fig. 7.
Bat3 relocalisation of GFP-Sed5 is independent of Sgt2. (A) Reticulocyte-lysate components eluted from recombinant TA proteins with or without a TA segment (see Fig. 2) were analysed for the presence of SGTA by immunoblotting. (B) Live-cell imaging of GFP-Sed5 in wild-type, Δsgt2 and Δmdy2sgt2 S. cerevisiae expressing full-length human Bat3. (C) Summary of potential interactions between TA proteins, Bat3, SGTA/Sgt2, Hsp70 and components of the TRC40/GET pathway (see Chang et al., 2010; Corduan et al., 2009; Costanzo et al., 2010; Favaloro et al., 2008; Jonikas et al., 2009; Sasaki et al., 2008; Schuldiner et al., 2008; Stefanovic and Hegde, 2007; Stelzl et al., 2005; Winnefeld et al., 2006). Solid lines indicate known or presumed physical interactions, the dashed line, a regulatory interaction and the dotted lines indicate possible contributions of Bat3 to TA-protein biogenesis.
Discussion
To better understand the pathways and components responsible for the biogenesis of TA proteins we used an affinity-binding approach to identify candidate cytosolic factors. A number of proteins preferentially associate with both Sec61β- and RAMP4-bearing intact tail-anchors, including all of the cytosolic factors previously implicated in TA-protein biogenesis (Abell et al., 2004; Abell et al., 2007; Favaloro et al., 2008; Rabu et al., 2008; Stefanovic and Hegde, 2007). We found an additional component of ~175 kDa that was substantially enriched in the presence of a tail anchor, and this protein was identified as Bat3. Bat3 has been implicated in a variety of biological processes, including the regulation of apoptosis (Desmots et al., 2008), Hsp70 stability (Corduan et al., 2009; Sasaki et al., 2008) and function of natural killer cells (Simhadri et al., 2008). Bat3 has no clear homology with TRC40, and its most obvious features are an N-terminal ubiquitin-like domain, a nuclear-localisation signal (NLS) and a C-terminal BAG domain (Kabbage and Dickman, 2008).
Strikingly, Bat3 depletion results in a substantial inhibition of the reticulocyte-lysate-dependent membrane integration of Sec61β, indicating that this component can influence the biogenesis of TA protein. Whether Bat3 removal also depletes other, as yet unidentified, cytosolic components that are important for TA-protein biogenesis remains to be established. However, the removal of Bat3 does not substantially alter TRC40 levels, nor is the integration of the TRC40-independent substrate Cytb5 affected by Bat3 removal. We conclude that the role of Bat3 is most likely restricted to TA proteins that are TRC40 clients (Colombo et al., 2009; Rabu et al., 2009; Rabu et al., 2008; Stefanovic and Hegde, 2007). Although the peak of integration-competent Sec61β chains co-migrate with the bulk of TRC40, strongly supporting a role in promoting membrane integration (Bozkurt et al., 2009; Favaloro et al., 2008; Stefanovic and Hegde, 2007), the bulk of Bat3 co-migrates with a pool of Sec61β chains that are poorly membrane integrated. Thus, although Bat3 can influence the fate of TA-protein substrates of the TRC40 pathway, its function might be spatially and temporally distinct from the TRC40-mediated delivery step that precedes membrane integration (Bozkurt et al., 2009; Favaloro et al., 2010). Interestingly, a recent proteomic analysis that used the ubiquitin-like-domain-containing protein UBL4A as bait identified a protein network containing both Bat3 and TRC40 (Asna-1), together with SGTA and C7orf20 [see table S4 in Sowa et al. (Sowa et al., 2009)]. Hence, Bat3 and TRC40 might associate transiently or share common interacting partners.
To place the role of Bat3 into a cellular context, we exploited S. cerevisiae mutants lacking various components of the GET pathway that is functionally equivalent to the mammalian TRC40 pathway, but is at present far better defined (Jonikas et al., 2009; Rabu et al., 2009; Schuldiner et al., 2008). The very clear outcome of this approach is that the expression of Bat3 in a variety of GET-pathway mutants results in the relocalisation of a model GET-pathway substrate, GFP-Sed5, to the nucleus. We show that the ability of Bat3 to redirect GFP-Sed5 to the nucleus relies on a previously defined nuclear-localisation signal present in Bat3, which is known to be functional in mammalian cells (Desmots et al., 2008; Manchen and Hubberstey, 2001). These data are reminiscent of the PEX19-mediated nuclear relocalisation of several peroxisomal membrane protein substrates, in this instance elicited by the introduction of an artificial NLS motif into PEX19, which was used to help identify class 1 peroxisomal-membrane proteins (Jones et al., 2004). The effect of Bat3 expression in S. cerevisiae strongly supports a model where the association of TA proteins with Bat3 influences their subsequent fate in vivo.
We found that the effect of Bat3 is comparable in the absence of any of known cytosolic and/or peripheral-ER-membrane components of the GET pathway (Get3, Get4 and Get5) (Jonikas et al., 2009; Rabu et al., 2009; Schuldiner et al., 2008). The simplest explanation of these data is that full-length Bat3 can associate with GFP-Sed5 independently of Get3, Get4 or Get5, with the resulting complex entering the nucleus by virtue of the NLS present in Bat3, thereby reducing the level of cytosolic GFP-Sed5 when the GET pathway is non-functional. This hypothesis is in good agreement with a ‘high-confidence’ interaction identified between Bat3 and syntaxin 5A, the human homologue of Sed5, using a yeast two-hybrid approach (Stelzl et al., 2005). Two recent publications identify the yeast protein Sgt2 as a component of the GET pathway for TA-protein biogenesis (Chang et al., 2010; Costanzo et al., 2010), providing an alternative model for our observations when Bat3 is expressed in yeast. Specifically, Bat3 interacts with the mammalian equivalent of Sgt2, SGTA (Winnefeld et al., 2006), providing a potential intermediary between the TRC40/GET pathway and Bat3 (Fig. 7C). Although we found that mammalian SGTA preferentially associates with hydrophobic TA segments, the Bat3-dependent relocalisation of TA proteins in yeast does not require Sgt2, and in fact the loss of Sgt2 alone is sufficient to enable Bat3-mediated relocalisation to occur. These data strongly support a role for Sgt2 in the yeast GET pathway (Chang et al., 2010; Costanzo et al., 2010), and indicate that SGTA might have a similar role in higher eukaryotes (Fig. 5A; Fig. 7C). However, the association of Bat3 with GFP-Sed5 in yeast, and its redirection to the nucleus does not require Sgt2. We conclude that either the association of Bat3 with TA proteins observed in yeast is direct, as previously suggested (Stelzl et al., 2005), or that the process involves some other functionally conserved component associated with or regulated by Bat3, for example Hsp/Hsc70 (Corduan et al., 2009; Sasaki et al., 2008).
Although we can only speculate about the precise role(s) of Bat3 during TA-protein biogenesis, the following scenarios provide a basis for further experimentation. Our current results suggest that Bat3 is not stably associated with other components of the TRC40 pathway (Fig. 5A; Fig. 7C); however, Bat3 might modulate or enhance particular steps in the TRC40 cycle for TA-protein delivery to the ER (Rabu et al., 2009). Hence, Bat3 could facilitate the formation of a productive TRC40-substrate ER-delivery complex (Rabu et al., 2009), or promote the interaction of the assembled complex with the ER membrane. If so, then the role of Bat3 can be circumvented by the co-expression of TRC40/Get3 and a TA-protein substrate in E. coli, because this results in a functional ER-delivery complex (Bozkurt et al., 2009; Favaloro et al., 2010). Alternatively, Bat3 could contribute to the recycling of the TRC40 complex by facilitating its release from the, as yet unidentified, mammalian ER-membrane receptor after delivery of TA protein has occurred (Rabu et al., 2009). Equally, the association of selected TA proteins with Bat3 might in fact represent an alternative and complementary fate to TRC40 binding. In this scenario, association with Bat3 could provide aberrant or misfolded TA proteins with an opportunity for refolding and return to the TRC40 pathway (Fig. 7C), most likely via the actions of Hsp/Hsc70 chaperones (Corduan et al., 2009; Sasaki et al., 2008). Removal of Bat3 from our in vitro system could cause the TRC40 pool to be titrated out by binding aberrant TA proteins to form complexes that are incapable of authentic ER delivery. Bat3 might also regulate the entry of aggregated or terminally misfolded TA proteins into a degradative pathway (Fig. 7C) (Auld et al., 2006), which is consistent with the presence of its N-terminal ubiquitin-like domain (Kabbage and Dickman, 2008).
Whatever the precise role of Bat3 during TA-protein biogenesis, the presence of a C-terminal BAG domain provides an opportunity for its regulation via several cellular components, including Hsp70 (Kabbage and Dickman, 2008). Intriguingly, the so-called BAG domain, found in various proteins, including Bat3, was first identified via its interaction with Bcl-2, a TA protein (Janiak et al., 1994) implicated in the regulation of apoptosis (Heath-Engel et al., 2008). Hence, one could speculate that the modulation of apoptosis and proliferation ascribed to Bat3 (Scythe) (Desmots et al., 2005) might reflect a BAG-domain-dependent interaction with TA proteins such as Bcl-2. Interestingly, our previous studies suggest that, similarly to Cytb5, Bcl2 is not an obligatory client of the generic TRC40-mediated pathway (Rabu et al., 2008), and on this basis it seems unlikely that Bat3 will influence the membrane integration of Bcl2 in a similar fashion to Sec61β. It is becoming increasingly apparent that the biogenesis of TA proteins is a remarkably complex and multifaceted process. Defining the precise function of Bat3 during this process and understanding this in the context of its various other cellular roles will be a major focus of our future efforts.
Materials and Methods
Materials
Bacterial expression vector, pHisTrx, was a gift from Richard Kammerer (University of Manchester, Manchester, UK). Rabbit polyclonal antisera recognising TRC40 (Asna-1) was a gift from Bernhard Dobberstein (ZMBH, Heidelberg, Germany) and the monoclonal anti-opsin tag antibody (Adamus et al., 1991) was provided by Paul Hargrave (Department of Ophthalmology, University of Florida, FL). Commercial antibodies were used to detect Hsp/Hsc70 and Hsp40 (Stressgen), SRP54 (BD Biosciences), Bat3 (Abcam), yeast Pgk1 (Invitrogen) and GFP (ab290, Abcam). Nuclease-treated rabbit reticulocyte lysate for in vitro translation was from Promega, rabbit reticulocyte untreated lysate was from Green Hectares.
Protein expression and purification
cDNAs encoding single cysteine variants of Sec61β, Sec61β lacking the tail anchor, RAMP4 and cytochrome b5, in each case including an in-frame opsin tag at the 3′ end (see supplementary material Fig. S1A), were subcloned into the pHisTrx expression vector and all constructs confirmed by DNA sequencing. The proteins were expressed in Escherichia coli strain BLR (DE3) pLysS cells by IPTG induction, and after harvesting cells were lysed in buffer A [50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 10 mM MgCl2, 10 mM imidazole, 5 mM 2-ME, 1 mM PMSF, 10 % (v/v) glycerol] supplemented with 0.2 mg/ml lysozyme, 15 U/ml DNase I, complete protease inhibitor cocktail (Roche) and 1% (w/v) dodecyl-β-D-maltopyranoside (DDM) for 1 hour at room temperature followed by three passes through a 25-gauge needle. Insoluble material was pelleted (30 min; 10,000 g at 4°C) and the His-tagged recombinant proteins isolated by binding to nickel-NTA resin and extensive washing with buffer B [50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol] supplemented with 0.1% (w/v) DDM. For the purification of recombinant RAMP4OPG, subsequent buffers contained 0.1% (w/v) DDM throughout the subsequent procedures. For Sec61βOPG and Cytb5OPG, the detergent was exchanged to 0.75% (w/v) octyl-β-D-glucopyranoside or 0.1% n-dodecyl-N,N-dimethylamine-N-oxide respectively, by extensive washing of the beads using buffer B supplemented with the appropriate detergent. For the Sec61βOPG variant lacking the tail anchor, Triton X-100 was used in place of DDM through out the procedure [1% (w/v) during cell lysis and 0.1% (w/v) during subsequent steps].
To release the recombinant proteins from the resin, beads resuspended in buffer B supplemented with the appropriate detergent were incubated with 15 U thrombin per ml of suspension overnight at room temperature. The resin was then isolated by centrifugation, and the thrombin either inactivated by the addition of PMSF or removed by incubating the supernatant with Benzamidine Sepharose (1 hour, 4°C). The resulting solution was then centrifuged at 100,000 g for 1 hour at 4°C to remove any aggregates. Protein concentration was estimated by absorbance at 280 nm and/or Coomassie Brilliant Blue staining of SDS-PAGE gels using lysozyme as a standard. Proteins were aliquoted, frozen in liquid N2 and stored at −80°C.
Membrane-integration reaction
Integration reactions comprised of 1.9 μM Sec61β-OPG, or 0.5 μM Cytb5-OPG, 5 μl sheep pancreatic microsomes (final concentration of ~3.5 OD280 per ml) (Kaderbhai et al., 1995) and rabbit reticulocyte lysate (Promega) made up to 30 μl, unless stated otherwise. The reactions were incubated at 30°C for 4 hours and then the membrane fraction recovered by centrifugation through a 140 μl HSC cushion (750 mM sucrose, 500 mM potassium acetate, 5 mM magnesium acetate, 50 mM HEPES-KOH, pH 7.9) for 10 minutes at 120,000 g and 4°C. Where appropriate, the membrane fraction was resuspended in 32 μl LSC buffer (250 mM sucrose, 100 mM potassium acetate, 5 mM magnesium acetate, 50 mM HEPES-KOH, pH 7.9), split in two and one half treated with Endoglycosidase H (New England Biolabs) as described by the supplier. All samples were solubilised in Laemmli buffer and resolved by SDS-PAGE before detection by immunoblotting.
Identification of cytosolic factors interacting with TA proteins
Recombinant proteins were immobilized via amino groups on 10 mg of UltraLink Biosupport (Pierce) according to manufacturer's instruction. The estimated concentration of bound proteins was: Sec61β-OPG, 0.9 mg/ml; Sec61β-OPG-TMD, 1.67 mg/ml; RAMP4-OPG, 0.99 mg/ml. Beads were stored in 800 μl buffer R (50 mM HEPES-KOH, pH 7.5, 40 mM potassium acetate, 5 mM MgCl2) with 0.1% (v/v) Triton X-100. For the pull-down experiment one third of the beads (about 25 μl volume) was washed with buffer R alone to remove detergent, then incubated with 800 μl of untreated rabbit reticulocyte lysate (Green Hectares, Oregon) supplemented with an energy regeneration system (1 mM ATP, 100 μM GTP, 10 mM creatine phosphate and 100 μg/ml creatine phosphokinase) for 2 hours at 4°C. The beads were washed three times with 1 ml buffer R and the remaining proteins eluted with 128 μl buffer R supplemented with 0.1% (v/v) Triton X-100. Eluted material was TCA precipitated, solubilised in Laemmli buffer and resolved by SDS-PAGE followed by Coomassie Brilliant Blue staining. Proteins that appeared to interact specifically with full-length TA proteins were excised and analysed by mass spectrometry following in-gel trypsin digestion.
Immunodepletion of cytosolic factors from rabbit reticulocyte lysate
20 μl (bead volume) of Protein-A-Sepharose was mixed with 120 μg rabbit anti-chicken antibody and incubated for 1 hour at 4°C. The beads were then washed with buffer R to remove any unbound antibody and 120 μl rabbit reticulocyte lysate (Promega), pre-incubated for 1 hour at 4°C with 5, 10 or 15 μg of chicken anti-Bat3 or chicken anti-SSBP1 control antibody was added as indicated. The beads and lysates were further incubated for 1 hour at 4°C, the Protein-A-Sepharose pelleted by centrifugation and the resulting immunodepleted lysates used in the membrane integration reaction as previously described. TRC40 (Asna-1) was similarly depleted by a 1 hour incubation of 120 μl rabbit reticulocyte lysate with 20 μl (bead volume) Protein-A-Sepharose-coated with anti-TRC40 antibody. Beads coated with an equivalent amount of non-related rabbit serum were used for control reactions.
Depletion of cytosolic factors from rabbit reticulocyte lysate by resin treatment
75 μl rabbit reticulocyte lysate (Promega) was incubated for 2 hours at 4°C with 30 μl of the indicated resins [50% (v/v) suspensions] previously equilibrated with buffer R. Unbound material was collected and analysed for cytosolic components of interest by immunoblotting or tested for function in a membrane-integration assay using recombinant Sec61β-OPG. For add-back experiments, 100 μl Cibacron Blue agarose [50% (v/v) suspension] was incubated with 250 μl of rabbit reticulocyte lysate (Promega) for 2 hours at 4°C, the beads were then washed three times in buffer R and bound material eluted with 100 μl buffer R supplemented with 1.5 M NaCl. After desalting 5 μl of the eluate or equivalent buffer control was mixed with 20 μl of the Cibacron-treated rabbit reticulocyte lysate, and a membrane integration-assay performed.
Sucrose-gradient centrifugation
Sec61β-OPG was synthesised in a 100 μl reaction volume in vitro, as previously described (Rabu et al., 2008) and the material fractionated by centrifugation through a sucrose gradient using a modified version of the procedure described (Stefanovic and Hegde, 2007). Thirteen individual fractions were collected from the gradient (1=top, 13=bottom) whereas pelleted material (P) was resuspended directly into the Laemmli buffer. A portion of fraction was mixed with the Laemmli buffer and, after SDS-PAGE, the location of distinct cytosolic factors was determined by immunoblotting and the distribution of the radiolabelled Sec61β-OPG visualised by phosphorimaging. In a parallel experiment, 50 μl of each fraction was mixed with canine pancreas microsomes (final concentration of 2.0 OD280 per ml), incubated for 50 minutes at 30°C and the membrane fraction isolated as described for membrane integration reaction. Following SDS PAGE and quantitative phosphorimaging, the relative proportion of the total N-glycosylated material in each fraction was determined to provide a measure of the capacity for membrane integration for the Sec61β-OPG chains present in each fraction.
Bat3 expression and analysis in S. cerevisiae
The full-length version of Bat3, isoform 2, was obtained from Origene and cloned into the yeast expression vector p416Met25 (Mumberg et al., 1994) via the XbaI and XhoI restriction sites. For the Bat3-N1 fragment that lacked the C-terminus, a stop codon was introduced in place of residue 676 of the wild-type sequence. The Bat3 ΔNLS mutant was generated as previously described (Manchen and Hubberstey, 2001). GFP-Sed5 was expressed from plasmid pRS315 (Weinberger et al., 2005). All yeast strains used were derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Brachmann et al., 1998). The respective deletion strains for GET3 (Δget3::KanR), GET4 (Δget4::KanR), GET5/MDY2 (Δget5/mdy2::KanR) and SGT2 (Δsgt2::KanR) were obtained from Euroscarf (Winzeler et al., 1999). A Δsgt2::KanR Δget5/mdy2::NatR double deletion was created using standard PCR-based replacement methods with plasmid pAG25 (Goldstein and McCusker, 1999).Yeast transformation and growth in synthetic complete medium lacking uracil and/or leucine followed well-established protocols (Ausubel et al., 1997), whereas total cell lysates for western blotting analysis were prepared as described (Yaffe and Schatz, 1984). Anti-Bat3 chicken antibody was used at 1:5000 and anti-Pgk1 mouse monoclonal at 1:2000; anti-chicken and anti-mouse horseradish-peroxidase conjugated secondary antibodies were used at 1:2500 and 1:5000, respectively. Immunofluorescence followed the method described (Roberts et al., 1991), except that cells were fixed for 1 hour in 4% formaldehyde. Attached spheroblasts were then incubated in ice-cold methanol for 6 minutes, followed by ice-cold acetone for 30 seconds. Anti-GFP and anti-Bat3 primary antibodies were diluted 1:1000, the respective anti-rabbit (Alexa Fluor 488, Invitrogen) and anti-chicken (Alexa Fluor 594, Invitrogen) secondary antibodies 1:500. Stained spheroplasts were mounted in SlowFade Gold antifade reagent containing DAPI (Invitrogen). Live-cell imaging of yeast cells was performed at room temperature in synthetic complete medium employing a DeltaVison restoration microscope equipped with a 100×, 0.35-1.5 Uplan Apo objective and a GFP filter set (Chroma 86006). The images were collected with a Coolsnap HQ camera (Photometrics). The same set-up was used to image immunostained spheroplasts for which stacks with Z optical spacing of 0.3 μm were acquired. Raw images were deconvolved using the additive algorithm of Softworx software.
Supplementary Material
Acknowledgments
This work was supported by a PhD studentship (P.L.) and Senior Fellowship (B.S.) from the Wellcome Trust. Mass spectrometry was performed by the biomolecular analysis core facility and microscopy in the bioimaging facility at the Faculty of Life Sciences. We thank Richard Kammerer for guidance regarding E. coli expression systems, Quentin Roebuck for technical assistance, Martin Pool, Lisa Swanton and Phil Woodman for their comments during manuscript preparation, and all of our colleagues who provided reagents and advice. Deposited in PMC for release after 6 months.
Footnotes
Note added in proof
A recent phylogenetic analysis (Borgese and Righi, 2010) suggests that the membrane insertion of TA proteins in prokaryotes either occurs through an unassisted pathway or is mediated by Hsp40 and Hsp70s.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/13/2170/DC1
References
- Abell B. M., Pool M. R., Schlenker O., Sinning I., High S. (2004). Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23, 2755-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abell B. M., Rabu C., Leznicki P., Young J. C., High S. (2007). Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 120, 1743-1751 [DOI] [PubMed] [Google Scholar]
- Adamus G., Zam Z. S., Arendt A., Palczewski K., McDowell J. H., Hargrave P. A. (1991). Anti-rhodopsin monoclonal antibodies of defined specificity: characterization and application. Vision Res. 31, 17-31 [DOI] [PubMed] [Google Scholar]
- Auld K. L., Hitchcock A. L., Doherty H. K., Frietze S., Huang L. S., Silver P. A. (2006). The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics 174, 215-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1997). Current Protocols in Molecular Biology New York: Greene Publishing Associates and Wiley-Interscience; [Google Scholar]
- Behrens T. W., Kearns G. M., Rivard J. J., Bernstein H. D., Yewdell J. W., Staudt L. M. (1996). Carboxyl-terminal targeting and novel post-translational processing of JAW1, a lymphoid protein of the endoplasmic reticulum. J. Biol. Chem. 271, 23528-23534 [DOI] [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368-375 [DOI] [PubMed] [Google Scholar]
- Borgese N., Righi R. (2010). Remote origins of tail-anchored proteins. Traffic [Epub ahead of print] doi: 10.1111/j.1600-0854.2010.01068.x [DOI] [PubMed] [Google Scholar]
- Bozkurt G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., et al. (2009). Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA 106, 21131-21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132 [DOI] [PubMed] [Google Scholar]
- Chang Y. W., Chuang Y. C., Ho Y. C., Cheng M. Y., Sun Y. J., Hsiao C. D., Wang C. (2010). Crystal structure of Get4/Get5 complex and its interactions with Sgt2, Get3 and Ydj1. J. Biol. Chem. 285, 9962-9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S. F., Longhi R., Borgese N. (2009). The role of cytosolic proteins in the insertion of tail-anchored proteins into phospholipid bilayers. J. Cell Sci. 122, 2383-2392 [DOI] [PubMed] [Google Scholar]
- Corduan A., Lecomte S., Martin C., Michel D., Desmots F. (2009). Sequential interplay between BAG6 and HSP70 upon heat shock. Cell. Mol. Life Sci. 66, 1998-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., et al. (2010). The genetic landscape of a cell. Science 327, 425-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross B. C., Sinning I., Luirink J., High S. (2009). Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 10, 255-264 [DOI] [PubMed] [Google Scholar]
- Desmots F., Russell H. R., Lee Y., Boyd K., McKinnon P. J. (2005). The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25, 10329-10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F., Russell H. R., Michel D., McKinnon P. J. (2008). Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 283, 3264-3271 [DOI] [PubMed] [Google Scholar]
- Favaloro V., Spasic M., Schwappach B., Dobberstein B. (2008). Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 121, 1832-1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V., Vilardi F., Schlecht R., Mayer M. P., Dobberstein B. (2010). Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 123, 1522-1530 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553 [DOI] [PubMed] [Google Scholar]
- Gorlich D., Prehn S., Laskey R. A., Hartmann E. (1994). Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79, 767-778 [DOI] [PubMed] [Google Scholar]
- Heath-Engel H. M., Chang N. C., Shore G. C. (2008). The endoplasmic reticulum in apoptosis and autophagy: role of the BCL-2 protein family. Oncogene 27, 6419-6433 [DOI] [PubMed] [Google Scholar]
- Hu J., Li J., Qian X., Denic V., Sha B. (2009). The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One 4, e8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Potthoff B., Hollenberg C. P., Ramezani-Rad M. (2006). Mdy2, a ubiquitin-like (UBL)-domain protein, is required for efficient mating in Saccharomyces cerevisiae. J. Cell Sci. 119, 326-338 [DOI] [PubMed] [Google Scholar]
- Janiak F., Leber B., Andrews D. W. (1994). Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 269, 9842-9849 [PubMed] [Google Scholar]
- Jones J. M., Morrell J. C., Gould S. J. (2004). PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164, 57-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693-1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M., Dickman M. B. (2008). The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 65, 1390-1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaderbhai M. A., Harding V. J., Karim A., Austen B. M., Kaderbhai N. N. (1995). Sheep pancreatic microsomes as an alternative to the dog source for studying protein translocation. Biochem. J. 306, 57-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hartmann E., Rapoport T. A. (1993). A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72-75 [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., Rapoport T. A. (1995). Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Foguet M., Renz M., Seelig H. P., Glick B. S., Hauri H. P. (1995). A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golgi apparatus. Proc. Natl. Acad. Sci. USA 92, 5102-5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchen S. T., Hubberstey A. V. (2001). Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 287, 1075-1082 [DOI] [PubMed] [Google Scholar]
- Mateja A., Szlachcic A., Downing M. E., Dobosz M., Mariappan M., Hegde R. S., Keenan R. J. (2009). The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. (1994). Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767-5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C., Wipf P., Brodsky J. L., High S. (2008). A Precursor-specific Role for Hsp40/Hsc70 during Tail-anchored Protein Integration at the Endoplasmic Reticulum. J. Biol. Chem. 283, 27504-27513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C., Schmid V., Schwappach B., High S. (2009). Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 122, 3605-3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Raymond C. K., Yamashiro C. T., Stevens T. H. (1991). Methods for studying the yeast vacuole. Methods Enzymol. 194, 644-661 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Marcon E., McQuire T., Arai Y., Moens P. B., Okada H. (2008). Bat3 deficiency accelerates the degradation of Hsp70-2/HspA2 during spermatogenesis. J. Cell Biol. 182, 449-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri V. R., Reiners K. S., Hansen H. P., Topolar D., Simhadri V. L., Nohroudi K., Kufer T. A., Engert A., Pogge von Strandmann E. (2008). Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS One 3, e3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Hegde R. S. (2007). Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147-1159 [DOI] [PubMed] [Google Scholar]
- Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. (2005). A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957-968 [DOI] [PubMed] [Google Scholar]
- Suloway C. J., Chartron J. W., Zaslaver M., Clemons W. M., Jr (2009). Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. USA 106, 14849-14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger A., Kamena F., Kama R., Spang A., Gerst J. E. (2005). Control of Golgi morphology and function by Sed5 t-SNARE phosphorylation. Mol. Biol. Cell 16, 4918-4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnefeld M., Grewenig A., Schnolzer M., Spring H., Knoch T. A., Gan E. C., Rommelaere J., Cziepluch C. (2006). Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp. Cell Res. 312, 2500-2514 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906 [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. (1984). Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81, 4819-4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.