Abstract
Summary: Epigenetics, the study of heritable somatic phenotypic changes not related to DNA sequence, has emerged as a critical component of the landscape of gene regulation. The epigenetic layers, such as DNA methylation, histone modifications and nuclear architecture are now being extensively studied in many cell types and disease settings. Few software tools exist to summarize and interpret these datasets. We have created a toolbox of procedures to interrogate and visualize epigenomic data (both array- and sequencing-based) and make available a software package for the cross-platform R language.
Availability: The package is freely available under LGPL from the R-Forge web site (http://repitools.r-forge.r-project.org/)
Contact: mrobinson@wehi.edu.au
1 INTRODUCTION
Epigenetics is the study of the phenotypic changes unrelated to DNA sequence. Epigenomics is the large-scale study of epigenetics, with various genome-wide assays having been introduced in the past few years and with many epigenome mapping projects on the horizon (Jones et al., 2008; Nature editorial, 2010). DNA methylation is one of the best studied epigenetic marks and can be assayed genome-wide using restriction enzyme, bisulphite or enrichment-based approaches (reviewed in Laird, 2010). Another significant class of epigenetic regulators is histone modifications, typically studied using chromatin immunoprecipitation (ChIP) in combination with microarrays (ChIP-chip) or next-generation sequencing (ChIP-seq).
There are limited general tools available for the exploratory analysis and summarization of enrichment-based epigenomics data (see Table 3 of Laird, 2010). We present Repitools, a software package for the R environment that is focused on the analysis of enrichment-based epigenomic data. Examples are shown to illustrate the diversity of tools within the package; many further examples can be found in the comprehensive user's guide. The routines have been tested on Affymetrix and Nimblegen tiling microarrays and Illumina Genome Analyzer sequencing data; generic data types are used so that other platforms can be easily supported.
2 DATA SUMMARIZATION
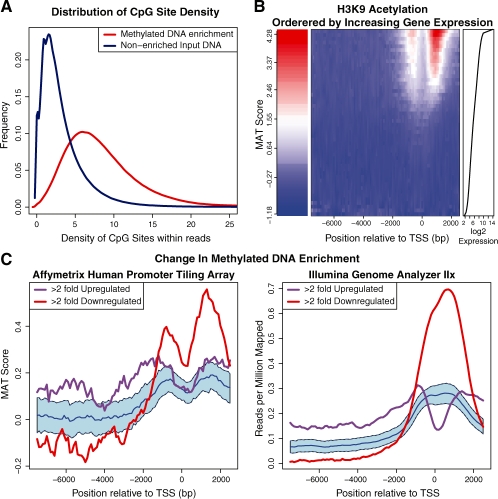
Various procedures for visualization are available within the package. For example, enrichmentPlot displays the distribution of enrichment across the whole genome for sequencing-based experiments. cpgBoxplots and cpgDensityPlot display microarray and sequencing results, respectively, for quality assessment of DNA methylation enrichment experiments. Figure 1A illustrates the cpgDensityPlot of a successful methylated DNA enrichment experiment using MethylMiner™ (Invitrogen, Carlsbad CA, USA) where, as expected, the CpG density of the enriched DNA population is heavily skewed to the right compared to the input DNA control.
Fig. 1.
Repitools visualization examples. (A) In cpgDensityPlot, each line is a single experiment's read distribution in terms of CpG density. (B) For binPlots, the middle panel displays a heatmap of summarized signal according to 50 expression level bins (rows), organized into 100 bp locations (columns) within promoters. The left panel gives the enrichment colour scale and the right panel displays the gene expression for each bin. (C) For significancePlots, the purple and red lines illustrate the median signal for the gene sets of interest. The blue line represents median signal of all remaining genes in the genome, while the blue shading illustrates a 95% confidence interval (example data taken from Coolen et al. (2010).
We have provided many ways to visualize and summarize promoter-level microarray or genome-wide epigenomic data. For example, given a table of annotation, the binPlots function summarizes median signal across points of interest (e.g. transcription start sites). We routinely use binPlots as a quality control step of new ChIP experiments where there is a previously known relationship between the interrogated chromatin mark and another metric, commonly gene expression. For example, Figure 1B clearly illustrates the positive association between gene expression levels (Affymetrix Gene 1.0 ST data) and the occurrence of H3K9 acetylation in the proximity of the corresponding promoters (Affymetrix Promoter 1.0R data). The routine handles tiling array or sequencing data as inputs, can accept alternative rankings for grouping and the display can be a plot with multiple lines, a heatmap or a 3D visualization.
Another useful strategy for summarizing sets of genes of interest is significancePlots. As illustrated in Figure 1C, significancePlots shows the distinct methylated DNA enrichment changes associated with genes whose expression is up- or down-regulated >2-fold between two samples, and how the profiles differ between array and high-throughput sequencing readout. For the comparison, a large number of random gene sets are taken to form the profile null distribution; median and confidence intervals are plotted. These plots show evidence that there is a clear enrichment of sequencing reads and hence, DNA methylation surrounding many genes are down-regulated in this comparison. Further data summaries are regularly added.
3 STATISTICAL PROCEDURES
The visualization procedures detailed above aggregate signal over a large number of promoters or regions of the genome. Often, it is of interest to focus on specific regions of the genome and summarize the signal observed at these regions (e.g. transcription start sites, exons, etc.). For example, an experimenter may be interested in promoter-level summaries of a particular epigenetic mark. The general purpose blocksStats procedure focuses on data for the specified genomic regions of interest. For microarray data, this involves the calculation of a probe-level score and applying a statistical test to the groups of probes within a specified distance from the region of interest. For sequencing data, we calculate statistics on aggregated read counts around the features of interest. Further details are available in the accompanying user's guide.
We also have procedures for untargeted analysis of epigenomic tiling array data. The regionStats function searches for a persistent change in signal in an untargeted fashion, similar in principle to model-based analysis of tiling arrays (Johnson et al., 2006), and therefore not relying upon annotation. Analogous procedures for sequencing data are in development.
4 ACCESSORY TOOLS
The package contains a number of useful tools in the spectrum of epigenomics. For example, in the context of CpG methylation, microarray probes or sequence reads are often affected by the local CpG density of the regions being interrogated. cpgDensityCalc is a procedure to calculate local CpG density according to a previous definition (Pelizzola et al., 2008). annotationLookup provides a framework for relating annotation (e.g. transcription start sites) information to probe positions on a tiling array. multiHeatmap is a general tool for creating adjacent heatmaps using separate colour scales. Additional included tools exist to access Nimblegen array quickly (e.g. readPairFile), access features of aroma.affymetrix objects (e.g. getProbePositionsDf) and aggregate sequencing reads according to proximity to annotation (e.g. annotationCounts). We expect further tools to be added and encourage others in the epigenomic community to contribute generally useful procedures.
5 DISCUSSION
There are relatively few tools currently available for the analysis of epigenomic data. We have developed Repitools, a software package for the R environment; it contains many useful functions for quality assessment, visualization, summarization and statistical analysis of epigenomics experiments. The package makes use of aroma.affymetrix and several Bioconductor packages for various preprocessing steps (Bengtsson et al., 2008; Gentleman et al., 2004) and may require an intermediate understanding of R for some features. A comprehensive user manual is available and examples can be run using supplied data. The analysis of large Affymetrix tiling array datasets is facilitated through the memory efficiency afforded by the aroma.affymetrix package (Bengtsson et al., 2008).
Funding: National Health and Medical Research Council (NH&MRC) project (427614, 481347) (M.D.R., C.S., D.S.) and Fellowship (S.J.C.), Cancer Institute NSW grants (CINSW: S.J.C., M.W.C., A.L.S.), and NBCF Program Grant (S.J.C.).
Conflict of Interest: none declared.
REFERENCES
- Bengtsson H, et al. Technical Report #745. Berkeley: Department of Statistics, University of California; 2008. aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. [Google Scholar]
- Coolen MW, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat. Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl Acad. Sci. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, et al. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Nature editorial. Time for the epigenome. Nature. 2010;463:587. doi: 10.1038/463587a. [DOI] [PubMed] [Google Scholar]
- Pelizzola M, et al. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]