Abstract
Circadian clocks organize behavior and physiology to adapt to daily environmental cycles. Genetic approaches in the fruit fly, Drosophila melanogaster, have revealed widely conserved molecular gears of these 24-h timers. Yet much less is known about how these cell-autonomous clocks confer temporal information to modulate cellular functions. Here we discuss our current knowledge of circadian clock function in Drosophila, providing an overview of the molecular underpinnings of circadian clocks. We then describe the neural network important for circadian rhythms of locomotor activity, including how these molecular clocks might influence neuronal function. Finally, we address a range of behaviors and physiological systems regulated by circadian clocks, including discussion of specific peripheral oscillators and key molecular effectors where they have been described. These studies reveal a remarkable complexity to circadian pathways in this “simple” model organism.
Keywords: peripheral clocks, pacemaker neurons, locomotor activity, feeding, mating
INTRODUCTION
The endogenous circadian system functions to organize behavior and physiology to adapt to and anticipate environmental changes in light, temperature, food, and mate availability. In addition, the circadian system temporally organizes molecular, cellular, and physiological processes relative to one another. Synergistic processes are timed to coincide, whereas mutually incompatible ones are temporally separated. The presence of circadian clocks in organisms from bacteria to mammals is evidence of their critical role in organism fitness (1).
Circadian rhythms are defined by four canonical properties. These oscillations persist, or free run, under constant conditions, indicating the presence of a self-sustaining clock. These clock-driven events recur approximately, but not precisely, every 24 h (circa = near, dia = day). These rhythms are yoked to, or entrained by, sun-driven changes in light and temperature. Although the phase of the clock can be sensitive to environmental changes, the period is remarkably stable over a wide temperature range, a phenomenon termed temperature compensation (2).
On the basis of these properties, circadian systems are organized into three main parts: the core clock, which keeps time; input pathways, which synchronize the clock to the environment; and output pathways, which transmit information to temporally organize behavior and physiology. Interestingly, these three parts are not mutually exclusive. Considerable progress had been made in our understanding of the first two components, which have been the subject of comprehensive reviews (3–5). Here we focus on output pathways to address how timing information is faithfully transmitted to control behavior and physiology.
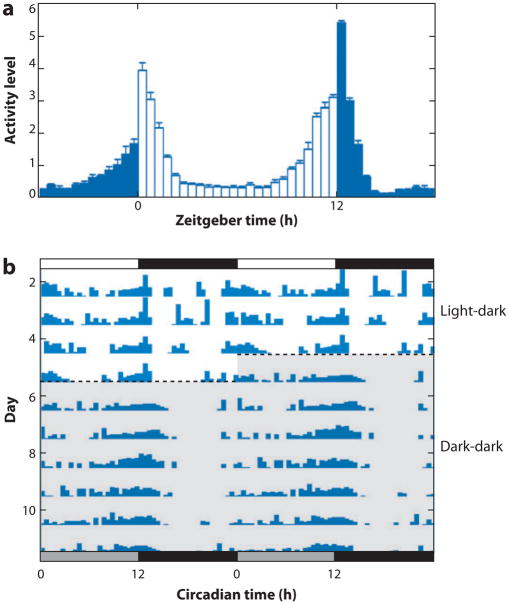
In Drosophila, circadian rhythms are typically assayed by monitoring locomotor activity using the Drosophila Activity Monitoring (DAM) system (TriKinetics; Waltham, MA). In this assay, individual flies are placed into glass tubes, and their activity measured via infrared beam breaks over several days. Under conditions of light-dark (LD) (usually alternating 12 h light and 12 h dark), flies show a morning activity peak (M) around the time of lights-on and an evening activity peak (E) around the time of lights-off (Figure 1). Flies gradually increase their locomotor activity in advance of both dark-to-light and light-to-dark transitions, a phenomenon termed anticipation. A presumed function of a circadian clock is to appropriately adjust behavior in advance of environmental changes such as sunrise or sunset. Flies also rapidly respond to these light transitions with substantial bursts of locomotor activity at both lights-on and -off transitions, termed masking because of their ability to mask underlying circadian behavior (Figure 1). To more clearly view circadian rhythms, flies are monitored in constant darkness (DD). In DD, flies display activity bouts at approximately the same time each day corresponding roughly to the evening activity peak and reflecting circadian clock function with a near-24-h period (Figure 1).
Figure 1.
Drosophila circadian locomotor behavior. (a) An averaged locomotor activity plot for wild-type flies. Light and dark bars indicate normalized activity levels during 12 h of light and 12 h of dark, respectively. Increases in activity are evident in advance of light-to-dark and dark-to-light transitions. (b) An activity plot, or actogram, for a single wild-type fly. The height of vertical bars indicates level of activity during a 30-min interval, or bin. Each horizontal line contains 48 h of activity data (double-plotted plot), with the second day of data on one line repeated on the first day of data on the following line to ease visualization of the circadian period. The transition from light-dark conditions (LD) to dark-dark conditions (DD) has been indicated by a dashed horizontal line. Under DD, activity bouts occur at the same time each day, indicating a free-running period of near 24 h.
MOLECULAR MECHANISMS OF DROSOPHILA CIRCADIAN CLOCKS
Genetic analysis of circadian behaviors, principally locomotor activity, has uncovered the transcriptional feedback loops that are central to biological timing. These feedback circuits consist of sequence-specific DNA binding proteins that stimulate transcription of their own repressors. Temporal delays between activation and repression generate 24-h oscillations in steady-state levels of many clock gene transcripts. Due to reference limitations, we refer the reader to recent reviews covering these topics in detail (3, 5, 6).
Briefly, the DNA-binding heterodimer CLOCK/CYCLE (CLK/CYC) binds to E-box sequences (usually CACGTG) in target promoters to activate gene expression (Figure 2). In the principal feedback loop, CLK/CYC activates transcription of period ( per) and timeless (tim), peaking during the late day. (By convention, proteins are indicated in all CAPS, whereas genes or transcripts are indicated in italics.) PER and TIM proteins accumulate and dimerize in the cytoplasm (early night), translocate to the nucleus (midnight), and bind CLK/CYC, inhibiting their DNA binding (7) and thus their transcriptional activation function (late night). Of note, PER and TIM may dissociate prior to entering the nucleus (8). Transcriptional feedback is an important determinant of circadian period (9). Importantly, similar feedback loops using similar components operate in mammalian clocks (1).
Figure 2.
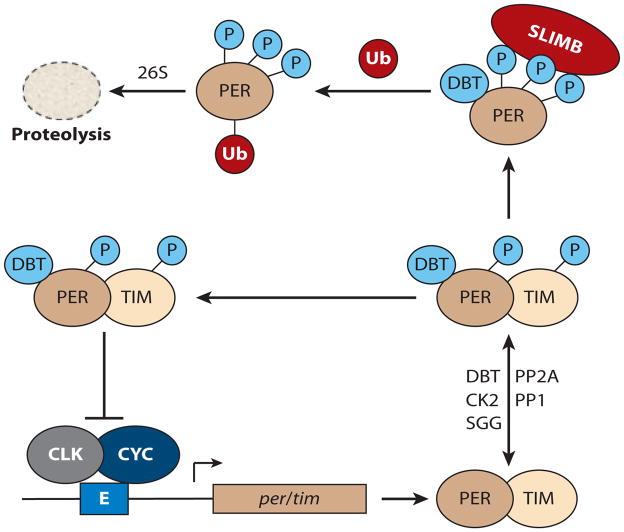
The core molecular clock in Drosophila. CLOCK/CYCLE (CLK/CYC) bind to E-box elements (E) contained in the promoters of period ( per) and timeless (tim). PER and TIM proteins are modified by the kinases DOUBLETIME (DBT), CASEIN KINASE 2 (CK2), and SHAGGY (SGG) and the phosphatases PROTEIN PHOSPHATASE 2A (PP2A) and PROTEIN PHOSPHATASE 1 (PP1). PER and TIM dimerize and transition to the nucleus, where they repress CLK/CYC activity. Phosphorylated PER and TIM also bind the E3 ubiquitin ligase SUPERNUMERARY LIMBS (SLIMB), which leads to ubiquitination and ultimately proteolysis by the 26S proteasome.
Posttranslational regulation of the inhibitory components imposes temporal delays between CLK/CYC transcriptional activation (late day) and PER/TIM repression (late night) (Figure 2). Temporal delays between activation and inhibition result in daily oscillations of CLK/CYC target transcription. PER, TIM, and CLK exhibit rhythms of phosphorylation state, with peak phosphorylation peaking during the late night/early day. PER is phosphorylated by CASEIN KINASE Iε (CK1ε)/DOUBLETIME (DBT) and CASEIN KINASE 2 (CK2). Phosphorylation by DBT and CK2 enhances PER repressor activity (10). TIM is phosphorylated by GLYCOGEN SYN-THASE KINASE 3β (GSK3β)/SHAGGY (SGG) and CK2 (11). Phosphorylated PER and TIM are substrates for the phosphatases PROTEIN PHOSPHATASE 2A (PP2A) and PROTEIN PHOSPHATASE 1 (PP1), respectively (12). Of note, one transcript isoform encoding a regulatory subunit of PP2A, twins, is under circadian regulation, suggesting a mechanism by which PP2A may direct PER phosphorylation rhythms. A nuclear PER-DBT complex also associates with and phosphorylates CLK, triggering its degradation and thus providing a mechanism for feedback inhibition (7, 13–15). Therefore, PER repression of CLK is likely mediated by the delivery of DBT to CLK, leading to its phosphorylation and reducing its DNA binding and half-life. Peak phosphorylation precedes protein disappearance, linking phosphorylation to PER/TIM/CLK protein half-life. The E3 ubiquitin ligase SUPERNUMERARY LIMBS (SLIMB) selectively associates with and ubiquitinates (at least) DBT-phosphorylated PER (16), triggering its degradation by the ubiquitin-proteasome pathway during the early day. The loss of PER relieves repression of CLK/CYC, permitting a new cycle of transcriptional activation.
To synchronize the internal clocks to the 24-h cycle of sunlight, Drosophila utilize the cell-autonomous blue-light photoreceptor CRYPTOCHROME (CRY) (3). Although we provide an overview here, we refer the reader to recent reviews on this subject for more details (3, 6). Light activates CRY, which then binds TIM, targeting it for degradation. Light-induced TIM degradation, in turn, explains how the clock responds to short light pulses. During the early night when TIM is accumulating, light-induced TIM degradation leads to a delay in TIM accumulation, explaining light-induced phase delays. In contrast, light pulses delivered during the late night advance TIM disappearance and explain light-induced phase advances. In addition to the function of CRY as a photoreceptor, CRY may also sustain peripheral clocks (17), perhaps by playing a role as a transcriptional repressor (18), and mediate the clock response to temperature (19). Besides CRY, Drosophila also employs a number of other light-sensing pathways, including the retina, ocelli, and the Hofbauer-Buchner eyelets, all of which convey environmental LD information to synchronize molecular clocks (3).
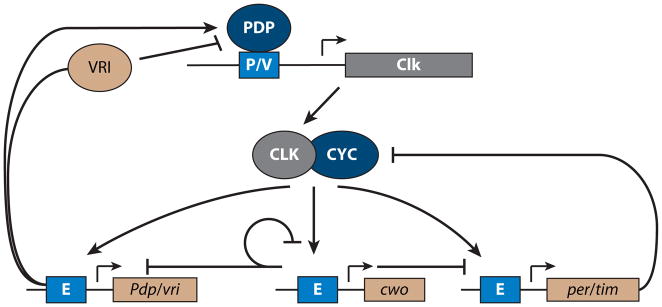
CLK/CYC also activate at least two other interdependent feedback loops that may be important for the phase and amplitude of core oscillators and rhythmic outputs (Figure 3). CLK/CYC directly activate the transcription of the basic zipper (bZip) activator Par domain protein 1 (Pdp1) and the bZip transcriptional repressor vrille (vri ), which, in turn, alternately activate and repress transcription of the Clk gene as well as cry (20). As a result of delayed accumulation of the PDP1 activator relative to the VRI repressor (20), Clk and cry transcript rhythms are antiphase to those of per/tim (they peak in the early day versus early night). The function of the second loop is unclear; altering the phase of Clk transcription has little effect on behavioral period or phase. Nonetheless, loss-of-function Pdp1 mutations, including one that selectively affects the rhythmic ε isoform, result in reduced Clk expression and, in the latter case, arrhythmic behavior (20, 21). CLK/CYC also activate a bHLH repressor, clockwork orange (cwo). CWO specifically binds the CLK/CYC target E-box and represses CLK/CYC-mediated transcription (22–24). In addition, CWO may also act as an activator (25). Loss of cwo results in low-amplitude, long-period molecular and behavioral rhythms, suggesting a role of cwo in sustaining robust rhythmicity (22–25). The mammalian homologs of cwo, Dec1 and Dec2, also appear to have clock functions and a role in regulating sleep amount (26, 27). Although the utility of multiple loops remains ambiguous, this intricate architecture highlights Clk as a focal point for feedback regulation. Indeed, ectopic Clk expression is sufficient to induce molecular circadian rhythms (28).
Figure 3.
Additional feedback loops in the Drosophila molecular clockwork. In addition to the principal PERIOD/TIMELESS (PER/TIM) feedback loop, CLOCK/CYCLE (CLK/CYC) also activate CLOCKWORK ORANGE (CWO), PAR DOMAIN PROTEIN 1 (PDP1), and VRILLE (VRI). CWO feeds back to repress CLK/CYC activation by binding to E-box elements (E). PDP1 activates the transcription of the Clk gene, whereas VRI competes with PDP1 binding to repress Clk expression. P/V domain denotes PDP/VRI-binding sites.
A salient aspect of fruit fly circadian timekeeping is the fact that most features of the fruit fly’s molecular clockwork are conserved in mammals, including humans. For example, individuals affected with familial advanced sleep phase syndrome (FASPS) exhibit an advanced phase of sleep-wake rhythms and shortened circadian period that is inherited in a Mendelian dominant manner. Mutations in the human PER2 and CK1δ genes, both orthologs of fly circadian genes per and Dbt, respectively, are responsible for this advanced sleep phase (29, 30). These data argue that the basic architecture and core components of circadian clocks can be traced back to the shared ancestor of flies and humans hundreds of millions of years ago.
A CIRCADIAN PACEMAKER NEURAL NETWORK MEDIATES DISTINCT ASPECTS OF LOCOMOTOR BEHAVIOR
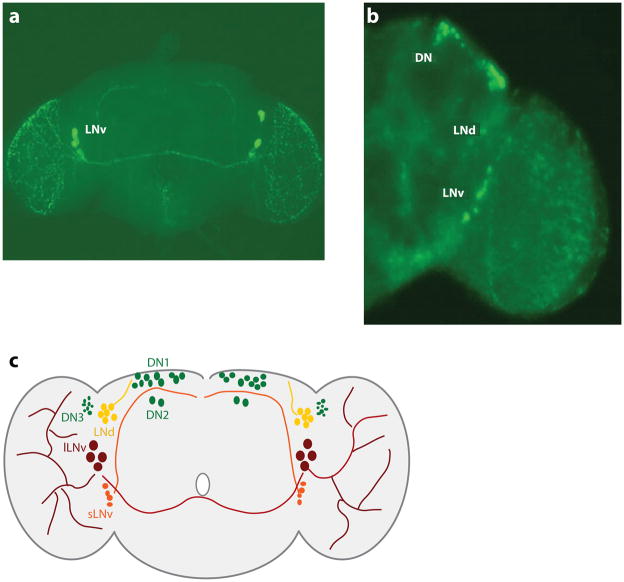
Although fly genetics has been instrumental in improving our understanding of molecular mechanisms of circadian clock function, genetic approaches have also led to remarkable advances in revealing the network architecture important for circadian behavior. Molecular clocks are located in only ~150 neurons (out of ~250,000) in the Drosophila brain (Figure 4). These neurons are part of a dedicated neural network that drives circadian behavior (please see reviews in References 3, 4, and 6). Distinct subsets of these 150 neurons serve different aspects of circadian behavior, particularly driving wakefulness/locomotor activity at different times of day and under different environmental conditions. These 150 neurons can be divided into the small and large ventral lateral neurons (sLNv and lLNv, respectively), the dorsal lateral neurons (LNd), three groups of dorsal neurons (DN1, DN2, and DN3) and the lateral posterior neurons (LPN) (3). The LNv are especially important for morning anticipation and so are termed morning (M) cells, whereas the LNd/DN1 underlie evening anticipation and thus are termed evening (E) cells. The circadian pacemaker network may arguably be the most well-studied and well-understood CNS network governing behavior in Drosophila.
Figure 4.
The circadian pacemaker network in adult Drosophila brains. (a) Immunofluorescence of the circadian neuropeptide PIGMENT-DISPERSING FACTOR (PDF) illustrates the divergent projections stemming from ventral lateral neurons to other neural loci in the central nervous system. The small ventral lateral neurons (sLNv) send projections dorsally (toward the top of the image), whereas the large ventral lateral neurons (lLNv) send projections contralaterally and extensively into the optic lobes (on the right and left of the image). (b) tim in-situ hybridizations reveal the spatial orientation of pacemaker neurons in the central nervous system. Labels denote the location of the ventral lateral neuron (LNv), dorsal lateral neuron (LNd), and dorsal neuron (DN) clusters. Images in both panels a and b were provided by Ela Kula-Eversole. (c) Schematic of the Drosophila circadian pacemaker network. sLNv (orange) send projections dorsally. lLNv (maroon) send projections contralaterally and into the optic lobes (left and right sides of the schematic). LNd are indicated in yellow, and three groups of dorsal neurons are indicated in green.
The neuropeptide PIGMENT-DISPERSING FACTOR (PDF) is the critical effector for the ~20 LNv. PDF expression is largely restricted to these neurons in the adult brain. The small subgroup (sLNv) sends projections dorsally toward the DN1 and a number of putative downstream circuits, including the pars intercerebralis (PI), a cluster of neuroendocrine cells functionally and developmentally analogous to the mammalian hypothalamus, and the mushroom bodies (MB), analogous to the mammalian hippocampus and a critical regulator of fly sleep-wake (31, 32). Rhythms in the dorsal sLNv terminal arborizations and the PDF neuropeptide levels are evident, although pdf RNA levels do not show similar fluctuations (33, 34). Notably, PDF rhythms are not required for robust circadian behavior (35). Ablation of PDF neurons as well as genetic loss of pdf ( pdf 01) or its receptor, pdfr, both result in a loss of morning anticipation and a phase advance of the evening peak in LD and the first day of DD (36–39). These behavioral rhythms slowly damp in DD. The loss of behavioral rhythms is accompanied by reduced amplitude and desynchronized or complete loss of molecular rhythms within clock neurons. Thus, PDF is released from the LNv but also feeds back to impact the clocks in both PDF and non-PDF neurons. Bath application of PDF activates cyclic AMP synthesis in most groups of clock neurons (40). A major functional target of the PDF neurons appears to be the ~30 evening cells (LNd/DN1), mediating PDF effects on morning and evening behavior in LD as well as circadian period (41, 42). PDF may actually both speed and slow target clocks (42, 43). MAPK activation in the dorsal brain may in part mediate these effects (44).
The large subset of LNv (lLNv) makes extensive projections into the optic lobes, a site of visual input, and contralaterally toward the LNv and optic lobe (36, 45, 46). These optic lobe projections may also be important for rhythms observed in visual input circuits (47). The lLNv are also important loci for sleep/arousal (48–51) and may mediate the increase in sleep observed in flies raised socially (52). Furthermore, these PDF varicosities in the optic lobe are also sensitive to social experience and sleep loss (52).
The sLNv appear to be especially important for driving rhythms under constant darkness conditions. Overexpression of sgg can speed up the molecular clock. Selective expression in the LNv results in speeding up the clock in the sLNv and also in the LNd, DN1, and DN3 neurons as well as the behavioral rhythm (53). These results suggest that the sLNv are especially important for driving free-running (DD) rhythms. The evening cells (LNd and/or subsets of DNs) are relevant to determining the phase (rather than the period) of DD behavior (53). Under constant light conditions, rhythms in wild-type flies damp. However, in mutants of the cry photoreceptor or in flies overexpressing per, rhythms persist. In these flies, subsets of evening cells (rather than morning cells) apparently drive behavioral rhythms in LL (54, 55).
Although LL and DD are clearly unnatural environmental conditions, circadian clocks are crucially important for adaptive seasonal responses to the long and short days of summer and winter, respectively. Under long days, the evening clocks appear to dictate the phase of morning clocks (56). In contrast, under short days, the morning clocks dictate the phase of evening clocks (56). One possible explanation is that light activates the output of evening cells while inhibiting output of the morning cells (54). In addition, reduced temperature can phase-advance behavior and molecular rhythms by thermosensitive splicing of a per 3′ UTR intron, and natural variation in these splice sites between different drosophilids may underlie their adaptation to different thermal environments (57). Thus, the coupled morning and evening oscillators can respond to seasonal changes in photoperiod and temperature to time the activity of the morning and evening oscillators. Under short days/cool temperatures, Drosophila can enter into diapause to improve survival. Naturally occurring variation at the tim locus that alters interactions with cry appears to be important for the tendency to enter this state (58, 59). These data suggest a role for clock genes in seasonal adaptation.
In addition to the pacemaker neural network, associated brain glial clocks are also important contributors to locomotor activity rhythms. Subsets of glia rhythmically express PER (60). Additionally, per mosaic analysis revealed that PER-expressing glial cells are sufficient to establish weak behavioral rhythms (60). The close proximity of PER+ glial cells with respect to LN suggests a functional relationship between glia and pacemaker cells in mediating locomotor rhythms (45, 61). Below we discuss the role of the output gene ebony in mediating glial clock function in circadian behavior.
CIRCADIAN CLOCKS COMMUNICATE TEMPORAL INFORMATION THROUGH THE RHYTHMIC EXPRESSION AND FUNCTION OF CLOCK-CONTROLLED GENES
Interdependent transcriptional feedback loops act as the internal gears for circadian clocks. To regulate the myriad rhythmic processes, these core timers must convey time information to output pathways. One mechanism by which temporal information is transmitted is through the rhythmic transcription of clock-controlled genes (CCGs). To identify such genes, Drosophila, as well as a number of other organisms, has been subjected to genome-wide gene expression analyses using DNA microarrays.
Researchers reported hundreds of rhythmically expressed genes representing a wide range of predicted gene functions (62–68). In addition, the temporal expression profiles exhibited by these CCGs are well distributed across circadian times, suggesting that they may mediate time-specific biochemical and cellular functions throughout the day. The limited overlap among these reports has led to reanalysis of the existing data to extract more robust datasets with some success (67, 69). The function of these rhythmically expressed genes in temporally modulated behaviors has been established for only a handful of genes, such as cwo. One possibility is that rhythmic genes have been identified from whole heads where the majority of clock gene expression can be attributed to the eye, whereas locomotor rhythms are driven by a relatively small number of neurons and glia.
Output genes have also been identified using other approaches, including other types of screens for clock-controlled transcripts and functional analysis of candidate genes. Some of these genes do not exhibit rhythmic changes in steady-state transcript levels. Some of these genes are rhythmic at the protein level, and others have no evident gene expression rhythm but may display a rhythm at a functional level (44, 65, 70–76). The diversity of genes and mechanisms suggests multiple pathways for molecular clocks to drive cellular rhythms.
Although a large number of tentatively rhythmic genes have been identified, only a small number have been confirmed as rhythmic, and even fewer have been shown to be functionally important in rhythmic behaviors. Of these genes, a number are not acting purely as output genes, such as Pdp1, vri, and cwo, which feedback and influence various aspects of Clk function. The number of genes with principal output roles is relatively small. Although Pdp1 is important for activating Clk levels in larval and adult pacemaker neurons (20, 21), additional data reveal a role downstream of core oscillators in regulating circadian behavior (21, 76). For example, Pdp1 RNAi knockdown or dominant-negative expression in circadian neurons results in reduced behavioral rhythmicity without significant molecular changes in the core clock (76, 77). Rescue of CLK expression in a Pdp1ε mutant restores core clock function but not behavioral rhythms. These studies strongly suggest that Pdp1ε rhythms are important for both sustaining CLK levels and independently regulating circadian output. Thus, transcriptional cascades may be a defining architecture for circadian output pathways.
In the circadian neural network, ion channel genes may be important transcriptional targets of these output pathways. Electrophysiological studies in the LNv, which are relatively difficult due to the small size of Drosophila neurons, have revealed that spontaneous activity peaks and resting membrane potential are more depolarized in the early morning (78, 79). The ion channel SLOWPOKE (SLO), a calcium-sensitive potassium channel, and its putative regulator Slowpoke Binding Protein (SLOB) are rhythmically expressed at the RNA and protein levels (65, 80–82). slo mutants display circadian behavioral changes with limited effects on molecular rhythms, suggesting an output role (65, 80). Of note, a mammalian SLO homolog is also rhythmically regulated and important for clock output (83). The precise site of slo function in the Drosophila pacemaker network or elsewhere remains unknown.
Other ion channels have also been implicated in circadian behavior, although it is not yet clear if they are clock-regulated. The NARROW ABDOMEN (NA) channel encodes a likely cation leak current that may be activated by neuropeptide/neurotransmitter receptors on the basis of the function of its highly conserved mammalian homolog NALCN (84, 85). na mutants display poor circadian rhythms in the absence of strong effects on core molecular clocks (73). Importantly, these effects can be rescued by na expression in pacemaker neurons (73). na mutants display elevated PDF levels in dorsal terminals, suggesting a role in coupling molecular clocks to neuronal outputs (73). Similarly, using a membrane-tethered spider toxin, investigators demonstrated a role for voltage-gated sodium channels in mediating clock effects on PDF (86). The SHAW potassium leak channel also appears to play an important role in pacemaker neuron function (87). In addition to conveying circadian timing to neuropeptide release, changes in membrane excitability may also feedback onto the molecular clock (88).
Cyclic AMP–regulated transcription is also rhythmic and has been implicated as a regulator of core clocks and outputs. Cyclic AMP levels and cyclic AMP response element (CRE)-driven transcription are both rhythmic (89, 90). Mutations of dCREB2 reduce CRE-driven transcription and alter the amplitude and/or period of molecular and behavioral rhythms, suggesting a contribution to core clock rhythms (89). Endocytic pathways may also regulate circadian period through protein kinase A (PKA) (91). Genetic loss of a catalytic subunit of PKA results in locomotor activity arrhythmicity with intact eclosion rhythms, suggesting a role in circadian output (70). Similarly, genetically mediated up- or downregulation of the cyclic AMP pathway alters sleep homeostasis independently of effects on circadian rhythms, implying a role in sleep/wake output pathways (92). These studies support a role of cyclic AMP signaling in circadian output, but the precise cellular and molecular mechanisms remain to be determined.
A molecular effector of glial function in circadian rhythms is the beta-alanyl conjugase synthetase ebony. ebony transcript and protein are expressed with a circadian rhythm including in PER-expressing glia (61, 62). ebony mutants exhibit poor locomotor activity rhythms, yet eclosion rhythms are intact, suggesting a specific output role (93). Glial ebony expression is sufficient to rescue ebony rhythm phenotypes (61). Rhythmic ebony production within glia may regulate dopaminergic circuits responsible for locomotor activity (61). Mutations in ebony also suppress the hyperactivity phenotypes observed in dopamine transporter mutants, suggesting a role in mediating dopaminergic neurotransmission (61). Of note, rhythmic ebony expression does not appear to be required for rhythms, although ebony activity may be rhythmic. Nonetheless, ebony represents an important output effector for glial rhythms and circadian behavior.
Considerable efforts have been devoted to understanding circadian locomotor activity rhythms in Drosophila. Genetic analysis has yielded remarkable insights into the gears of the core molecular clock and how these clocks are synchronized to the environments. Despite the application of genetic and genomic techniques, few bona fide output genes with in vivo circadian functions that regulate locomotor rhythms have been described. One possibility is that, given the multitude of rhythmic genes, obtaining single-gene mutants with strong phenotypes may be difficult to obtain and/or the rhythm of a single gene may not be required for rhythmic behaviors. In these cases, more detailed behavioral or physiological analyses may be necessary to tease out the roles of potential output genes.
CIRCADIAN REGULATION OF OTHER BEHAVIORS AND PHYSIOLOGY
Because of the automated nature of the DAM assay, most Drosophila chronobiologists have focused on locomotor activity rhythms. Here we detail an ever-expanding catalog of other behavioral and physiological parameters that are temporally organized by circadian clocks. These rhythms can be roughly divided into those largely governed by the circadian pacemaker network or associated glia in the central brain and those timed by peripheral clocks. These peripheral clocks, both non-CNS neural (largely sensory) and nonneural clocks, largely express the same PER-based core molecular program present in CNS neurons and regulate a host of rhythmic processes. Nonetheless, comparison of rhythmically expressed genes in head and body suggests tissue-specific control of circadian outputs (65). In many cases, these clocks demonstrate rhythmic expression that can persist as explanted cells using circadian bioluminescent reporters (94, 95). Although there is evidence that central brain pacemaker neurons can drive the rhythmic release of humoral factors (96), as is observed in mammals, whether these factors synchronize or entrain peripheral clocks is uncertain. Instead, peripheral clocks are primarily synchronized to the external LD cycle through the cell-autonomous expression and action of the blue-light photoreceptor CRY (3).
Sleep
Like circadian behavior, sleep in Drosophila is also monitored using the DAM system (97). Five minutes of continuous inactivity, i.e., no beam breaks, is conventionally defined as sleep. During sleep, flies exhibit reduced activity/movement and reduced responsiveness to sensory stimulation. In addition, sleep deprivation results in a subsequent increase in sleep amount and intensity, indicating homeostatic regulation. Pharmacological and genetic evidence suggests that the underlying mechanisms of Drosophila sleep are similar to those in mammals (97).
As in other animals, sleep is also under circadian control (98, 99). Flies bearing dominant-negative or null alleles of circadian transcriptional activators Clk and cyc exhibit less overall sleep than do wild-type flies (100). The molecular and cellular basis of these mutant effects is not yet known. As in rats, prolonged sleep deprivation (days) results in death (101, 102). cyc mutants, but not other circadian clock mutants, are hypersensitive to these lethal effects, and the heat shock stress pathway partially mediates this hypersensitivity (102). Nonetheless, this finding likely reflects a nonclock role for cyc.
Studies suggest that the large subset of the LNv circadian pacemaker neurons promotes arousal. Activation of the lLNv by selective channel expression disrupts nighttime sleep, whereas ablation increases sleep (48–50). GABAergic inputs inhibit the arousal-promoting function of these neurons (49, 51), yet the source of these inputs remains unknown. Preliminary data suggest that non-PDF evening cells may oppose the effects of the lLNv (50). Future work will be necessary to fully understand the role of circadian neurons (e.g., non-PDF neurons) and their molecular effectors in sleep-wake regulation.
Behavioral Responses to Light: Diurnality, Masking, and Phototaxis
Most animals live in rhythmic environments governed by daily cycles of sunlight, which can have direct effects on behavior, i.e., masking effects. The term masking is derived from the observation that behavioral effects of light can obscure underlying circadian rhythms (103). These responses are critical in determining whether an animal is primarily day-active (diurnal) or night-active (nocturnal), which has important implications for the evolution of its sensory systems, the nature of its metabolism, and its choice of mates for reproduction.
Drosophilae respond to the lights-on signal with an increase in activity consistent with their diurnal nature but paradoxically respond to a lights-off signal also with an increase in activity (Figure 1). These masking responses do not require a functioning circadian system (104). Nonetheless, the magnitude of the lights-on response exhibits daily oscillations (105). Flies display a reduced locomotor response to light during the subjective day in constant darkness and an enhanced response during the subjective night (105). Interestingly, whereas per01 and tim01 mutants retain normal behavioral responses to light, Clk and cyc mutants display a reduced or absent response to lights-on but an intact response to lights-off (106–108). In addition, mutants of the na ion channel, important for circadian output of pacemaker neurons, also phenocopy Clk and cyc mutants (109). These results suggest an intimate connection between behavioral light response and the circadian clock program.
Compound eyes are cellular candidates for mediating masking rhythms. Eyes consist of 800 units termed ommatidia, each containing eight rhodopsin-expressing photoreceptor neurons (R1–R8). Each photoreceptor neuron houses a functional circadian clock (60, 110). In fact, these cells account for the majority of molecular clock gene expression in the adult head (111). Rhythms in visual pigment and in electroretinogram sensitivity have been noted as potential circadian outputs (112), yet the role of the compound eyes remains unclear (104, 113, 114). Regardless, suppression of clock function in the visual system did not abolish the masking rhythm, suggesting that clock function in the brain, perhaps in circadian pacemaker neurons, may be sufficient (105). The precise cellular and molecular basis of these photic response rhythms remains to be determined.
Another behavioral light response under circadian regulation is larval phototaxis. Drosophila larvae are negatively phototactic, i.e., they avoid light. This behavior is under circadian regulation, peaking during the late night/early morning (115). Mutants affecting the positive limb of the transcriptional feedback loop (i.e., Clock and cycle mutants) are highly photophobic, whereas mutants affecting the negative limb of the transcriptional feedback loop (i.e., period and timeless mutants) are less responsive to light (115). The ecological purpose of this rhythm is unclear. Not surprisingly, how flies respond to light is tuned to circadian time.
Learning and Memory
Fruit flies exhibit many forms of learning and memory, including visual, olfactory, spatial, and courtship conditioning (116). In aversive olfactory memory, flies are trained to discriminate between two odors after one has been paired with an electric shock punishment. A single aversive training trial can induce a short-term memory (STM) that lasts for minutes after training. The ability of flies to form short-term associative memories appears to be under the influence of the circadian clock, with peak performance occurring during the early nighttime under both LD and DD conditions (117). In both per01 and tim01 mutants, the circadian modulation of STM was abolished (117). The molecular and cellular bases of these rhythms and whether they extend to other forms of memory remain unknown.
Eclosion
The first behavior used to monitor Drosophila circadian rhythms is also the first adult fly behavior: emergence from the pupal case, or eclosion. Adult flies have a tendency to emerge from their pupal casing during the early morning hours when the environment is moist, as newly emerged flies lose water more rapidly than mature flies and wing expansion can fail when conditions are too dry (2). As an individual ecloses only once in its lifetime, circadian eclosion rhythms are measured using population assays.
A key mediator of circadian regulation of eclosion rhythms is a peripheral clock in the prothoracic glands (PGs). In Drosophila, the PG assesses the growth and size of the developing pupae to determine when they are ready for eclosion. Indeed, rhythmic oscillations of both PER and TIM in LD and DD are evident in the PG just prior to eclosion (94, 118, 119). Interestingly, disruption of the PG molecular clock by tim overexpression degraded normal eclosion rhythms, whereas tim rescue in neurons, including pacemaker neurons, failed to rescue eclosion rhythms (119). Furthermore, ablation of PDF+-expressing lateral neurons disrupted the PG clock and rhythmic eclosion (119). Taken together, these results suggest a hierarchical relationship similar to that observed in mammals in which peripheral clocks are synchronized or driven by central pacemaker neurons.
Circadian eclosion rhythms may also involve crustacean cardioactive peptide (CCAP) neurons. These neurons are important, but are not required, for eclosion rhythms (120). In addition, they rhythmically express an RNA-binding protein, LARK (121), which is important for timing eclosion without altering the core clock (122). LARK is rhythmic at the protein level, but not at the RNA level, suggesting posttranscriptional control of output (123). A mammalian homolog of lark appears to regulate Period1 posttranscriptionally (124). These studies suggest that CCAP neurons/LARK are part of a clock-driven circuit important for rhythmic eclosion. LARK may also cooperate with the RNA-binding protein FMR (Fragile X mental retardation) protein (125), which is required for robust locomotor activity rhythms in both flies and mammals (74, 75, 126, 127).
Feeding and Metabolism
Similar to observations in rodents, circadian clocks in flies help coordinate rhythmic feeding behavior and regulate proper energy consumption and metabolism. Food consumption in Drosophila consistently occurs at specific times of the day (primarily during the morning), and this rhythmic behavior persists under constant darkness (128).
Several feeding- and metabolism-related parts of the fly harbor circadian clocks. These parts include the fat bodies (the fly homolog of fat, liver, and the immune system), involved in fuel storage and energy balance; the antennae and maxillary palp, involved in food/odor detection; proboscis (the fly mouth part), involved in taste and feeding; and the gastrointestinal tract, involved in digestion and nutrient absorption. The fat body clock is an important contributor to metabolism and feeding. Targeted disruption of clock function in the fat body results in reduced glycogen storage, increased sensitivity to starvation, and elevated food consumption (128). A key molecular mediator of fat body function is the rhythmically expressed ligand carrier protein takeout (to) (129). to mutants also display increased food intake and starvation sensitivity (72, 130).
Chemosensation
Possible contributors to feeding rhythms are rhythms in odor detection. The antennae and maxillary palps enable the fly to detect chemical odorant stimuli from its surroundings. Small hairs that encompass much of the surface area of these organs, called sensilla, contain olfactory receptor neurons that allow chemical signals to be transformed into an electrical output. Electroantennogram (EAG) responses, electro-physiological assessments of antennal neuron responses to chemical odorants, cycle under LD and DD conditions, with a peak response during night (131). Interestingly, unlike behavioral rhythms, this DD antennal rhythm depends on CRY, suggesting a core clock function in the periphery for this photoreceptor (17).
Several pieces of evidence suggest that clock function in antennal neurons can act independently from pacemaker neurons to regulate rhythmic olfactory function. Through use of a transgenic per line in which PER cycling is retained only in pacemaker cells and not in peripheral cells, rhythmic olfactory response was abolished, even though flies retained rhythmic locomotor behavior (131), thereby suggesting a clear dissociation between locomotor and olfactory rhythms. Additionally, EAG response rhythms were disrupted when a dominant-negative Clk or cyc transgene was overexpressed exclusively within antennal neurons (132). Rescue of cyc expression in antennal neurons restored rhythmic EAG amplitude (132). Thus, intact molecular clocks within the antennal neurons are both sufficient and required for rhythmic behavioral output and can function independently from pacemaker cells. Of note, the protein kinase G protein–coupled receptor kinase 2 appears to be an important mediator of these rhythms (133). Nonetheless, it is not yet clear how these rhythms contribute to feeding or other behaviors.
Courtship and Mating
Fruit flies display a highly stereotyped courtship ritual. Courtship and mating behavior is driven by circadian clocks and depends on clock genes (134–136). This courtship/mating rhythm is monitored by insemination (134, 136) or close proximity of males and females, the latter likely a reflection of courtship behavior (135). Increased mating/courtship are not a mere consequence of increased locomotor activity rhythms, as the trough of the mating rhythm around dusk corresponds to the evening peak of locomotor activity (134, 135). Depending on the assay used, this rhythm requires rhythmic females (134) or rhythmic males (135).
A number of distinct clocks are important for mating rhythms. Expression of a neurotoxic polyglutamine-expanded protein in tim-expressing cells, presumably including neurons and fat body cells, affects courtship behavior (137). Neither rescue of locomotor activity rhythms by per rescue in the brain LN nor selective rescue of olfactory rhythms was sufficient to rescue the close-proximity rhythm (135). The fat body is also an important contributor to male courtship behavior. to is expressed in a sex-specific manner in the fat body (138). Feminization of male to-expressing cells through expression of the female-specific sex determination gene transformer causes reduced male courtship behavior (138, 139). The presence of TO protein in the hemolymph, the fly’s circulatory system, suggests that the fat bodies may coordinate reproductive behaviors with the brain by means of TO signaling (139).
Another potential contributor to male courtship rhythms is the cytochrome P450 family member sxe1. sxe1 is much more strongly expressed in male flies than in female flies (140). Its transcript and protein are under circadian clock control, with protein levels peaking during the night (141). RNAi-mediated knockdown and disruption of sxe1 reduced male courtship levels but did not abolish circadian regulation (141). sxe1 is expressed in nonneuronal cells in association with sensory bristles, including those involved in chemosensation and potentially important for courtship behavior (141). sxe1 may also be expressed in the fat body (140). Although the focus has been on male-driven reproduction-related rhythms, females also display a circadian rhythm in egg-laying behavior (142).
Immunity
Fruit flies deploy a highly conserved innate immune system to protect themselves against pathogens. Microarray studies initially identified rhythms in innate immunity genes (63, 65). Flies tend to be more susceptible to bacterial infections during the early daytime hours than during the nighttime (143). Loss-of-function per01 mutant flies appear to be immunodeficient, exhibiting increased mortality rates following bacterial infection, regardless of the time of infection (143, 144). Other clock mutants display infection resistance. Additionally, antimicrobial peptides (AMPs), namely Peptidoglycan recognition protein SA and drosocin, exhibit a clock-dependent increase in expression immediately following bacterial infection that is greater during the nighttime than during the daytime (143). An important source of AMPs is the fat body. However, it is not yet clear if the fat body clock or other cellular clocks are responsible for immune rhythms.
CONCLUSIONS AND FUTURE DIRECTIONS
The list of rhythms that flies display is ever-expanding. Some rhythms that we did not have space to discuss include those in cuticle deposition (145), in susceptibility to oxidative stress (146, 147), and in synaptic bouton size at the neuromuscular junction (148). Although the cellular basis of these rhythms is also unclear, they are likely driven by peripheral clocks as well. In addition, clocks are present in a range of organs beyond those mentioned above, such as the Malpighian tubules, which serves a similar function as do the kidneys (149). The larger point is that, although the focus has been on locomotor activity rhythms, Drosophila exhibits a rich repertoire of circadian rhythms reflecting the diversity of its physiological systems. The application of genetics, which has been so successful in elucidating the core components of molecular clocks, will be valuable in understanding the functional significance of peripheral clocks as well as how these clocks convey timing information to orchestrate a multitude of cellular processes important for circadian rhythms. Given the highly conserved nature of animal circadian systems, the fly should continue to provide insights into our own circadian biology.
SUMMARY POINTS.
Molecular genetics in Drosophila has revealed transcriptional feedback loops and rhythmic phosphorylation as key timekeeping mechanisms of circadian clocks.
An elaborate interconnected network of just 150 neurons encodes temporal information to mediate circadian locomotor activity.
Microarray and candidate approaches have been used to define rhythmically expressed genes that potentially transmit temporal information to control cellular outputs. Thus far, only a few rhythmic genes have been shown to be functionally important for circadian output.
Flies display a wide range of circadian rhythms in behavior and physiology, including sleep, responses to light, learning and memory, eclosion, feeding, chemosensation, mating, and immunity. Peripheral clocks, sometimes independently of brain clocks, play an important role in many of these rhythms.
FUTURE ISSUES.
How do molecular clocks transmit temporal information to regulate cellular output, physiology, and behavior?
What is the functional significance of peripheral clocks?
What is the cellular and molecular basis of rhythms beyond locomotor activity?
Acknowledgments
We apologize to those whose primary work we were unable to cite due to space and reference limitations. R.A. is supported by NIH R01NS059042 and R01NS052903. B.Y.C. is supported by F31NS062551. We thank Valerie Kilman for helpful comments and Ela Kula-Eversole and Valerie Kilman for providing images in Figure 4.
- Circadian
a rhythmic biological process that recurs approximately every 24 h (from Latin: circa = near, dia = day)
- Free run
when a biological rhythm persists in the absence of external time cues or does not synchronize to external time cues
- Entrainment
the synchronization of a rhythmic biological process to external (light-dark cycles) or internal (a circadian pacemaker) oscillations
- DAM system
Drosophila Activity Monitoring system
- LD
light-dark
- Anticipation
a change in behavior or physiology occurring in advance of an environmental transition
- Masking
when an external factor (e.g., light) acutely affects behavior or physiology in a clock-independent manner, interfering with the observation of underlying rhythms (e.g., locomotor behavior)
- DD
constant darkness
- per
period gene
- Peripheral clock
a circadian clock beyond the clock in the pacemaker neural network of the lateral and dorsal neurons in Drosophila and the suprachiasmatic nuclei in mammals. These can include other neural clocks and nonneural clocks in other organs
- Clk
Clock gene
- cry
cryptochrome gene
- LNv
ventral lateral neurons
- LNd
dorsal lateral neurons
- Pacemaker
a cell or group of cells that is capable of generating endogenous rhythms in the absence of external time cues
PIGMENT-DISPERSING FACTOR
- CCG
clock-controlled gene
- Subjective day/night
the interval during constant darkness that corresponds to the day/night phase under prior light-dark cycling conditions
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pittendrigh C. On temperature independence in the clock-system controlling emergence time in Drosophila. Proc Natl Acad Sci USA. 1954;40:1018–29. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubruille R, Emery P. A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol. 2008;38:129–45. doi: 10.1007/s12035-008-8035-y. [DOI] [PubMed] [Google Scholar]
- 4.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 5.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–55. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins B, Blau J. Even a stopped clock tells the right time twice a day: circadian timekeeping in Drosophila. Pflüg Arch. 2007;454:857–67. doi: 10.1007/s00424-006-0188-9. [DOI] [PubMed] [Google Scholar]
- 7.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–33. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–29. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- 9.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kivimae S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meissner RA, Kilman VL, Lin JM, Allada R. TIMELESS is an important mediator of CK2 effects on circadian clock function in vivo. J Neurosci. 2008;28:9732–40. doi: 10.1523/JNEUROSCI.0840-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, Sathyanarayanan S, Sehgal A. Posttranslational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–18. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression and circadian clock function. Mol Cell Biol. 2007;27:5014–28. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–13. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim EY, Edery I. Balance between DBT/CKIε kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci USA. 2006;103:6178–83. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–72. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, et al. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–17. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- 18.Collins B, Mazzoni EO, Stanewsky R, Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr Biol. 2006;16:441–49. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M. PER-TIM interactions with the photoreceptor cryptochrome mediate circadian temperature responses in Drosophila. PLoS Biol. 2007;5:e146. doi: 10.1371/journal.pbio.0050146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–41. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 21.Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, et al. An isoform-specific mutant reveals a role of PDP1ε in the circadian oscillator. J Neurosci. 2009;29:10920–27. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, et al. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–89. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–86. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richier B, Michard-Vanhee C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms. 2008;23:103–16. doi: 10.1177/0748730407313817. [DOI] [PubMed] [Google Scholar]
- 26.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–44. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Jones CR, Fujiki N, Xu Y, Guo B, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–66. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 29.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–43. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, et al. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–44. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 31.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–56. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 32.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–60. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 33.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–13. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kula E, Levitan ES, Pyza E, Rosbash M. PDF cycling in the dorsal protocerebrum of the Drosophila brain is not necessary for circadian clock function. J Biol Rhythms. 2006;21:104–17. doi: 10.1177/0748730405285715. [DOI] [PubMed] [Google Scholar]
- 36.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 37.Hyun S, Lee Y, Hong ST, Bang S, Paik D, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–78. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 38.Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–27. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, et al. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–19. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–37. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lear BC, Zhang L, Allada R. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol. 2009;7:e1000154. doi: 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi C, Fortin JP, McCarthy E, Oksman L, Kopin AS, Nitabach MN. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr Biol. 2009;19:1167–75. doi: 10.1016/j.cub.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, et al. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–56. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 45.Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:612–16. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park D, Griffith LC. Electrophysiological and anatomical characterization of PDF-positive clock neurons in the intact adult Drosophila brain. J Neurophysiol. 2006;22:22. doi: 10.1152/jn.00117.2006. [DOI] [PubMed] [Google Scholar]
- 47.Gorska-Andrzejak J, Keller A, Raabe T, Kilianek L, Pyza E. Structural daily rhythms in GFP-labelled neurons in the visual system of Drosophila melanogaster. Photochem Photobiol Sci. 2005;4:721–26. doi: 10.1039/b417023g. [DOI] [PubMed] [Google Scholar]
- 48.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–45. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–82. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105:19587–94. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19:386–90. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–8. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–42. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 54.Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoleru D, Nawathean P, de la Paz Fernandez M, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–19. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 57.Low KH, Lim C, Ko HW, Edery I. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron. 2008;60:1054–67. doi: 10.1016/j.neuron.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316:1898–900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 59.Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–98. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- 60.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–49. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–47. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–71. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 63.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–78. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 64.Lin Y, Han M, Shimada B, Wang L, Gibler TM, et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–67. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–19. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–52. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 67.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Majercak J, Kalderon D, Edery I. Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Mol Cell Biol. 1997;17:5915–22. doi: 10.1128/mcb.17.10.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rouyer F, Rachidi M, Pikielny C, Rosbash M. A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 1997;16:3944–54. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–56. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 73.Lear BC, Lin JM, Keath JR, McGill JJ, Raman IM, Allada R. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48:965–76. doi: 10.1016/j.neuron.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 74.Dockendorff T, Su H, McBride S, Yang Z, Choi C, et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–84. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 75.Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, et al. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–35. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- 76.Benito J, Zheng H, Hardin PE. PDP1ε functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–47. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim C, Lee J, Koo E, Choe J. Targeted inhibition of Pdp1ε abolishes the circadian behavior of Drosophila melanogaster. Biochem Biophys Res Commun. 2007;364:294–300. doi: 10.1016/j.bbrc.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 78.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2007;99:976–88. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fernandez Mde L, Chu J, Villella A, Atkinson N, Kay SA, Ceriani MF. Impaired clock output by altered connectivity in the circadian network. Proc Natl Acad Sci USA. 2007;104:5650–55. doi: 10.1073/pnas.0608260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaramillo AM, Zeng H, Fei H, Zhou Y, Levitan IB. Expression and function of variants of slob, slowpoke channel binding protein, in Drosophila. J Neurophysiol. 2006;95:1957–65. doi: 10.1152/jn.00427.2005. [DOI] [PubMed] [Google Scholar]
- 82.Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, et al. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 2004;5:3. doi: 10.1186/1471-2202-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meredith AL, Wiler SW, Miller BH, Takahashi JS, Fodor AA, et al. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci. 2006;9:1041–49. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–83. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 85.Lu B, Su Y, Das S, Wang H, Wang Y, et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–44. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu Y, Cao G, Pavlicek B, Luo X, Nitabach MN. Phase coupling of a circadian neuropeptide with rest/activity rhythms detected using a membrane-tethered spider toxin. PLoS Biol. 2008;6:e273. doi: 10.1371/journal.pbio.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hodge JJ, Stanewsky R. Function of the Shaw potassium channel within the Drosophila circadian clock. PLoS One. 2008;3:e2274. doi: 10.1371/journal.pone.0002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–95. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 89.Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–87. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levine JD, Casey CI, Kalderon DD, Jackson FR. Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron. 1994;13:967–74. doi: 10.1016/0896-6273(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 91.Kilman VL, Zhang L, Meissner RA, Burg E, Allada R. Perturbing dynamin reveals potent effects on the Drosophila circadian clock. PLoS ONE. 2009;4:e5235. doi: 10.1371/journal.pone.0005235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 93.Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- 94.Liu X, Lorenz L, Yu QN, Hall JC, Rosbash M. Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 1988;2:228–38. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- 95.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–35. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 96.Handler AM, Konopka RJ. Transplantation of a circadian pacemaker in Drosophila. Nature. 1979;279:236–38. doi: 10.1038/279236a0. [DOI] [PubMed] [Google Scholar]
- 97.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–79. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 99.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–37. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 100.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 101.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–84. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 102.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 103.Mrosovsky N. Masking: history, definitions, and measurement. Chronobiol Int. 1999;16:415–29. doi: 10.3109/07420529908998717. [DOI] [PubMed] [Google Scholar]
- 104.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- 105.Lu B, Liu W, Guo F, Guo A. Circadian modulation of light-induced locomotion responses in Drosophila melanogaster. Genes Brain Behav. 2008;7:730–39. doi: 10.1111/j.1601-183X.2008.00411.x. [DOI] [PubMed] [Google Scholar]
- 106.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 107.Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–75. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–14. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 109.Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12:2152–58. doi: 10.1016/s0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- 110.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunore-activity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–62. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–98. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen DM, Christianson JS, Sapp RJ, Stark WS. Visual receptor cycle in normal and period mutant Drosophila: microspectrophotometry, electrophysiology, and ultrastructural morphometry. Vis Neurosci. 1992;9:125–35. doi: 10.1017/s0952523800009585. [DOI] [PubMed] [Google Scholar]
- 113.Rieger D, Stanewsky R, Helfrich-Forster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18:377–91. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- 114.Klarsfeld A, Malpel S, Michard-Vanhee C, Picot M, Chelot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–77. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mazzoni EO, Desplan C, Blau J. Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron. 2005;45:293–300. doi: 10.1016/j.neuron.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 116.Siwicki KK, Ladewski L. Associative learning and memory in Drosophila: beyond olfactory conditioning. Behav Processes. 2003;64:225–38. doi: 10.1016/s0376-6357(03)00137-2. [DOI] [PubMed] [Google Scholar]
- 117.Lyons LC, Roman G. Circadian modulation of short-term memory in Drosophila. Learn Mem. 2009;16:19–27. doi: 10.1101/lm.1146009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Emery IF, Noveral JM, Jamison CF, Siwicki KK. Rhythms of Drosophila period gene expression in culture. Proc Natl Acad Sci USA. 1997;94:4092–96. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–33. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- 120.Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–56. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, McNeil GP, Hilderbrand-Chae MJ, Franklin TM, Schroeder AJ, Jackson FR. Circadian regulation of the lark RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45:14–29. doi: 10.1002/1097-4695(200010)45:1<14::aid-neu2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 122.Newby LM, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135:1077–90. doi: 10.1093/genetics/135.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 124.Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104:1859–64. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, et al. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci. 2008;28:10200–5. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, et al. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–72. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 127.Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.So WV, Sarov-Blat L, Kotarski CK, McDonald MJ, Allada R, Rosbash M. takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol Cell Biol. 2000;20:6935–44. doi: 10.1128/mcb.20.18.6935-6944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007;210:1424–34. doi: 10.1242/jeb.02755. [DOI] [PubMed] [Google Scholar]
- 131.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–78. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 132.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–49. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 133.Tanoue S, Krishnan P, Chatterjee A, Hardin PE. G protein-coupled receptor kinase 2 is required for rhythmic olfactory responses in Drosophila. Curr Biol. 2008;18:787–94. doi: 10.1016/j.cub.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci USA. 2001;98:9221–25. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–51. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr Biol. 2003;13:140–45. doi: 10.1016/s0960-9822(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 137.Kadener S, Villella A, Kula E, Palm K, Pyza E, et al. Neurotoxic protein expression reveals connections between the circadian clock and mating behavior in Drosophila. Proc Natl Acad Sci USA. 2006;103:13537–42. doi: 10.1073/pnas.0605962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–92. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lazareva AA, Roman G, Mattox W, Hardin PE, Dauwalder B. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3:e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujii S, Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–63. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fujii S, Toyama A, Amrein H. A male-specific fatty acid omega-hydroxylase, SXE1, is necessary for efficient male mating in Drosophila melanogaster. Genetics. 2008;180:179–90. doi: 10.1534/genetics.108.089177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Howlader G, Paranjpe DA, Sharma VK. Non-ventral lateral neuron-based, non-PDF-mediated clocks control circadian egg-laying rhythm in Drosophila melanogaster. J Biol Rhythms. 2006;21:13–20. doi: 10.1177/0748730405282882. [DOI] [PubMed] [Google Scholar]
- 143.Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18:195–99. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shirasu-Hiza MM, Dionne MS, Pham LN, Ayres JS, Schneider DS. Interactions between circadian rhythm and immunity in Drosophila melanogaster. Curr Biol. 2007;17:R353–55. doi: 10.1016/j.cub.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 145.Ito C, Goto SG, Shiga S, Tomioka K, Numata H. Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:8446–51. doi: 10.1073/pnas.0800145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci USA. 2007;104:15899–904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mehnert KI, Beramendi A, Elghazali F, Negro P, Kyriacou CP, Cantera R. Circadian changes in Drosophila motor terminals. Dev Neurobiol. 2007;67:415–21. doi: 10.1002/dneu.20332. [DOI] [PubMed] [Google Scholar]
- 149.Hege DM, Stanewsky R, Hall JC, Giebultowicz JM. Rhythmic expression of a PER-reporter in the Malpighian tubules of decapitated Drosophila: evidence for a brain-independent circadian clock. J Biol Rhythms. 1997;12:300–8. doi: 10.1177/074873049701200402. [DOI] [PubMed] [Google Scholar]