SUMMARY
In metazoa, nuclear pore complexes (NPCs) assemble from disassembled precursors into a reforming nuclear envelope (NE) at the end of mitosis, and into growing intact NEs during interphase. Here, we show by RNAi-mediated knockdown that ELYS, a nucleoporin critical for the recruitment of the essential Nup107/160 complex to chromatin, is required for NPC assembly at the end of mitosis, but not during interphase. Conversely, the transmembrane nucleoporin POM121 is critical for the incorporation of the Nup107/160 complex into new assembly sites specifically during interphase. Strikingly, recruitment of the Nup107/160 complex to an intact NE involves a membrane curvature-sensing domain of its constituent, Nup133, which is not required for post-mitotic NPC formation. Our results suggest that, in organisms with open mitosis, NPCs assemble by two distinct mechanisms to accommodate cell cycle-dependent differences in NE topology.
INTRODUCTION
NPCs are the exclusive channels of nucleo-cytoplasmic transport in eukaryotic cells. These multiprotein assemblies have an estimated mass of ~60 MD (Hetzer et al., 2005; Tran, and Wente, 2006) and are embedded in the double lipid bilayer of the NE. Each NPC assembles from ~30 different nucleoporins (Nups), present in multiple copies, totaling ~500 polypeptides (Alber et al., 2007; Beck et al., 2004; Cronshaw et al., 2002). NPCs consist of a NE-embedded scaffold surrounding the central channel, largely composed of the Nup107/160 and Nup93/Nup205 complexes (Figure 1A). The Nup107/160 complex has been shown to be an early and essential player in NPC formation both in vitro and in vivo (Harel et al., 2003; Walther et al., 2003a). In vertebrates it consists of nine polypeptides (Nup160, Nup133, Nup107, Nup96, Nup85, Nup43, Nup37, Seh1 and Sec13), assembled in a Y-shaped complex (Lutzmann et al., 2002). Its members are primarily composed of β-propellers and α-solenoids (Brohawn et al., 2009), a protein fold composition shared exclusively with other membrane coating protein complexes including clathrin coats and the COPII coatomer of the ER/Golgi (Alber et al., 2007; Brohawn et al., 2008; Devos et al., 2004). Furthermore, several of the scaffold Nups in yeasts and vertebrates possess an ALPS-like motif shown to target curved membranes in vitro (Drin et al., 2007). Attached to the NPC core are the cytoplasmic and nuclear rings from which eight filaments and the nuclear basket emanate. Many peripheral Nups contain phenylalanine-glycine (FG)-repeats that interact with nuclear transport receptors, providing a selective barrier for diffusion of macromolecules (Rabut et al., 2004; Weis, 2003).
Figure 1. POM121 and ELYS have non-redundant roles in NPC assembly.
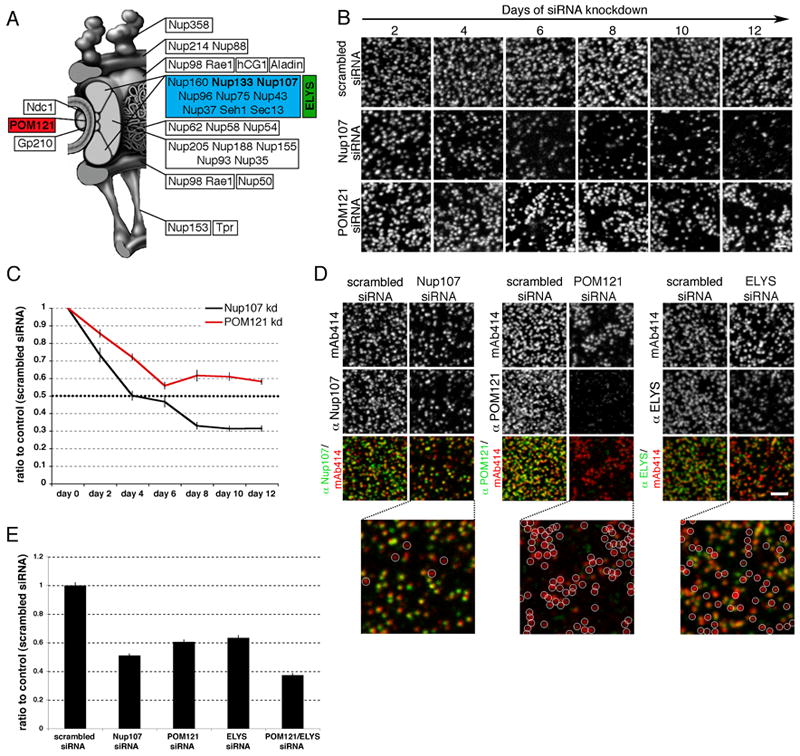
(A) Schematic of NPC composition. (B) U2OS cells were treated repeatedly with scrambled, POM121 or Nup107 siRNA oligos for 12 days, fixed at indicated time points and stained with mAb414. (C) Quantification of mAb414 immunofluorescence (representing total NPCs per nucleus) over time, graphed as a ratio to control levels, N>25 per time point. (D) Immunofluorescence staining of nuclear surfaces using mAb414 and antibodies against Nup107, POM121 or ELYS in U2OS cells treated with siRNA oligos for 4 days against the indicated Nup. White circles indicate NPCs lacking either Nup107, POM121 or ELYS. (E) Quantification of mAb414 immunofluorescence in U2OS cells treated with siRNA oligos against indicated Nups, N>26 nuclei per condition. All error bars are standard error. Scale bars 2 μm. See also Figures S1 and S2.
Relatively little is known about NPC biogenesis in metazoa, which occurs during two different cell cycle phases. The first pathway occurs post mitosis and involves the ordered recruitment of ER membranes and disassembled NPC components to chromatin (Anderson and Hetzer, 2008b; Dultz et al., 2008; Walther et al., 2003b). In vitro studies using Xenopus egg extracts revealed NPC assembly during NE reformation is initiated by recruitment of the Nup107/160 complex (Belgareh et al., 2001; Harel et al., 2003; Walther et al., 2003b) to chromatin; a step mediated by the DNA-binding Nup ELYS/Mel28 (Franz et al., 2007; Gillespie et al., 2007; Rasala et al., 2006). This is followed by recruitment of ER membranes, containing the transmembrane Nups POM121 and Ndc1, and subsequent incorporation of Nup155 and Nup53 (Antonin et al., 2008).
The second pathway requires targeting and insertion of newly synthesized Nups to an intact interphase NE and it is unclear if this process is distinct from post-mitotic assembly. In mammalian cells, only three transmembrane Nups have been identified: POM121, gp210, and Ndc1 (Chial et al., 1998; Hallberg et al., 1993; Mansfeld et al., 2006; Stavru et al., 2006b). While gp210 is not expressed in all cell types (Eriksson et al., 2004) and thus unlikely to play a role in NPC biogenesis, RNAi-mediated silencing of POM121 and Ndc1 has been shown to negatively affect NPC assembly (Antonin et al., 2005; Mansfeld et al., 2006; Stavru et al., 2006a; Stavru et al., 2006b). Furthermore, the appearance of POM121 has been shown to be an early step in NPC assembly both in vivo and in vitro (Dultz et al., 2008; Rasala et al., 2008), and essential for NE formation in vitro (Antonin et al., 2005). Other studies, however, suggest POM121 might be dispensable for NPC formation (Stavru et al., 2006). These apparently contradicting studies imply the role of transmembrane Nups in NPC biogenesis, while still undefined, may be redundant.
Here we show that incorporation of NPC components into an intact NE occurs by a mechanism that specifically requires POM121 and a membrane curvature-sensing domain in Nup133. Neither of these components was required for post-mitotic NPC formation. Interestingly, recruitment of the Nup107/160 complex to new assembly sites in interphase does not involve ELYS, which is specifically required for its recruitment to chromatin at the end of mitosis.
RESULTS
The nucleoporins ELYS and POM121 have non-redundant functions in NPC assembly
Studying potential differences between post-mitotic and interphase NPC assembly is complicated by a subset of extremely long-lived Nups difficult to efficiently deplete by RNA interference (Rabut et al., 2004) such as the essential Nup107/160 complex (Figure 1A) (D’Angelo et al., 2009; Rabut et al., 2004). To improve the extent of Nup107/160 complex depletion, we repeatedly transfected cells with siRNA oligos specific for Nup107 for a total of 12 days. Cells were stained at two day time points with mAb414, an antibody that recognizes Nup358, Nup214, Nup153 and Nup62 (Davis and Blobel, 1987) (Figure 1B). While the overall NPC number in control cells, treated with scrambled oligos, remained constant, Nup107 knockdown resulted in a strong reduction of NPC density (Figure 1B, S2A). Measuring the total fluorescence intensity of each nucleus, which accurately reflected the doubling of NPC number in interphase (Maul, 1971)(Figure S1A), we detected a sharp decrease of NPC numbers to ~30% in Nup107-depleted cells (Figure 1C), correlating with a reduction in Nup107 protein levels to <25% (Figure S1B, S1C). These results show that successive RNAi treatments over prolonged periods of time allow efficient depletion of scaffold Nups and confirm the Nup107/160 complex is required for NPC assembly.
We used this experimental strategy to test if other NPC components implicated in early steps of pore assembly behave similarly. We first focused on POM121, a transmembrane Nup shown to be required for NE formation in vitro (Antonin et al., 2005). Although cells treated with siRNA oligos against POM121 also exhibited reduced NPC levels (Figure 1B, S2A), total NPC numbers plateaued at only ~50–60% of control levels (Figure 1C) and endogenous Nup107 protein (required for NPC assembly) was reduced to ~60% in POM121 depleted cells. However, in these conditions, levels of POM121 were reduced to <8% (Figures S1B, S1C), suggesting that NPCs assembled in the absence of this protein. To directly test this possibility, we stained POM121 depleted cells with mAb414 and α-POM121 antibodies and found that the majority of NPCs did not contain detectable POM121 signal (Figure 1D, middle panels).
Another NPC component shown to be critical for NPC assembly is ELYS (Franz et al., 2007; Rasala et al., 2008). We found that knockdown of ELYS exhibited a similar phenotype to that of POM121, reducing total NPCs while allowing the formation of ELYS negative NPCs (Figure 1D, S2A, S2B). Double knockdown of both proteins had an additive inhibitory effect on NPC assembly (Figure 1E), indicating POM121 and ELYS play distinct roles in pore assembly. To confirm NPCs detected in knockdown cells were indeed fully formed NPCs, we performed TEM as well as immunofluorescence analysis with a variety of Nups representing multiple subcomplexes of the NPC, including Nup214 which is incorporated in later steps of NPC assembly (Dultz et al., 2008). We found no difference in overall NPC structure or composition (Figure S2B, S2C, S2D).
ELYS is specifically required for post-mitotic NPC assembly
Consistent with previous findings (Rasala et al., 2008), we found depletion of ELYS (Figure S3A) strongly inhibited recruitment of the Nup107/160 complex to chromatin, visualized by 3GFP-Nup133, during post-mitotic NPC assembly (Figure 2A, 2B). This implies that the observed ELYS-independent formation of NPCs likely occurs in interphase. To directly test this we specifically measured interphase NPC formation by comparing NPC levels in G1 versus G2 in synchronized cells (Figure S1A). Cells treated with ELYS RNAi exhibited a strong reduction in G1 NPCs, yet NPC doubling in interphase was not affected (Figure 2C). Supporting previous findings (Franz et al., 2007)(Rasala et al., 2006), we observed mis-localization of Nups to cytoplasmic annulate lamellae (Figure S3B, S3C, S3D), consistent with a role for ELYS in targeting the Nup107/160 complex to post-mitotic chromatin.
Figure 2. ELYS is required specifically during post-mitotic NPC assembly.
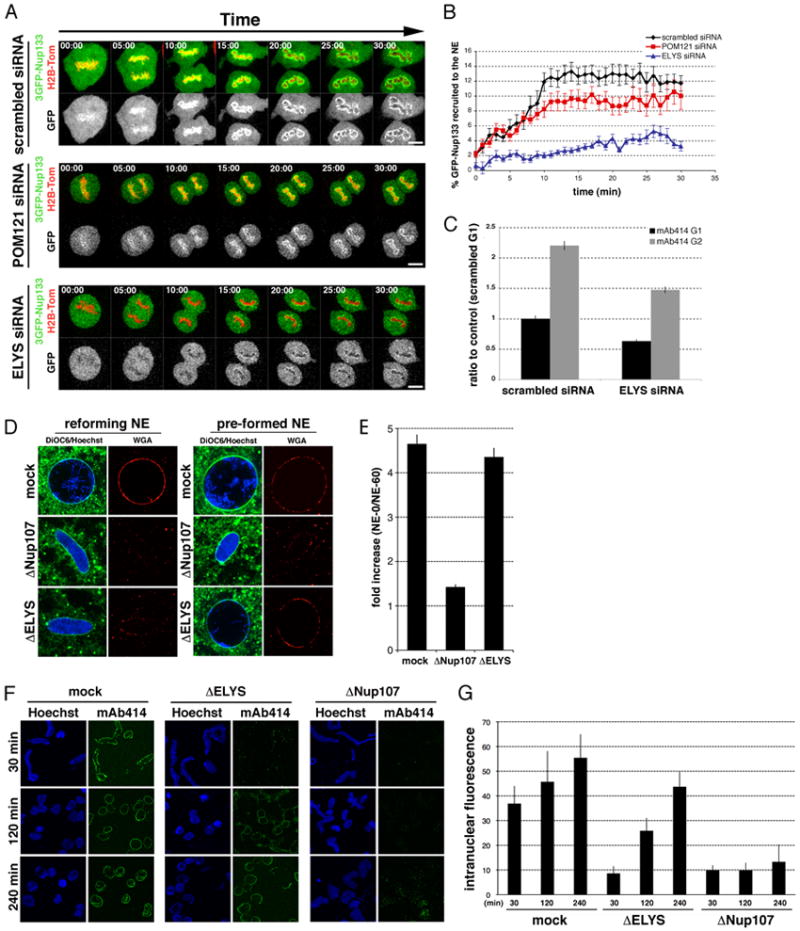
(A) U2OS cells were transfected with 3GFP-Nup133 and H2B-tomato together with either scrambled, ELYS or POM121 specific siRNA oligos and imaged from metaphase for 120 min at the rate of 1 frame per minute. The onset of chromosome segregation marks t=0 (elapsed min are indicated for each frame). Scale bar 5 μm. (B) Quantification of GFP signal around chromatin over time, representing the Nup107/160 complex, to the reforming NE (details in Methods); N>12. (C) Quantification of total mAb414 fluorescent signal during G1 and G2 in U2OS cells transfected with control and ELYS siRNA oligos; N>98 nuclei per condition. (D) In vitro nuclear assembly reactions using mock-, ELYS- or Nup107-depleted cytosol and either demembranated sperm heads (reforming NE) or washed, NE-enclosed sperm chromatin (pre-formed NE) were incubated for 60 min. Nuclei were incubated with DiOC6 (green) and fluorescent WGA (red) to visualize membranes and NPCs. (E) Quantification of total fluorescence, representing NPC number; N>150 nuclei. All error bars represent standard error. (F) In vitro nuclear assembly reactions using mock-, ELYS- or Nup107-depleted cytosol were incubated for 240 min. Reactions were fixed and stained with mAb414 at indicated time points. (G) Quantification of total intranuclear fluorescence from time points illustrated in (F) shows efficient NPC insertion over time in ELYS- but not Nup107-depleted nuclei. See also Figure S3.
To confirm that depletion of ELYS had not prevented cell cycle progression and thus the associated insertion of NPCs, cyclin A-positive cells in each knockdown condition were quantified and compared to control cells (Figure S3F, S3G).
Next we tested the specific role of ELYS in post-mitotic NPC formation in a system that does not rely on RNAi-mediated knock down. We used the Xenopus egg extract system, in which NPC formation into the reforming NE (post-mitotic assembly) can be analyzed when sperm chromatin is mixed with isolated membranes and cytosol (Anderson and Hetzer, 2007; Walther et al., 2003b). In the same system, nuclei can be pre-formed around sperm chromatin and NPC insertion into intact nuclei (interphase assembly) is monitored (D’Angelo et al., 2006). Using this approach, we first immuno-depleted either the Nup107/160 complex or ELYS from cytosol (Figure S3E) and then performed NPC assembly reactions. Consistent with previous reports (Rasala et al., 2006; Walther et al., 2003a), the depletion of either the Nup107/160 complex or ELYS inhibited NPC assembly in a reforming NE (Figure 2D, left). In contrast, when NEs were pre-formed in the presence of a limiting amount of cytosol (Anderson et al., 2008), washed and then incubated with the depleted extracts, the absence of the Nup107/160 complex prevented further NPC insertion whereas ELYS depletion did not (Figure 2D, 2E). Furthermore, a time course experiment performed in ELYS-depleted cytosol revealed that very few NPCs were detected in early time points (reforming NE), however, after extended incubation NPC levels matched control (mock-depleted) numbers (Figure 2F, 2G). In contrast, nuclei formed with Nup107/160 depleted cytosol were unable to insert new NPCs. Taken together these results suggest that ELYS is dispensable for interphase NPC insertion.
POM121 is rate limiting for interphase NPC assembly
We next focused on POM121, since like ELYS, while its depletion resulted in a general decrease in total NPCs, it appeared dispensable for NPC formation (Figure 1B, 1C, 1D). Consistent with our finding that ELYS and POM121 have non-redundant functions, we found that the knockdown of POM121 did not interfere with recruitment of a 3GFP-Nup133 reporter to chromatin (Figure 2A, 2B), and thus did not seem to be required for early events in post-mitotic NPC assembly. We noticed, however, that following closed NE formation (~12 min; Anderson et al., 2009), levels of Nup133 at the NE were reduced in cells depleted of POM121 (Figure 2B). This indicated that POM121 might be preferentially required for NPC assembly into an intact NE. To test this, we depleted POM121 and examined total pore numbers in synchronized cells. Despite the efficient knockdown of POM121, cells exhibited normal NPC numbers in G1, but an almost complete inhibition of pore insertion during interphase (Figure 3A). Notably, both mAb414 reactive Nups and Nup107 were no longer recruited to the NE, suggesting POM121 has an early role in interphase NPC assembly (Figure 3A).
Figure 3. POM121 plays an early and essential role in interphase NPC assembly.
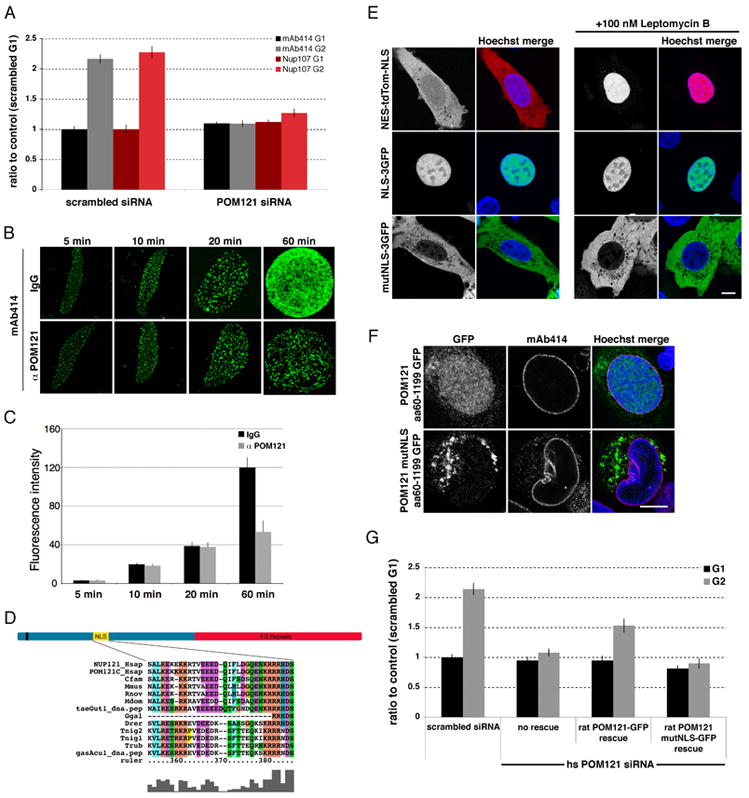
(A) Quantification of total mAb414 or α-Nup107 fluorescent signal during G1 and G2 in U2OS cells transfected with control or POM121 siRNA oligos. N>73 nuclei per condition. (B) In vitro nuclear assembly reactions were performed in the presence of rabbit IgG or an inhibitory α-POM121 antibody. NPCs were visualized using mAb414 antibody. (C) Quantification of total fluorescence of conditions and time points illustrated in (B) shows a specific block in interphase NPC assembly in the presence of the α-POM121 antibody (D) Schematic of POM121 topology and NLS motif sequence. (E) POM121 wtNLS and mutNLS sequence fused to 3GFP (green) or NES-tdTom-NLS (red) were transfected in U2OS cells and imaged with and without treatment with Leptomycin B for 2.5 hrs. Scale bar 5μm. (E) The cytoplasmic domain of POM121 (aa60-1199) wtNLS and POM121mutNLS was fused to GFP and expressed in U2OS cells. (F) Quantification of total mAb414 fluorescent signal during G1 and G2 in control U2OS cells (scrambled) and in cells depleted of the endogenous human POM121 and rescued with either wt or mutNLS rat POM121; N>30 nuclei per condition. All error bars are standard error. See also Figure S4.
To further analyze the interphase-specific role of POM121 in NPC formation, we performed an in vitro time course experiment monitoring nuclear assembly by incubating sperm chromatin with cytosol in which the ER network was pre-formed (Anderson and Hetzer, 2007). In the presence or absence of inhibitory α-POM121 antibodies, NPCs assembled at similar rates in both conditions at early time points. In contrast, NPC insertion during NE expansion (i.e. after ~ 20 min when the NE is closed) was specifically inhibited by the POM121 antibody (Figure 3B, 3C, data not shown).
Interestingly, we identified a conserved bipartite nuclear localization signal (NLS) in POM121 (aa292–317) (Figure 3D) and wondered if this motif is critical for NPC assembly. We first tested if the NLS sequence was functional in mediating nuclear import by fusing the POM121 NLS sequence to 3GFP. While a mutant version of the 3GFP-NLS localized in the cytoplasm, the reporter protein with the original NLS sequence accumulated in the nucleus (Figure 3E). Similar results were obtained with the wild type and mutant NLS sequences in the context of the cytoplasmic domain of POM121 (aa60–1199) (Figure 3F). To ask if the NLS is critical for POM121 function we generated the same mutations in the NLS of the full-length protein (POM121mutNLS). The NLS mutant protein co-localized with NPCs (Figure S4A) and exhibited similar residence times at the NPC as determined by fluorescence recovery after photobleaching (FRAP)(Figure S4B), indicating POM121mutNLS incorporates into NPCs. As endogenous POM121 protein was present in these experiments, we depleted human POM121 and rescued with either the wild type or NLS mutant rat protein (Figure S4C). The specific block in interphase NPC assembly was rescued with the wild type rat POM121 but not the NLS mutant (Figure 3G) suggesting the NLS is critical for POM121 function in interphase NPC insertion. The observed incorporation of POM121mutNLS is therefore likely to have occurred during post-mitotic NPC formation, further highlighting cell-cycle dependent differences in NPC assembly.
POM121 precedes the Nup107/160 complex at new assembly sites during interphase
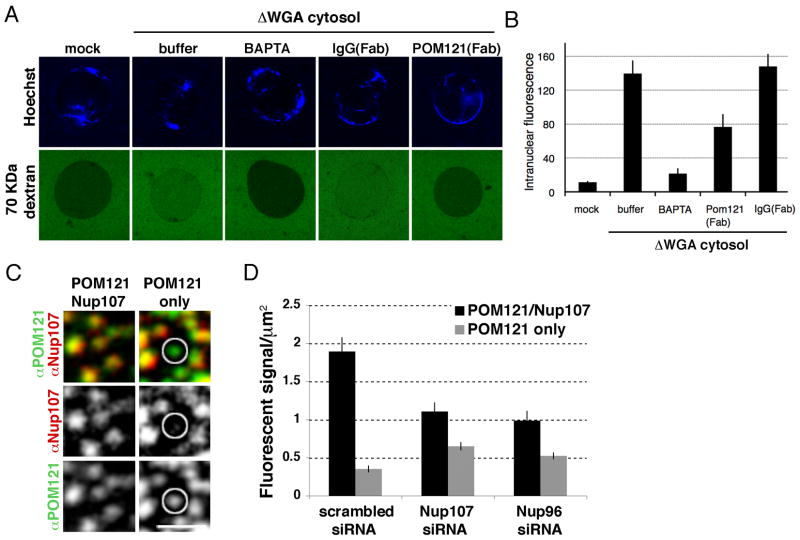
Our results suggest that POM121 precedes incorporation of the Nup107/160 complex and functions as an early player in interphase NPC assembly. To test this, we turned to an in vitro assay that functionally monitors INM/ONM fusion (Dawson et al., 2009). In brief, glycosylated Nups, critical for establishing the permeability barrier of the NPC, are depleted from egg cytosol. Nuclei assembled in these extracts contain a closed NE but are permeable for diffusion of molecules <150 kDa. Thus, the formation of new NPCs can be detected by influx of a normally excluded, fluorescently labeled, 70-kDa dextran. When we preassembled nuclei for 60 min and added either mock- or WGA-depleted cytosol in the presence of the calcium chelator BAPTA, a known inhibitor of NPC formation, the dextran influx was strongly inhibited (Figure 4A, 4B), suggesting the INM and ONM had not fused. Next, we added WGA-depleted cytosol in the absence or presence of inhibitory Fab fragments of an α-POM121 antibody (Figure S4F). In reactions with control antibodies including Nup107, Nup96, ELYS and Ndc1 (data not shown), nuclear influx of the dextran was detected 5 min after the addition of WGA-depleted cytosol. In contrast, no influx was observed in the presence of the α-POM121 antibody, providing evidence that POM121 may be involved in the formation of a pre-pore/hole in an intact NE (Figure 4A, B). Importantly, when the POM121 antibody was added after new holes had formed in the NE, the dextran was not excluded, indicating that the antibody did not plug the formed holes but had prevented their formation (Figure S4D, S4E).
Figure 4. POM121 precedes the Nup107/160 complex during interphase NPC assembly.
(A) Assembled nuclei were incubated with mock- or WGA-depleted cytosol for 30 min in the presence of buffer, BAPTA, IgG or POM121 Fab fragments. Fluorescently labeled 70 kDa dextran (green) was added to probe for INM-ONM fusion. (B) Quantification of intranuclear 70 kDa dextran signal in conditions described in (B), N>150 nuclei per condition. (C) Immunofluorescence staining of the nuclear surface using specific antibodies against POM121 and Nup107. Circle indicates NPC intermediate containing POM121 but not Nup107, scale bar 1 μm. (D) Quantification of NPC intermediates in control (scrambled) and Nup107 or Nup96 RNAi-treated cells, n = 30 fields. See also Figure S4.
Our results indicating POM121 precedes the Nup107/160 complex at new assembly sites (Figure 3A) suggests the latter is not involved in the fusion of the INM and ONM. Consistent with this idea, we did detect 70 KDa dextran influx (INM and ONM fusion) in nuclei assembled from Nup107/160 complex depleted egg extracts (data not shown). As interphase NPC assembly requires 30–60 min (D’Angelo et., 2006), we wondered if NPC intermediates could be detected that contain POM121 but not the Nup107/160 complex. To test this we stained dividing cells, which double their NPCs between G1 and G2, with antibodies against POM121 and Nup107. As expected, most fluorescent loci contained both Nups. However, we found many of the fluorescent sites analyzed contained POM121 signal alone (Figure 4C, 4D). Importantly, treatment with siRNA oligos against either Nup107 or Nup96 resulted in a significant increase in POM121 only signal, indicating true intermediates (Figure 4D). Together these results support the conclusion that POM121 precedes the Nup107/160 complex at forming NPCs during interphase.
The ALPS motif of Nup133 is a membrane curvature sensor in vivo
As POM121 recruitment and fusion of the INM and ONM would result in a local increase in membrane curvature, the membrane curvature sensor domain (ALPS) of Nup133 presented a potential mechanism for targeting of the Nup107/160 complex to new NPC assembly sites. To test this, we mutated amino acid I254 in mNup133 (Figure 5A) to an aspartate (D), a non-hydrophobic residue, shown to impair the ALPS function of hNup133 in vitro (Drin et al., 2007). As a control we mutated I254 to valine (V), which should not change its ability to interact specifically with highly curved membranes. Using a recently developed Single Bead Affinity Detection (SINBAD) assay (Schulte et al., 2008), we confirmed that the His-tagged fragments of Nup133 wild type and the Nup133 IV mutant motifs bound specifically to highly curved 30 nm (but not 400 nm) liposomes, whereas the Nup133 ID mutant did not (Figures 5B, S5A, S5B).
Figure 5. The ALPS domain of Nup133 senses high-curvature membranes in vitro and in vivo.
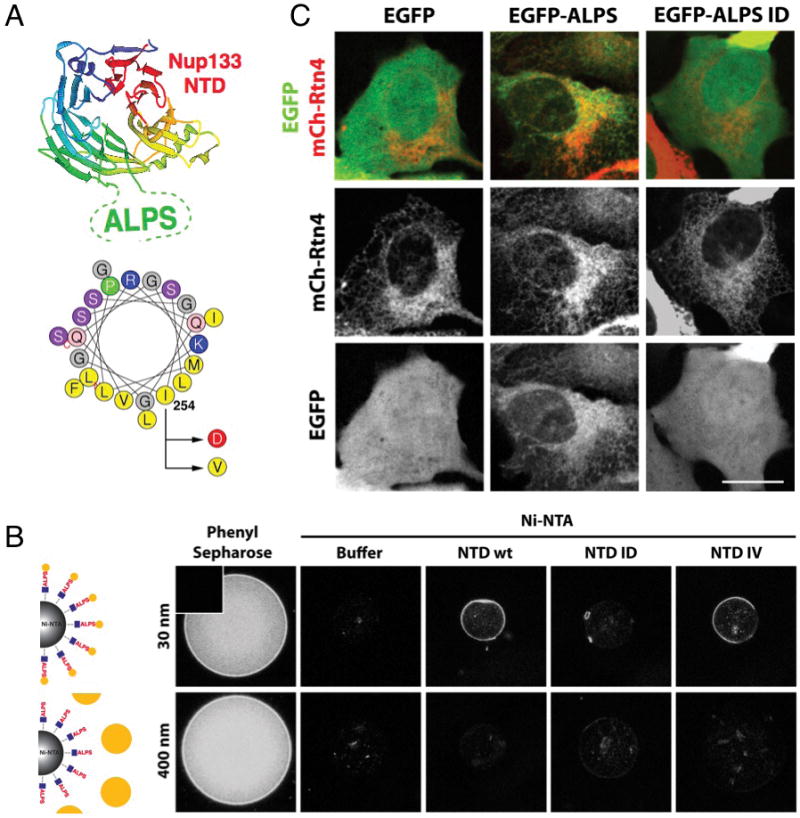
(A) Schematic of the ALPS domain of hNup133 within the structure of the beta-propeller (top) and representation of the ALPS motif of mNup133 on a helical wheel (bottom). The residue substitutions used in this study are indicated with arrows. (B) Ni-NTA magnetic beads were coated with equivalent amounts of His tagged recombinant NTD of mNup133 wt, ID or IV, or buffer as a control, and incubated with fluorescent liposomes of different diameter (30 or 400 nm). Phenyl Sepharose beads were used as a positive control; insert in top left corner shows a bead without liposomes. Beads were imaged by confocal microscopy. (C) U2OS cells were co-transfected with mCherry-Rtn4 (red) and EGFP wt, EGFP-ALPS or EGFP-ALPS ID (green) and imaged live 24h after transfection. Scale bar 2μm. See also Figure S5.
To analyze if the Nup133 ALPS domain functions as a curved membrane sensor in vivo, we introduced the motif into an exposed loop of the fluorescent protein EGFP (Figure S5C). As expected, EGFP displayed a diffuse nucleo-cytoplasmic localization with no indication of membrane association (Figure 5C, S5D). In striking contrast, the EGFP-ALPS fusion protein localized to membrane structures co-localizing with mCherry-Reticulon 4, a marker for tubular ER (De Craene et al., 2006; Voeltz et al., 2006) (Figure 5C, S5D). The EGFP-ALPS ID fusion had a localization identical to EGFP (Figure 5C,S5D). These results reveal the remarkable capacity of the ALPS motif of Nup133 to target a soluble protein to highly curved membranes, indicating this motif is a functional membrane curvature sensor in vivo.
The ALPS motif of Nup133 is required for proper localization of Nup107
To ask whether the ALPS domain of Nup133 plays a role in targeting or incorporation of the Nup107/160 complex to the NE, we ectopically expressed GFP-Nup133 wt, the Nup133 ID or IV mutant in U2OS cells (Figure S6A). While Nup133 wt and Nup133 IV localized specifically to the NE and NPCs, the mutant Nup133 ID, defective in sensing curved membranes, mislocalized to the cytoplasm (Figure 6A). Importantly, the Nup133 ID mutant was recruited to kinetochores, colocalizing with mitotic Nup107 (Figures S6B, S6C), indicating proper protein folding. In addition to the signal in the cytoplasm, we observed that GFP-Nup133 ID partially co-localized with mAb414 staining (Figure 6B), indicating an ability to incorporate into NPCs. Since GFP-Nup133 wt, ID and IV are expressed at similar levels, we anticipated the ID mutant would be inserted in the NE less efficiently than the wild type protein. To test this, we co-expressed Nup133 wt fused to a red fluorescent tag (3KR-mNup133) with either GFP-mNup133 wt, ID or IV, and measured the ratio of the GFP-tagged proteins (green) to wild-type Nup133 (red) at the NE (Figure 6C) in digitonin-permeabilized cells. We found this ratio to be lower in the Nup133 ID mutant, indicating that its incorporation into NPCs is less efficient.
Figure 6. The ALPS domain of Nup133 is critical for the proper localization of Nup133 and Nup107 to the NE.
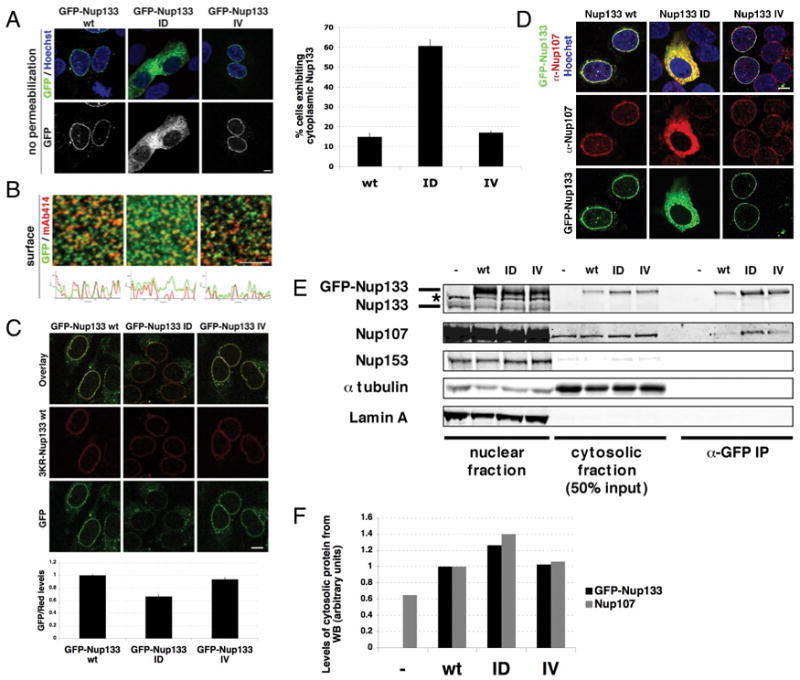
(A) U2OS cells were transfected with GFP-mNup133 wt, GFP-mNup133 ID or GFP-mNup133 IV (green) for 72h, fixed and stained with Hoechst (blue). The percentage of cells exhibiting cytoplasmic GFP signal 72h after transfection with either GFP-mNup133 wt, ID or IV is shown in the right panel. (B) U2OS cells were transfected as in (A), fixed and stained with mAb414. Histograms show co-localization of GFP and mAb414 signal at the nuclear surface (lower panels). (C) U2OS cells were co-transfected with mNup133 wt fused to a red fluorescent tag (3KR-Nup133) and either GFP-mNup133 wt, ID or IV for 72h. Plasmamembranes were permeabilized with digitonin 72h after transfection and fixed. The ratio of green and red fluorescence signals at the NE were quantified and normalized to the ratio of GFP-mNup133 wt/3KR-mNup133 wt (lower panel). (D) U2OS cells were transfected as in (A), then fixed and stained with α-Nup107 (red). (E) U2OS cells were transfected with GFP-mNup133 wt, ID or IV and collected after 48h. Cytosolic fractions were obtained by hypotonic cell lysis and nuclei were lysed in denaturing buffer. GFP fusion proteins were immuno-precipitated from the cytosolic fraction with an α-GFP antibody; untransfected cells served as a control. The nuclear, cytosolic fractions and immunoprecipitates were analyzed by Western Blotting using specific antibodies against Nup133, Nup107, Nup153 (mAb414), tubulin (cytosolic marker) and lamin A (nuclear marker). (F) Quantification of GFP-Nup133 and Nup107 levels in the cytosolic fractions, normalized to tubulin levels. All scale bars 10 μm. See also Figure S6.
As an important fraction of the ALPS mutant GFP-Nup133 ID was observed in the cytoplasm, we wanted to test if this affected localization of endogenous Nup107. Cells were transfected with GFP-Nup133 wt, ID or IV and stained with antibodies against Nup107. Interestingly, only cells expressing GFP-Nup133 ID showed Nup107 signal in the cytoplasm (Figure 6D). We confirmed that GFP-Nup133 ID is enriched in the cytosol when compared to the GFP-Nup133 wt and IV mutants (Figure 6E). We also observed a slight increase in Nup107 signal in the cytoplasm of cells expressing Nup133 ID, and detected an interaction between GFP-Nup133 ID and Nup107 in the cytosol (Figure 6E, 6F). Together these data indicate that the ALPS motif of Nup133 is critical for proper targeting of the Nup107/160 complex to the NE.
ALPS-dependent targeting of Nup133 is required for NPC assembly during interphase
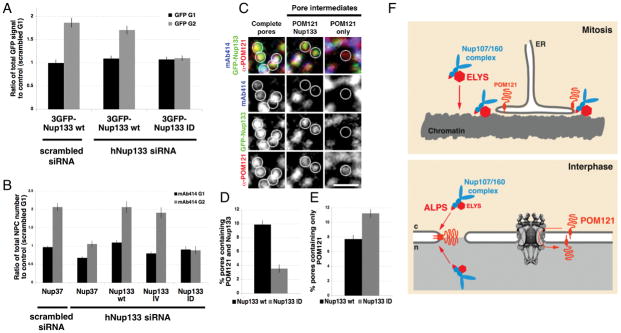
To investigate the potential role of the ALPS domain in post-mitotic and interphase NPC assembly, we depleted endogenous hNup133 and expressed RNAi-resistant GFP-tagged versions of mNup133 wt and the ID or IV mutants (Figure S7A). Using time-lapse microscopy we found that in the absence of endogenous Nup133, both 3GFP-Nup133 wt and the 3GFP-Nup133 ID mutants were recruited to the reforming NE with similar kinetics during anaphase (Figure S7B, S7C). Consistent with the proposed role of the ALPS in targeting Nup133 to INM and ONM fusion sites, recruitment of the ID mutant was specifically inhibited during interphase. In contrast, the signal of GFP-mNup133 wt was comparable to control conditions (Figure 7A). Measuring total mAb414 fluorescence in G1 and G2 revealed that cells expressing mNup133 ID, but not mNup133 wt or the IV mutant, were specifically defective in interphase NPC assembly (Figure 7B). Nup37, also in the Nup107/160 complex, failed to rescue the Nup133 depletion phenotype, suggesting an effect specific to Nup133 and that the Nup133 ALPS motif is required for interphase but not post-mitotic NPC formation.
Figure 7. The Nup133 ALPS domain is required for interphase NPC assembly.
(A) Quantification of total GFP signal at the NE during G1 and G2 in control cells (scrambled siRNA + 3GFP-mNup133 wt) and in cells depleted of endogenous human Nup133 and rescued with either 3GFP-mNup133 wt or mNup133 ID. Cells were permeabilized with digitonin before fixation; N>60 nuclei per condition. (B) U2OS cells were co-transfected with either control or hNup133 specific siRNA oligos and GFP-hNup37; or the hNup133 specific siRNA oligo and either GFP-mNup133 wt, GFP-mNup133 IV or GFP-mNup133 ID. Total mAb414 fluorescent signal (representing total pore numbers) in G1 and G2 under these conditions was quantified and graphed as a ratio to G1 control levels; N>50. (C) Immunofluorescence staining of the nuclear surface using mAb414 and POM121 specific antibodies in U2OS cells depleted of endogenous Nup133 and rescued with GFP-mNup133 wt. Circles indicate examples of fully assembled (complete) and intermediate NPCs containing either POM121 only or POM121 and GFP-mNup133 without mAb414. Scale bar 1μm. (D) & (E) Percentage of pores containing POM121 and GFP signal without mAb414 (D) or only POM121 signal (E) in cells depleted of endogenous Nup133 and rescued with either GFP-mNup133 wt or GFP-mNup133 ID. (D) p-value < 10−12, n > 50. (E) p-value = 2.75×10−5; n > 50. All error bars represent standard error.
(F) Model of post-mitotic and interphase NPC assembly. In mitosis, ELYS (red) is rate limiting for the recruitment of the Nup107/160 complex (blue) to chromatin, while the NE reforms from the mitotic ER (upper panel). During interphase POM121 (orange) is required for NPC assembly from both sides of an intact NE. The ALPS motif of Nup133 is critical for targeting of the Nup107/160 subcomplex to sites of ONM and INM fusion (lower panel).
See also Figure S7.
In cells depleted of endogenous Nup133 and rescued with GFP-Nup133 (wt or ID), we identified NPC intermediates containing only POM121 and also POM121 and GFP-Nup133 without mAb414 (Figure 7C), supporting the idea that POM121 precedes the Nup107/160 subcomplex at sites of forming NPCs. Replacement of endogenous Nup133 by the ALPS mutant decreased the number of sites containing POM121/Nup133 and, as expected, increased sites containing only POM121 (Figure 7D, E). These experiments provide additional evidence for mechanistic differences between post-mitotic and interphase assembly.
DISCUSSION
We provide the first evidence that interphase NPC assembly into an intact NE is distinct from post-mitotic NPC formation in a reforming NE (Figure 7F). It is best to view the two NPC assembly pathways in light of the differences in NE topology at the end of mitosis and in interphase. During NE reformation, ER membrane sheets enclose chromatin, (Anderson and Hetzer, 2008a; Anderson et al., 2009; Lu et al., 2009) which at that time is freely accessible to disassembled NPC components (Walther et al., 2003a; Walther et al., 2003b). Consistent with previous reports (Franz et al., 2007; Gillespie et al., 2007; Rasala et al., 2006) ELYS appears to mediate the rate-limiting step of Nup107/160 complex recruitment to chromatin. Since in the absence of ELYS, NPCs can form in the ER as annulate lamellae, it appears that its major role is to spatially determine sites of NPC formation at the surface of chromatin. Another potentially critical aspect in evaluating the role of ELYS in NPC assembly is the cell cycle difference in the rate of NPC formation. While pores form in a matter of minutes between anaphase and telophase, interphase assembly is a much slower process that can last up to an hour (D’Angelo et al., 2006). Thus the rapid targeting of the Nup107/160 complex and possibly other NE components by ELYS (Rasala et al., 2008) could be rate-limiting specifically during this cell cycle stage. Interestingly, ELYS is conserved among metazoa (Rasala et al., 2006) with no obvious homolog identified in yeast, which undergo a closed mitosis, further supporting the idea of a post-mitotic specific function for ELYS in NPC assembly.
Incorporation of the Nup107/160 complex to an intact NE does not depend on ELYS and thus chromatin association of Nups is unlikely to be rate limiting in a compartmentalized interphase cell. To accommodate the difference in NE topology, interphase assembly seems to involve a distinct set of events, beginning with fusion of the inner and outer nuclear membranes. As INM and ONM fusion does not occur in the presence of an inhibitory POM121 antibody, POM121 appears to play a critical role, either directly or indirectly, in this process leading to subsequent recruitment of the Nup107/160 complex. Several lines of evidence indicate that during interphase, POM121 is present at forming pores before the Nup107/160 becomes incorporated. First, we readily detected NPC intermediates containing POM121 but not the Nup107/160 complex. Secondly, depletion of POM121 inhibits recruitment of both endogenous Nup107 and 3GFP-Nup133 to the NE, specifically in the context of a closed NE. Additionally we show that an inhibitory antibody against POM121 does not affect NPC insertion in the context of NE reformation in vitro, but specifically impairs new pore insertion during nuclear expansion. This contradicts a previous study showing POM121 is required for NE reformation in vitro (Antonin et al., 2005). However, in this study the NE formation assays were performed with membrane vesicles, whereas we recapitulated nuclear assembly with a preformed ER network, more accurately reflecting the in vivo situation (Anderson et al, 2007). It is thus likely that the defect observed by Antonin et al was related to a deficiency in membrane targeting or fusion, rather than a role of POM121 in post-mitotic pore assembly.
We have previously shown that NPC insertion into an intact NE occurs de novo from both sides of the NE (D’Angelo et al., 2006). Thus, it is interesting that a functional NLS in POM121 is specifically required for interphase NPC formation. Our results suggest that POM121 is delivered to the INM through existing NPCs, similar to the NLS-mediated transport of other INM proteins (King et al., 2006). This requirement for active transport might explain why POM121 is rate-limiting in interphase, but not at the end of mitosis when the NE is not closed and proteins have free access to the forming INM. While the exact role of POM121 in the INM/ONM fusion process remains to be determined, it is tempting to speculate that this transmembrane Nup is present both at the INM and ONM in order to participate in membrane bending, possibly in coordination with reticulons, also implicated in NPC assembly (Dawson et al., 2009).
It was recently suggested that the ALPS motif in metazoan Nup133 might have a role in NPC assembly (Guttinger et al., 2009). In support of this idea and in agreement with the specific topology of forming new pores in an intact NE, we found a membrane curvature-sensing motif in Nup133 mediates recruitment of the Nup107/160 complex to assembly sites during interphase. There is no consensus on the structural organization of the Nup107/160 complex in the NPC (Alber et al., 2007; Brohawn et al., 2008; Debler et al., 2008) and therefore it remains to be determined if the ALPS can interact with the highly curved pore membrane and play a structural role in the assembled NPC. Alternatively, the ALPS domain may be critical for the NPC assembly process in sensing curved membranes, ensuring that new NPCs are specifically assembled at sites of INM and ONM fusion. Consistent with this idea, X-ray crystallography shows the ALPS motif in the N-terminal domain of hNup133 is well exposed (Boehmer et al., 2008) and exhibits a high degree of conservation in metazoa (Berke et al., 2004).
ALPS domains have been identified in other proteins and shown to act as sensing domains for highly curved membranes (Drin et al., 2007). Though we cannot exclude the possibility that the ALPS domain of Nup133 is involved in bending, we do not think this is the case. First, the biophysical properties of the loop are designed for interaction with curved membranes, which are characterized by a looser packing of the lipids. (Antonny, 2006; Drin et al., 2007). While we did not observe a dramatic induction of curvature in cells over-expressing EGFP-ALPS, initial ONM and INM fusion intermediates exhibit a complex membrane topology with both positive and negative curvature of the membrane (Antonin, 2009), and it is conceivable that the ALPS motif of Nup133 can sense the positively curved edges of these intermediates, and potentially promote their progression towards fusion. Finally, membrane curvature sensing provides an elegant solution to the incorporation of scaffold Nups from both sides of the NE.
Since NPCs are critical for cell proliferation and homeostasis, very few Nup mutations have been associated with diseases or phenotypes, likely due to lethality of such mutations. However, mouse Nup133 was recently shown to be critical for embryonic development of the neuronal lineage (Lupu et al., 2008). In this study, Nup133-null neural progenitor cells did not efficiently differentiate into terminally differentiated neurons. Although the molecular basis of this developmental defect is unclear, our data suggest that in the absence of Nup133 the overall pore number might be lower in these animals. Since many neurons have been shown to contain high pore numbers (Hetzer et al., 2005), likely due to their dependency on rapid and reliable nucleo-cytoplasmic transport, development of the nervous system might be particularly vulnerable to low NPC numbers. This hypothesis remains to be tested and may provide new insights into the role of pore number in development and disease.
MATERIALS AND METHODS
DNA constructs
Most DNA constructs were cloned using the Gateway system from Invitrogen (Table S1). pDONR207 (Invitrogen) was modified by adding 2GFP into engineered restriction sites. rPOM121NLS and mNup133 were generated by PCR and cloned into pDONR207-2GFP; and then recombined with the pDEST53 to obtain final 3GFP fusions. The NES-tdTomato-NLS construct was obtained by adding NES (LQLPPLERLTL) with a N-terminal linker (GGGG) and NLS (PPKKKRKVQ) with the same linker to the C-terminus of tdTomato by PCR. This fusion was recombined into pCDNA6.2/C-Lumio. The ALPS domain of hNup133 (aa 247-269) was inserted by PCR between residues I172 and G175 of EGFP-N1 (Clontech). Empty vector was used as a control. Evrogen pKillerRed-C was modified by adding two KillerReds at the Xno-HindIII-Pst restriction sites. mNup133 was generated by PCR and cloned into this vector.
Two rounds of site directed mutagenesis were used to mutate the NLS of rPOM121 from KKKR-KRRR to AAAA-AAAA.
mNup133 I254D and mNup133 I254V mutants were generated by site directed mutagenesis. H2B-tdTomato was a gift from Tony Hunter’s lab.
Antibody production, Western blotting and Immunohistochemistry
Western blots were analyzed using the Odyssey colorimetric infrared detection system. Blots were stained with fluorescent secondary antibodies emitting at 680nm and/or 800nm (Invitrogen/Rockland). Primary antibodies are listed in Table S2. Fab fragments of the α-xPOM121 antibody were generated using the Pierce Micro Preparation kit.
Cell culture and transfection
U2OS cells were grown in DMEM with 10% FBS with 1× antibiotic-antimycotic (Invitrogen). Cells were plated on coverslips or in 6 well plates 12 to 24h prior to transfection with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Either 1 or 5μg of DNA plasmids or, 50 or 250 pmol of siRNA duplexes were used for transfection of cells plated in 24 or 6 well plates, respectively. For increased siRNA knockdown efficiency, cells were transfected again 48h later. Cells were fixed or lysed 24–48 hours after the second transfection. siRNA oligo duplexes are from Invitrogen (Table S3).
For synchronization, cells were treated with Nocodazole (0.6 μg/ml) for 18 hrs. Mitotic cells were collected by shake off, washed in PBS and plated on coverslips in fresh medium. For G1 and G2 time points, cells were fixed and immunostained 5 and 19 hours later. For the Leptomycin B experiment U2OS were treated with 100 nM LMB for 2.5 hrs 24 hrs post transfection. Cells were washed in PBS, fixed in 4% PFA and stained with Hoechst dye.
Cell fractionation and immuno-precipitation
U2OS cells were plated in one 6-well plate per condition (2.105 cells per well) and transfected with GFP-mNup133 wt, GFP-mNup133 ID or GFP-mNup133 IV for 48h. Cells were collected in PBS, pelleted and resuspended in 150 μl Hypotonic buffer (Table S4). Cells were left 15 min on ice and lysed with a 27.5 gauge needle until 90% broken. Nuclei were pelleted by centrifugation (2,000g for 5 min) and resuspended in denaturing buffer (Table S4). Residual membranes were cleared by ultracentrifugation (200,000g for 30 min). 100mM NaCl and 1% Triton, final concentrations, were added to cytosolic fractions. Immunoprecipitation was performed on precleared lysates, using GFP-Trap beads (Allele, 2h at 4°C). Beads were washed 3X in Hypotonic buffer with 100mM NaCl and 1% Triton, resuspended in loading buffer and analyzed by Western Blotting.
Image analysis and statistics
Western blot bands were quantified and normalized to loading controls using Odyssey software.
To determine total NPC number, cells were immunostained with mAb414. For each nucleus, the area and average fluorescence intensity were measured from maximum projections of 20 confocal z-series spanning the entire nucleus, using either Image J or Adobe Photoshop. Numerical data were analyzed and summarized graphically using Excel (Microsoft). Data for each experiment were collected from at least three independent experiments and combined for statistical analysis.
To quantify recruitment of 3GFP-Nup133 during post-mitotic assembly, the contour of chromatin was traced using Adobe Photoshop and transformed into a fringe. Due to the heterogeneity in expression levels, the acquisition parameters and level adjustments differed between cells and thus the recruitment of 3GFP-Nup133 was quantified as the ratio of protein at the nuclear periphery to total protein, using the equation below: % 3GFP-Nup133 recruited to the nuclear periphery = fringe area × (avg fringe fluroescence − avg cytoplasm fluorescence)/[section area × (avg section fluorescence − background fluorescence)]. Live cell imaging was performed as described (Anderson et al., 2009).
Xenopus egg extract preparation and immunodepletion
Extract and sperm chromatin preparation, fluorescent labeling of membranes, and immunofluorescence were performed as previously described (Hetzer et al., 2000; Walther et al., 2003b). Immunodepletion of cytosol was performed by 3 cycles of 10 min incubation with either α-xELYS or α-xNup107 antibodies (Table S2) coupled to protein A Pansorbin cells (Calbiochem). Cleared cytosol was used to perform in vitro nuclear assembly reactions as previously described (Lohka and Masui, 1983). Depletion was confirmed by Western blotting.
SinBAD
The 9His-NTD of Nup133 wt, I254D or I254V were produced in 100 mL cultures of BL21(DE3) induced with 0.5 mM IPTG for 3h at 37°C. Cells were lysed in 10 mL of PBS, 1% Igepal and sonicated on ice. Lysates were centrifuged for 20 min at 14000 rpm (Sorvall, SS-35) and incubated with 200 μl of Ni-NTA beads (Qiagen) for 1 hr at 4°C. Elution was performed in 50mM Tris pH7.5, 150mM NaCl, 500mM Imidazole. The buffer was exchanged using NAP-10 columns (GE Healthcare) against 50mM Tris pH8.0, 150mM NaCl. Aliquoted proteins were stored at −80°C.
Liposomes were prepared as follows: a mixture of NBD-phosphatidylcholine (Avanti Polar Lipids) and unlabeled egg phosphatidylcholine (Sigma) was hydrated overnight in S250 buffer (Table S4) at a final concentration of 20mg/ml with a weight ratio of 1:40. After 5 freeze/thaw cycles, the vesicle suspension was extruded sequentially through polycarbonate filters with pore sizes of 0.4, 0.2, 0.1, 0.05 and 0.03 μm using a Mini-Extruder (Avanti Polar Lipids). Liposomes were stored at room temperature for up to 24h.
For the interaction assay, magnetic Ni-NTA beads (Qiagen) were incubated with 2μg of purified protein or PBS for 10 min at room temperature. Beads were washed and incubated with 20 μl of a 1:10 dilution of the liposome preparation for 10 min at room temperature. Beads were washed and imaged by confocal microscopy. Fluorescence intensity was quantified with Adobe Photoshop.
EM and FRAP experimental procedures are described in the Supplemental material.
Supplementary Material
Acknowledgments
We thank the Hunter lab for antibodies and the H2B-tomato reporter construct, and the Hetzer lab for critical discussion and reading of the manuscript, in particular Roberta Schulte for help with the Xenopus egg extract depletion experiments. We also thank Stephane Richard for help with design of the EGFP-ALPS fusion protein. This work was supported by Award Number R01GM057438 from the National Institute Of General Medical Sciences and a Core Cancer Grant (CA14195).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008a;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. The life cycle of the metazoan nuclear envelope. Curr Opin Cell Biol. 2008b;20:386–392. doi: 10.1016/j.ceb.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W. Nuclear envelope: membrane bending for pore formation? Curr Biol. 2009;19:R410–412. doi: 10.1016/j.cub.2009.03.053. [DOI] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The Integral Membrane Nucleoporin pom121 Functionally Links Nuclear Pore Complex Assembly and Nuclear Envelope Formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Antonny B. Membrane deformation by protein coats. Current opinion in cell biology. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JR, 3rd, Ferro-Novick S, Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature structural & molecular biology. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson C, Rustum C, Hallberg E. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 2004;572:261–265. doi: 10.1016/j.febslet.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Cisternal Organization of the Endoplasmic Reticulum during Mitosis. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Maul GG. On the octagonality of the nuclear pore complex. J Cell Biol. 1971;51:558–563. doi: 10.1083/jcb.51.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R, Talamas J, Doucet C, Hetzer MW. Single bead affinity detection (SINBAD) for the analysis of protein-protein interactions. PLoS One. 2008;3:e2061. doi: 10.1371/journal.pone.0002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006a;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Nautrup-Pedersen G, Cordes VC, Gorlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J Cell Biol. 2006b;173:477–483. doi: 10.1083/jcb.200601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.