Abstract
Viral particles in stool samples from wild-living chimpanzees were analysed using random PCR amplification and sequencing. Sequences encoding proteins distantly related to the replicase protein of single-stranded circular DNA viruses were identified. Inverse PCR was used to amplify and sequence multiple small circular DNA viral genomes. The viral genomes were related in size and genome organization to vertebrate circoviruses and plant geminiviruses but with a different location for the stem–loop structure involved in rolling circle DNA replication. The replicase genes of these viruses were most closely related to those of the much smaller (∼1 kb) plant nanovirus circular DNA chromosomes. Because the viruses have characteristics of both animal and plant viruses, we named them chimpanzee stool-associated circular viruses (ChiSCV). Further metagenomic studies of animal samples will greatly increase our knowledge of viral diversity and evolution.
INTRODUCTION
Viral metagenomics is an effective method for identifying previously uncharacterised viruses through recognition of encoded protein sequences related to those of known viruses (Edwards & Rohwer, 2005; Breitbart et al., 2002; Delwart, 2007; Allander et al., 2001). Studies of faecal samples from mammals using viral metagenomics have recovered numerous bacteriophages, plant viruses and eukaryotic viruses (Breitbart et al., 2003, 2008; Blinkova et al., 2009; Kapoor et al., 2008a, 2009; Li et al., 2009; Victoria et al., 2009; Finkbeiner et al., 2008; Zhang et al., 2006; Nakamura et al., 2009; Chiu et al., 2008).
Viruses with small (<10 kb) circular DNA genomes, either single- or double-stranded, have been found to infect vertebrates as well as plants (Fauquet et al., 2005). The single-stranded circular viral genomes infecting vertebrates consist of the highly diverse Anellovirus genus (including the species torque teno virus, torque teno mini virus and small anellovirus/torque teno midi virus) (Okamoto, 2009) and the Circoviridae family (consisting of the Gyrovirus and Circovirus genera) (Todd et al., 2001a; Fauquet et al., 2005). Double-stranded circular animal DNA viruses consist of the Papillomaviridae and the Polyomaviridae families (Fauquet et al., 2005). Single-stranded circular genome viruses infecting plants include the Geminiviridae and Nanoviridae families. Recently a novel group of small (1.7–2.3 kb) single-stranded (ss)DNA circular genomes, distantly related to gyrovirus chicken anemia virus (CAV), was characterized in sea turtles (Ng et al., 2009). Single-stranded circular viral genomes are thought to replicate through a rolling-circle mechanism resembling that used by bacterial plasmids, possibly reflecting an evolutionary link between these small circular genomes (Cheung, 2006; Gibbs et al., 2006). Despite different host specificities, animal circoviruses and plant geminiviruses and nanoviruses share certain protein motifs in the replicase (rep) gene, suggesting evolution from a common ancestor. A host switch from plant to animal, possibly involving recombination with a picornavirus-like sequence has been postulated (Gibbs & Weiller, 1999). Circoviruses include known porcine and avian pathogens (Todd, 2004; Segales & Domingo, 2002; Neumann et al., 2007) while geminiviruses and nanoviruses include known plant pathogens (Nawaz-ul-Rehman & Fauquet, 2009).
Despite similarities in their replicase proteins, nanoviruses, geminiviruses and circoviruses differ in genome size and organization. Circoviruses have monopartite genomes of 1.8–2.3 kb, while nanoviruses have segmented genomes, including at least six circular ssDNA chromosomes, of about 1 kb each (Chu & Helms, 1988; Harding et al., 1991; Katul et al., 1997). Geminiviruses may be composed of one or two circular genomes of about 2.6–3.0 kb each, with a genome organization that is highly reminiscent of animal circoviruses (Niagro et al., 1998).
We describe here circular DNA genomes in chimpanzee stool samples, which encode replicase-like proteins most closely related to those of nanoviruses, but with a gene organization closer to that of circoviruses and geminiviruses.
RESULTS
Detection, amplification and sequence analysis of novel circular DNA genomes
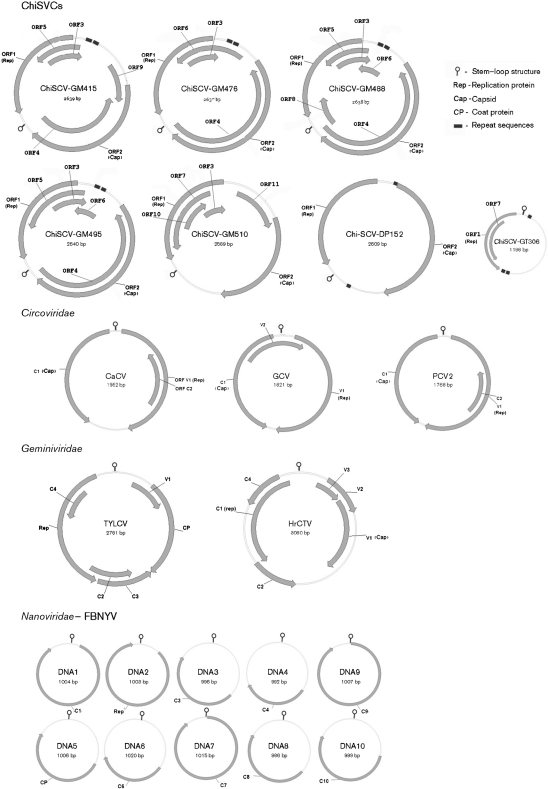
Viral particles in three stool samples (GM488, GM495 and GM510) of three chimpanzees (Ch-065, Ch-046 and Ch-080, respectively) from Gombe National Park, Tanzania, were partially purified using filtration and nuclease digestion of non-viral capsid protected naked host nucleic acids (Edwards & Rohwer, 2005; Breitbart et al., 2002; Delwart, 2007; Allander et al., 2001). Total viral nucleic acids were then extracted and the RNA was converted to cDNA. To identify RNA and DNA viruses, both cDNA and DNA were then amplified by random PCR (see Methods). Amplicons were subcloned into plasmids and 50 inserts were sequenced from each sample. blastx analyses identified three closely related short fragments that were distantly related to the replicase gene of both animal circoviruses and plant nanoviruses. Because both circovirus and nanovirus genomes are circular, inverse PCR was performed using outward pointing primers based on the shotgun-derived sequences. This approach generated amplicons that were 2000–2300 bp in length which were then sequenced by primer walking. Circular genome sequences were then assembled and the open reading frame (ORF) locations are shown in Fig. 1.
Fig. 1.
Genomic organization of ChiSCVs and representative members of the families Circoviridae, Geminiviridae and Nanoviridae. Circoviridae: goose circovirus (GCV) GenBank accession no. AAN37984, porcine circovirus 2 (PCV2) ABY82498 and canary circovirus (CaCV) AF246618. Geminiviridae: tomato yellow leaf curl virus (TYLCV) AF071228, horseradish curly top virus (HrCTV) U49907. Nanoviridae: faba bean necrotic yellows virus (FBNYV) DNA1, NC_005558; DNA2, NC_003560; DNA3, NC_003561; DNA4, NC_003562; DNA5, NC_003563; DNA6, NC_003564; DNA7, NC_003565; DNA8, NC_003566; DNA9, NC_003567; DNA10, NC_003559. The genome organization of GCV, PCV2, BFDV, TYLCV and HrCTV has been redrawn based on GenBank annotations and as described by Klute et al. (1996), Morilla et al. (2005), Todd et al. (2001a, b) and Phenix et al. (2001).
The genomes contained two major ORFs, including a putative replicase gene (ORF1, based on 18–20 % identity with the replicase of the nanoviruses subterranean clover stunt virus and faba bean necrotic yellow virus) and an ORF of unknown function (ORF2). We thus named this divergent group of viruses chimpanzee stool-associated circular viruses (ChiSCVs). The three genomes were called ChiSCV-GM488, ChiSCV-GM495 and ChiSCV-GM510.
Screening of stool and tissue genomic DNA samples for ChiSCV
To examine the frequency of the newly identified ChiSCVs, we screened 57 additional chimpanzee stool samples by nested PCR specific for ChiSCV. Including the three samples in which ChiSCV was originally discovered, 11 of 60 stool samples contained ChiSCV viral sequences (18 %). Based on mitochondrial (mt)DNA analysis, these represented seven different chimpanzees (Table 1). ChiSCV sequences were found in nine faecal samples (GM415, GM841, GM488, GM491, GM1062, GM1199, GM495, GM476 and GM510) from five Gombe chimpanzees (Ch-006, CH-045, Ch-046, Ch-065 and Ch-080). In addition, one faecal sample from Cameroon (DP152) and one from the Republic of the Congo (GT306) were ChiSCV-sequence-positive using degenerate PCR primers to anneal to both ChiSCV and circoviruses (see Methods). Of the seven animals shedding ChiSCV, three were seropositive for chimpanzee simian immunodeficiency virus (SIVcpz), all from Gombe, while the other four positive animals and the 46 ChiSCV-negative animals were SIVcpz sero-negative (except for one animal from primate centre 2) (Keele et al., 2009).
Table 1.
Detection of ChiSCV in wild-living and captive chimpanzees
| Faecal samples (chimpanzees) | ChiSCV-positive faecal samples (chimpanzees) | |
|---|---|---|
| Tanzania | 11 (7) | 9 (5) |
| Cameroon | 12 (12) | 1 (1) |
| Central African Republic | 2 (2) | 0 |
| Democratic Republic of the Congo | 12 (12) | 0 |
| Republic of the Congo | 2 (2) | 1 (1) |
| Rwanda | 2 (2) | 0 |
| Uganda | 4 (4) | 0 |
| Primate Center 1 | 8 (8) | 0 |
| Primate Center 2 | 7 (4) | 0 |
| Total number of samples (chimpanzees) | 60 (53) | 11 (7) |
To examine whether chimpanzees were systemically infected with ChiSCV, we tested necropsy samples from one of the chimpanzees with a positive faecal sample (Ch-045) who had died of trauma-related causes (Keele et al., 2009). None of five brain, spleen and lymph node samples were ChiSCV-positive. We also tested peripheral blood mononuclear cell DNA from five different captive chimpanzees. Again, none were ChiSCV-positive using both specific or degenerate primers. Finally, we screened a total of 662 human stool samples from the USA (Minnesota), South Asia (Afghanistan and Pakistan) and Africa (Nigeria and Tunisia) using both sets of primers, but found no evidence of ChiSCV-like viral sequences. Together, these results indicated that the new circular viruses were present in chimpanzee (but not human) faecal samples from a number of different locations in sub-Saharan Africa.
Genome organization of ChiSCVs
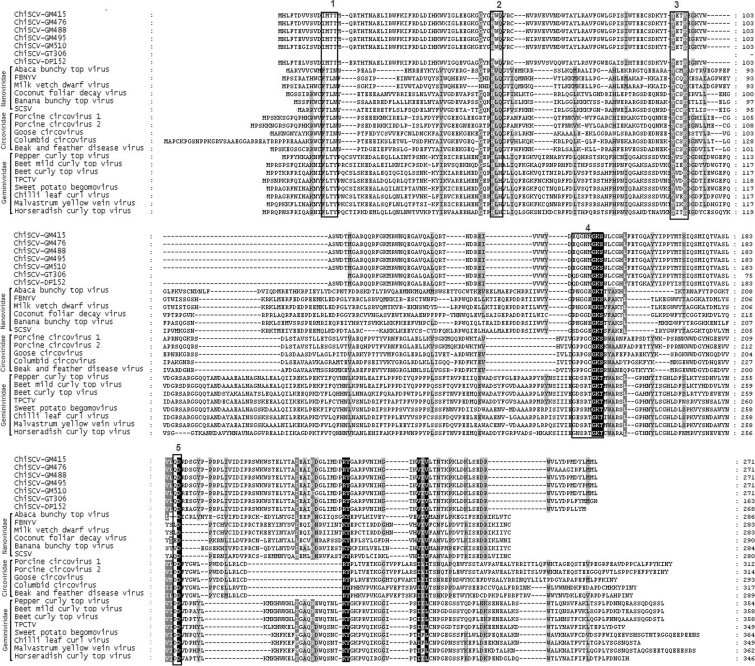
Inverse PCR was used to amplify full-length genomes from stool samples of all seven ChiCSV-positive chimpanzees (Table 1). This approach yielded genomes of 2589–2640 nt for ChiSCV-GM415, -GM476, -GM488, -GM495 and -GM510 from Gombe National Park and ChiSCV-DP152 from Cameroon. In contrast, ChiSCV-GT306 from the Republic of the Congo yielded an amplicon of only 1198 nt (Fig. 1). A protein alignment showed that some of the residues conserved among plant and vertebrate viral replicases were also found in the ChiSCV replicases (Fig. 2). Because alignments of replicase ChiSCV sequences with anelloviruses were the most problematic and blast analyses yielded the weakest E scores, further comparisons to anelloviruses were not pursued. Genomes from four Gombe chimpanzees Ch-006, Ch-045, Ch-046 and Ch-065 (ChiSCV-GM415, -GM476, -GM495 and -GM488) were most similar to each other, sharing 98–99 % of their nucleotide sequences (Table 2). The fifth ChiSCV-GM510 Gombe strain from chimpanzee Ch-080 was only 60–61 % identical to the other Gombe viruses (Table 2).
Fig. 2.
Alignment of the ORF1 replicase protein of ChiSCVs and representative members of the families Nanoviridae, Circoviridae and Geminiviridae. GenBank accession nos are given on Fig. 5(a). Amino acid motifs conserved between rolling-circle replication Rep proteins of different viruses are labelled 1–5 (boxed). Amino acid conservation of 100, >80 and >60 % is highlighted in black, dark grey and light grey, respectively.
Table 2.
Nucleotide sequence identity (%) among full-length ChiSCV genomes
| GM488 | GM495 | GM415 | GM476 | GM510 | DP152 | GT306 | |
|---|---|---|---|---|---|---|---|
| GM488 | 100 | ||||||
| GM495 | 98 | 100 | |||||
| GM415 | 98 | 99 | 100 | ||||
| GM476 | 98 | 98 | 98 | 100 | |||
| GM510 | 60 | 61 | 61 | 60 | 100 | ||
| DP152 | 55 | 55 | 55 | 54 | 81 | 100 | |
| GT306 | 67 | 67 | 67 | 66 | 64 | 69 | 100 |
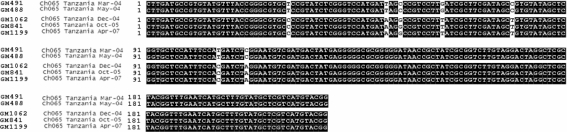
For four stool samples, all from the same Gombe chimpanzee (Ch-065), inverse PCR failed to amplify the circular genomes. The partial replicase PCR products from these latter samples were aligned with the genome sequence derived from a previously amplified Ch-065 stool sample (ChiSCV-GM488). Interestingly, the multiple partial replicase sequences from Ch065 fell into two groups, according to their time of sampling. ChiSCV-GM491 (March 2004) and ChiSCV-GM488 (May 2004) sequences were identical to each other, but differed from ChiSCV-GM1062 (December 2004), ChiSCV-GM841 (October 2005) and ChiSCV-GM1199 (April 2007) sequences in seven nucleotides (Fig. 3).
Fig. 3.
Alignment of partial ChiSCV replicase sequences amplified from consecutively collected stools samples from a single chimpanzee (Ch-065).
Replicase and putative capsid genes
Analysis of the ChiSCV genomes (Fig. 1) revealed two primary ORFs, in opposite orientations. ORF1 was 279–281 aa in length in most ChiSCV genomes, but only 163 aa in the smaller ChiSCV-GT306 genome (Figs 1 and 2). When the ChiSCV ORF1 was compared with the replicase gene of plant nanoviruses, the amino acid identity ranged from 15–24 %; decreasing identities were obtained for circoviruses, geminiviruses and gyroviruses, respectively (Table 3). The putative capsid gene, ORF2, encoded proteins of 352 to 420 aa in length. Based on blastx, ORF2 shared no significant similarity to any proteins listed in GenBank. A cluster of basic amino acids described in the capsid proteins of some circular ssDNA viruses (Ng et al., 2009; Niagro et al., 1998; Wilson et al., 1987; Rohde et al., 1990) was not found in ORF2 of ChiSCV. The putative capsid protein of ChiSCV included only 6–9 % basic amino acids (arginine and lysine) and only 1–5 arginines among the first 50 aa. Protein family analysis using the SVM-Prot software (Cai et al., 2003a, b) suggested that ORF2 proteins belonged to either coat protein or zinc-binding functional families, with an 84–96 % probability of correct classification.
Table 3.
Amino acid identity (%) among replicase proteins of nanoviruses, circoviruses, geminiviruses, gyrovirus and ChiSCV strains
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| ChiSCV | |||||||
| 1. ChiSCV-GM415 | 100 | ||||||
| 2. ChiSCV-GM476 | 100 | 100 | |||||
| 3. ChiSCV-GM488 | 100 | 100 | 100 | ||||
| 4. ChiSCV-GM495 | 99 | 99 | 99 | 100 | |||
| 5. ChiSCV-GM510 | 97 | 97 | 97 | 96 | 100 | ||
| 6. ChiSCV-GT306 | 95 | 95 | 95 | 95 | 95 | 100 | |
| 7. ChiSCV-DP152 | 88 | 88 | 88 | 87 | 88 | 95 | 100 |
| Nanovirus | |||||||
| Abaca bunchy top virus | 17 | 17 | 17 | 17 | 17 | 20 | 18 |
| FBNYV | 19 | 19 | 19 | 18 | 18 | 23 | 20 |
| Milk vetch dwarf virus | 19 | 19 | 19 | 18 | 19 | 24 | 20 |
| Coconut foliar decay virus | 16 | 15 | 16 | 16 | 15 | 19 | 17 |
| Banana bunchy top virus | 21 | 20 | 21 | 21 | 19 | 24 | 21 |
| SCSV | 20 | 20 | 20 | 20 | 19 | 21 | 22 |
| Circovirus | |||||||
| Porcine circovirus 2 | 16 | 16 | 16 | 15 | 16 | 15 | 17 |
| Porcine circovirus 1 | 14 | 14 | 14 | 14 | 14 | 13 | 16 |
| Goose circovirus | 17 | 16 | 17 | 16 | 16 | 16 | 17 |
| Columbid circovirus | 13 | 13 | 13 | 13 | 13 | 12 | 14 |
| Beak and feather disease virus | 14 | 14 | 14 | 14 | 14 | 13 | 15 |
| Geminivirus | |||||||
| Pepper curly top virus | 15 | 15 | 15 | 15 | 14 | 15 | 14 |
| Beet mild curly top virus | 13 | 13 | 13 | 14 | 12 | 15 | 13 |
| Beet curly top virus | 16 | 16 | 16 | 17 | 15 | 18 | 16 |
| Tomato pseudo-curly top virus | 13 | 13 | 13 | 14 | 12 | 13 | 14 |
| Sweet potato begomovirus | 14 | 14 | 14 | 15 | 13 | 14 | 14 |
| Chilli leaf curl virus | 14 | 14 | 14 | 15 | 13 | 13 | 13 |
| Malvastrum yellow vein virus | 13 | 13 | 13 | 14 | 12 | 12 | 12 |
| Horseradish curly top virus | 14 | 14 | 14 | 14 | 13 | 14 | 14 |
| Gyrovirus | |||||||
| Chicken anemia virus | 7 | 7 | 7 | 7 | 6 | 9 | 8 |
All ChiSCV genomes, except for DP152 and the shorter GT306 sequences, encoded an additional 3–5 ORFs (Fig. 2) with no significant homology to any previously reported proteins. ORFs given the same numerical designation were >60 % identical in their protein sequences, except for the putative capsid proteins of ORF2 which, in some cases, could not even be aligned. ORF3, 104–127 aa in length, was found in five viral genomes (ChiSCV-GM415, -GM476, -GM488, -GM495 and -GM510) in a conserved location and orientation. ORF4, 327–421 aa in length, was found in genomes ChiSCV-GM415, -GM476, -GM488 and -GM495. ORF5, 138–210 aa in length, was found in genomes ChiSCV-GM415, -GM488 and -GM495. OFR6, 105–210 aa in length, was found in genomes ChiSCV-GM476, -GM488 and -GM496. The short ChiSCV-GT306 genome did not encode ORF2, but contained ORF7 (111 aa in length), which also was found in GM510 (212 aa in length). ORF8 (104 aa), ORF9 (143 aa) and ORF10 (117 aa) were found only once in ChiSCV-GM488 and ChiSCV-GM510.
Intergenic regions, stem–loop structure and repeated sequences
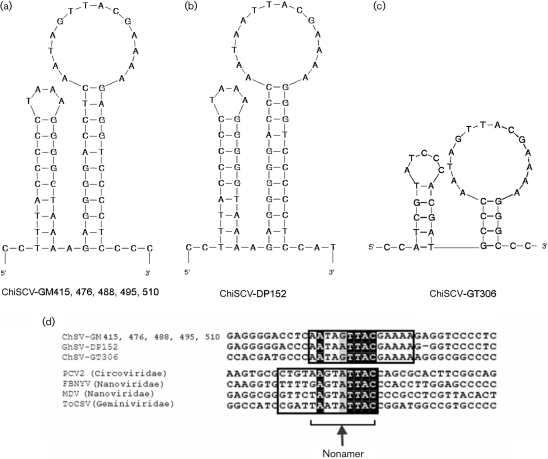
All ChiSCVs, except the small ChiSCV-GT306, contained two putative intergenic regions: one located between the initiation codons of the two major ORFs and another located downstream of their stop codons. A stem–loop containing a conserved nonanucleotide loop sequence, known to act as a binding site for the replicase protein in circoviruses (Steinfeldt et al., 2001), is located in the upstream intergenic region of circoviruses, nanoviruses and geminiviruses (Fig. 1). In ChiSCVs, a homologous stem–loop and nanonucleotide sequence were found, but at a different location in the downstream intergenic region (Figs 1 and 4). The nonanucleotide sequence was conserved among ChiSCV genomes and was identical at 5 of 9 nucleotides with the same region in nanoviruses, geminiviruses and circoviruses (Fig. 4). In the stem region, the ChiSCV-DP152 genome contained 3 of 11 base pairs that differed from the more common stem sequence seen in the other ChiSCV (excluding the small ChiSCV-GT306). Base pair complementarity was maintained through compensating changes in the annealing strand, supporting the hypothesis of a stem structure in this region. The major loop of all ChiSCV genomes was highly conserved (13 of 14 bases), even in the small ChiSCV-GT306 genome with a much-reduced stem. Another highly conserved, potential hairpin structure was also found immediately upstream of the stem–loop structure of the large ChiSCV genomes (Fig. 4).
Fig. 4.
Putative stem–loop and hairpin structures of (a) ChiSCV-GM415, -GM476, -GM488, -GM495, -GM510, (b) ChiSCV-DP152 and (c) ChiSCV-GT306. (d) Alignment of loop nonanucleotide sequences from ChiSCVs, circoviruses, nanoviruses and a geminivirus. Nucleotides that are 100% conserved between ChiSCV and other circular ssDNA viruses are highlighted in black. Boxed regions indicate loop sequences.
A TATAA box was identified in all ChiSCV genomes, located 26 bases upstream of the start codon of the replicase gene, except in the small ChiSCV-GT306 genome and in the more divergent DP152 genome, in which it was located 253 and 82 bp upstream of the replicase gene, respectively.
Several direct tandem repeats were found in all ChiSCV genomes, except GM510 (Fig. 1). Two direct repeats of 14 bp were found in ChiSCV-GM476, -GM495, -GM415 and -GM488. These repeats had the same sequence (AGGTCGTATGGAAG) and were located 113 bp upstream of the TATAA box. ChiSCV-GT152 had two widely separated repeated sequences (AGAAGGTACTAC). ChiSCV-GT306 had three repeated sequences (CCCCCTCCATC).
Phylogenetic analysis of ChiSCVs
Phylogenetic analysis of the complete replicase protein sequences showed that all ChiSCV variants were closely related to one another and formed an independent group that clustered separately from viruses in the Nanoviridae, Geminiviridae and Circoviridae families (Fig. 5a). Within the ChiSCV group, DP152 from Cameroon had a basal position, while viruses from Tanzania (GM488, GM495, GM415, GM476 and GM510) were more closely related. In terms of replicase amino acid sequence similarity, the ChiSCV replicase proteins were most similar to nanoviruses (15–24 % pairwise similarity) (Table 3). Geminiviruses and circoviruses were more divergent, exhibiting only 12–17 % protein sequence similarities each. Interestingly, a different topology was observed when capsid sequences were compared. Phylogenetic analysis of ChiSCV capsid protein sequences yielded trees in which the ChiSCV sequences clustered together into two groups: Tanzanian sequences (GM488, GM415, GM495 and GM476) and Congolese and Cameroonian sequence (GM510 and DP152) (Fig. 5b).
Fig. 5.
Phylogenetic analyses of (a) the ORF1 replicase genes of ChiSCVs and members of the families Geminiviridae, Nanoviridae and Circoviridae and (b) the ORF2 (putative capsid gene) of ChiSCVs. The GenBank accession no. of each sequence is shown on the figure.
Recombination between ChiSCVs
The different groupings obtained in phylogenetic analyses based on capsid versus replicase proteins suggest that recombination might have occurred within this group of viruses. A SimPlot analysis of full-length genomes of ChiSCV using GM510 as the reference (excluding GT306 because of its much shorter genome) showed that GM510 is more similar to DP152 in ORF2, but more similar to other ChiSCV genomes in ORF1, as expected from a recombination event (Supplementary Fig. S1, available in JGV Online).
Comparative sequence analysis of ORF1 and replicase regions of circoviruses, nanoviruses and geminiviruses
Alignment of the replicase proteins of ChiSCVs with those encoded by nanoviruses, circoviruses and geminiviruses identified several highly conserved amino acid motifs known to function in rolling circle replication and dNTP binding (Fig. 2) (Phenix et al., 2001; Bassami et al., 1998; Hattermann et al., 2003). All ChiSCVs (except short ChiSCV-GT306) had identical rolling-circle replication motifs. ChiSCV-DP152 and ChiSCV-GT306 differed from all other ChiSCV replicase genes in dNTP binding motifs. Finally, the replicase sequence of ChiSCV-DP152, an outlier in phylogenetic analyses (Fig. 5a), differed from all other ChiSCV replicase genes in rolling-circle replication motifs 1, 2 and 3 (Fig. 2).
DISCUSSION
In this study, we used metagenomic and pan-PCR approaches to identify novel viruses in chimpanzee faecal samples and characterized several circular DNA viral genomes related in sequence and genomic organization to both plant and animal viruses. Whether these genomes are single-stranded, like circoviruses, geminiviruses and nanoviruses, or double-stranded, like papillomaviruses, is not known. The viral genomes included two long ORFs encoded on opposite strands, a feature shared with members of the Circoviridae and Geminiviridae families. ChiSCV ORF 1 is 15–24 % similar in amino acid sequence to the replicase of the multi-segmented plant viruses of the Nanoviridae family but only 12–17 % similar to the replicases of circoviruses and geminiviruses. No region of significant sequence similarity to any anellovirus or bacterial plasmids was detected. The ChiSCV ORF2 presumably encodes a structural capsid protein, but blast searches of sequences in GenBank failed to identify any sequences with significant similarity. The ChiSCV genomes include short direct repeats, also reported in circoviruses, geminiviruses, anelloviruses and gyroviruses, and in sea turtle tornovirus 1. ChiSCV genomes therefore share features with plant nanoviruses and geminiviruses, as well as with animal circoviruses and gyroviruses. The presence of a stem–loop structure associated with rolling-circle replication (Vega-Rocha et al., 2007a, b), located between the 3′ ends of the two major ORFs, is unique to this new group of viruses. The combination of novel genomic features, limited similarity to the replicase gene product of other viruses, and the presence of ORFs without known homologues indicates that the ChiSCV variants may represent a new viral family.
In terms of genome size (2589–2639 bp), ChiSCVs are intermediate between geminiviruses and circoviruses (Fig. 1). Moreover, the various ChiSCV genomes differed in their size and structure; for example, GT306 was much smaller than six other variants, and DP152 differed in its 5′ intergenic spacing. These results suggest extensive genome size heterogeneity within this new group of viruses.
Some ChiSCV variants were nearly identical. For example, four of the five genomes identified in stool samples from Gombe chimpanzees were 98–99 % similar in nucleotide sequence (Table 2). GM510 (a fifth Gombe isolate) was very similar in its replicase gene sequence to the other Gombe strains (97 % protein identity), but very different in its capsid protein sequence, strongly suggesting recombination with a DP152-like ChiSCV genome (Fig. 5b). Recombination is common among both geminiviruses (Lefeuvre et al., 2009; Varsani et al., 2008; Padidam et al., 1995, 1999) and circoviruses (Ma et al., 2007; Heath et al., 2004). A large proportion (3 of 5) of the ChiSCV-shedding chimpanzees from the Gombe were sero-positive for SIVcpz (Keele et al., 2009). It is conceivable that SIVcpz-induced immunodeficiency (Keele et al., 2009) increased susceptibility or chronicity of infection with ChiSCV. It is also possible that since the ChiSCV PCR primers used were based on the viral genomes originally derived from Gombe chimpanzees, these primers efficiently detected local strains of ChiSCV but failed to amplify divergent strains in other chimpanzee communities. Further analyses of stool samples from different chimpanzee communities should uncover the geographical range and diversity of this new group of viruses.
The host of the newly identified ChiSCV strains remains to be determined. The presence of these viruses in chimpanzee stool suggests several possibilities. Either chimpanzees were productively infected with these viruses which may replicate in the digestive track, or the viruses infected plants that were ingested and excreted by chimpanzees (Victoria et al., 2009; Zhang et al., 2006). Still another possibility is that ChiSCV represents a consumed animal virus, since chimpanzees are known to hunt and eat other mammals. The detection of ChiSCVs in members of the same chimpanzee community may reflect virus transmission within that community or their consumption of the same infected plants or animals. Analysis of five consecutive stool samples from a single ape collected over several years revealed two different variants over time (Fig. 3). This observation might be explained by viral evolution or chronic infection followed by reinfection with a new strain. Consumption of plants containing different geminiviruses or other plant viruses may also account for the transition from one ChiSCV variant to another in this animal. Ultimately, tissue culture amplification in animal cells, detection in chimpanzee tissues or positive ChiSCV antibody tests will be needed to conclusively identify chimpanzees as the host species of these viruses.
Based on the estimated number of mammal species (n>5000), a greater characterization of their viruses is likely to uncover a very large amount of mostly still uncharacterized viral diversity. The identification of a potential new viral family after minimal shotgun sequencing (150 plasmid inserts) from three chimpanzee stools suggests a much larger reservoir of viral diversity than currently known. Given recent technical advances in massive parallel sequencing and bioinformatics, an in-depth exploration of this animal viral diversity is now within reach.
METHODS
Virus samples.
Chimpanzee faecal samples were selected from existing banks of specimens collected for molecular epidemiological studies of SIVcpz and chimpanzee simian foamy virus (Keele et al., 2006; Liu et al., 2008; Santiago et al., 2003; Worobey et al., 2004). Samples were collected between 1998 and 2007. The majority of samples (n=45) were collected from wild chimpanzees in Cameroon (n=12), the Central African Republic (n=2), the Republic of the Congo (n=2), the Democratic Republic of the Congo (n=12), Uganda (n=4), Rwanda (n=2) and Tanzania (n=11) (Table 1). All of these were subjected to mtDNA analysis to confirm their species and subspecies origin, and to identify individuals with different mtDNA haplotypes.
Nine ChiSCV-positive faecal samples from five chimpanzees were collected from habituated chimpanzees in Gombe National Park, Tanzania, including members of the Mitumba (Ch-065, Ch-046, Ch-080 and Ch-045) and Kasekela (Ch-006) communities. One of these (Ch-065) was sampled on five consecutive occasions between May 2004 and April 2007 (samples GM491, GM488, GM1062, GM841 and GM1199). Ch-006, Ch-045 and Ch-080 were naturally infected with SIVcpz (Keele et al., 2009).
Faecal samples (n=15) were also collected from 12 captive chimpanzees housed at primate facilities in the USA (Yerkes Regional Primate Center, Atlanta, Georgia; n=8) and the Netherlands (Biomedical Primate Research Centre, Rijswijk; n=7) (Table 1). All chimpanzee faecal samples were preserved in RNAlater as described previously (Keele et al., 2006).
Chimpanzee blood and tissue samples were obtained from six different individuals. Blood was collected from five captive chimpanzees as part of their annual health survey. Spleen and brain tissues as well as axillary, mesenteric and submandibular lymph nodes were obtained at necropsy from Ch-045 who died of trauma-related injuries (Keele et al., 2009). All studies were carried out in strict accordance with international guidelines for the ethical, scientific use and humane care of primates in research.
Human stool samples were obtained from the USA (Minnesota, n=186 including 50 % diarrhoea samples), South Asia (Afghanistan and Pakistan, n=107, all from children <15 years of age with non-poliovirus acute flaccid paralysis) and Africa (Nigeria and Tunisia, n=369 with 246 from children <15 years of age with non-poliovirus acute flaccid paralysis and 123 from healthy children in contact with acute flaccid paralysis patients). These samples were collected without a preservative and stored at −80 °C.
Viral particle nucleic acid purification was performed as described previously to enrich for viral particle protected DNA and RNA (Blinkova et al., 2009; Victoria et al., 2009, 2008; Kapoor et al., 2008a, b; Allander et al., 2001). Briefly, stool samples were thawed and resuspended in Hank's balanced salt solution and vortexed. Supernatant from samples centrifuged twice at 12000 r.p.m. (Eppendorf centrifuge 5415C) – to remove large particulate debris such as partially digested plant material – was filtered through a 0.45 μm filter (Millipore). Filtrate was treated with 14 U Turbo DNase (Ambion) and 2 μl 10 mg RNase A ml−1 at 37 °C for 2 h to digest non-particle-protected nucleic acids. Total nucleic acid was extracted from 140 μl filtrate, using a QIAamp viral RNA mini kit (Qiagen) and eluted into 50 μl water with 20 U recombinant RNase inhibitor (Roche).
Random amplification, subcloning and sequencing.
To generate cDNA, 10 μl total viral nucleic acid was mixed with 50 pmol (1 μl) of a random primer, primK (5′-GACCATCTAGCGACCTCCACNNNNNNNN-3′), denatured at 85 °C for 2 min and chilled on ice. A reaction mix (9 μl) containing 4 μl 5× Superscript III buffer (Invitrogen), 2 μl 100 mM DTT, 1.25 μl 10 mM dNTP solution, 0.75 μl DEPC-treated water and 1 μl superscript III reverse transcriptase (200 U) was added. The reaction was incubated at 25 °C for 10 min, 42 °C for 60 min and 70 °C for 5 min and chilled on ice for 2 min. For the second-strand cDNA synthesis, 0.5 μl 100 μM primK primer was added, and the reaction mix was incubated at 95 °C for 2 min and chilled on ice for 2 min. To extend primK, 2.5 U 3′–5′ exo- Klenow DNA polymerase (New England Biolabs) was added, and incubated at 37 °C for 60 min, followed by enzyme inactivation at 75 °C for 10 min. A 5 μl aliquot of the cDNA was used as template in a 50 μl PCR, including 1× AmpliTaq Gold DNA polymerase buffer (Applied Biosystems), 8 μl 25 mM MgCl2, 1.25 μl 10 mM dNTP, 5 μl 10 μM specific primer 5′-GACCATCTAGCGACCTCCAC-3′ and 2.5 U AmpliTaq Gold DNA polymerase L.D. (Applied Biosystems). The conditions for PCR were as follows: denaturation at 95 °C for 5 min, 5 cycles of 95 °C for 1 min, 55 °C for 1 min and 72 °C for 1.5 min, 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1.5 min, with an increase in extension time by 2 s per cycle, and a final extension at 72 °C for 10 min. Random PCR products were separated on a 1.5 % agarose gel, and the DNA smear ranging from 500 to 1500 bp was cut from the gel and extracted using the QIAquick gel extraction kit (Qiagen). The gel-extracted PCR product was ligated to the vector pGEMT-easy (Promega) and transformed into chemically competent Escherichia coli cells. Bacteria were plated on LB agar plates containing ampicillin, X-Gal and IPTG. Fifty white colony inserts were sequenced using the T-7 forward primer. Sequences were assembled in Sequencher 4.1 (Genecod), and a sequence similarity search was performed using blastx (http://www.ncbi.nlm.nih.gov/blast/).
Diagnostic PCR screening for ChiSCVs.
To assess the prevalence of ChiSCV, primers for nested PCR screening were designed. Both specific and degenerate nested PCR primer sets were used to screen samples. First, we used an alignment of ChiSCV replicase genes to design the following nested PCR primers: specifChiSCV-F1 (5′-TCCCGCGTTAGCGCTCAAA-3′), specifChiSCV-R1 (5′-TATTCGAGACAGGGCAGGC-3′), specifChiSCV-F2 (5′-TCCTTTACCCCCTAAAGGG-3′) and specifChiSCV-R2 (5′-ACTACATACCGCCGTACATGAC-3′). To create more degenerate primers, we used an alignment of replicase genes from both ChiSCVs and circoviruses, and we included mixed bases in the following primers: degenChiSCV-F1 (5′-GGIAACATYGGIAARWSITGG-3′), degenChiSCV-R1 (5′-GAGYTTRTGAAGYTTGGGYTT-3′), degenChiSCV-F2 (5′-CAGGCTTAYTAYATACCICCG-3′) and degenChiSCV-R2 (5′-CAGAGDATAGYTTGATGCC-3′). For the first round of nested PCR, 3 μl template DNA was mixed with 5 μl 10× ThermoPol Reaction buffer (New England Biolabs), 5 μl 10 μM dNTP, 2.5 μl of each 10 μM primer (specifChiSCV-F1 and specifChiSCV-R1, or degenChiSCV-F1 and degenChiSCV-R1), 1 μl Taq DNA Polymerase (New England Biolabs) and 31 μl DEPC-treated water. The PCR conditions were as follows: denaturation at 95 °C for 3 min, 5 cycles of 95 °C for 1 min, 54 °C for 1 min and 72 °C for 1 min, 35 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 45 s, and a final extension at 72 °C for 10 min. For the second round of nested PCR, the reaction mix included 1.5 μl of PCR product from the first round, 5 μl 10× ThermoPol reaction buffer (New England Biolabs), 5 μl 10 μM dNTP, 2.5 μl each 10 μM primer (specifChiSCV-F2 and specifChiSCV-R2, or degenChiSCV-F2 and degenChiSCV-R2), 1 μl Taq DNA Polymerase (New England Biolabs) and 32.5 μl DEPC-treated water. PCR conditions for the second round were identical to the first-round conditions. Products were visualized by electrophoresis on a 1.5 % agarose gel. PCR products of the expected size (365 bp for specific primers and 220 bp for degenerate primers) were purified using a Qiagen PCR purification kit and directly sequenced. GenBank accession numbers of the seven full ChiSCV genomes are GQ351272–GQ351278.
Phylogenetic analysis and protein function prediction.
Reference sequences of the rep gene from the Circoviridae, Geminiviridae and Nanoviridae families were obtained from NCBI. Sequence alignments were generated using the clustal w package (mega4) with the default settings and edited in genedoc software. Aligned sequences were trimmed to match the genomic region of the sequences obtained in this study and used to generate phylogenetic trees in mega4 using either neighbour-joining, maximum-likelihood or maximum-parsimony, with bootstrap values calculated from 1000 replicates.
Putative ORFs in the genome and circular genome architecture were predicted using Vector NTI 10.3.0 (Invitrogen) with the following conditions: minimum ORF size of 100 codons, start codons ATG and GTG, stop codons TAA, TGA and TAG. Across different ChiSCV genomes, ORFs with the same orientation and similar genome location that encoded proteins with amino acid similarity >60 % were considered homologous (except for some ORF2 putative capsid pairs whose predicted protein sequences were difficult to align).
Putative protein function was predicted using SVMProt, web-based software that predicts protein function based on an analysis of the physico-chemical properties of a protein generated from its sequence (http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi) (Cai et al., 2003a, b).
Repeats were analysed with tandem repeats finder (http://tandem.bu.edu/trf/trf.html) (Benson, 1999) and by aligning the sequence with itself using blast. To identify hairpin and stem–loop structures, nucleotide sequences were analysed with the Mfold web server (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi) (Zuker, 2003).
Recombination analysis.
Plots showing the similarity among the aligned nucleotide sequences across the length of the genome were generated using SimPlot, version 3.5.1 (http://sray.med.som.jhmi.edu/SCRoftware) (Lole et al., 1999). Similarity was calculated in each window of 400 nt by the Kimura two-parameter method. The overall transition/transversion ratio (Ts/Tv) was calculated using mega4 software. To assess potential recombinational relationships, aligned sequences were subsequently analysed by using the bootscanning method implemented in SimPlot.
Supplementary Material
Acknowledgments
We thank Dr Michael P. Busch, the Blood Systems Research Institute, and NHLBI grant R01HL083254 to E. L. D. for support for the laboratory-based studies. We thank Dr Gerardo R. Argüello Astorga of the Deptartment of Molecular Biology, IPICYT, San Luis Potosi, S.L.P. Mexico, for help with the replicase protein alignment. We also thank Cecile Neel, Aimee Mebenga, Innocent Ndong Bass and Eitel Mpoudi-Ngole for sample collection in Cameroon; the staff of Gombe Stream Research Centre including Hilali Matama, Simon Yohana, Gabo Paulo and Tofiki Mikidadi for collecting samples, and Dr Anthony Collins and Shadrack Kamenya for logistical support in Gombe National Park (Tanzania); David Morgan for sample collection in the Goualougo Triangle (Republic of the Congo); Martin Muller for sample collection in the Kyambura Gorge and Kibale National Park (Uganda); Nicole Gross-Camp for sample collection in the Nyungwe National Park (Rwanda); Michael A. Huffman for sample collection in Mahale Mountain National Park (Tanzania); the Cameroonian Ministries of Health, Environment and Forestry and Research for permission to collect samples in Cameroon; the Republic of Congo Ministry of Science and Technology and Ministry of Forest Economy for permission to collect samples in the Goualougo Triangle; the Tanzania National Parks, the Tanzania Commission for Science and Technology and the Tanzania Wildlife Research Institute for permission to conduct research in Gombe Stream and Mahale Mountain National Parks; the Uganda Wildlife Authority, the Uganda National Council for Science and Technology and the Makerere University Biological Field Station for permission to conduct research in Kibale; the Rwandan Office of Tourism and National Parks for permission to collect samples in Nyungwe National Park; the Department of Ecology and Management of Plant and Animal Resources (University of Kisangani) for authorization to collect samples in the Democratic Republic of the Congo; the staff of Yerkes and Rjiswijk Primate Research Centers for providing faecal and PBMC samples from captive chimpanzees; and Joann Schumacher-Stankey for demographic records of Gombe chimpanzees. This work was supported by grants from National Institutes of Health (R01 AI50529, R01 AI58715 and R01 AI44596), the US Fish and Wildlife Great Ape Conservation Fund, the Arcus, Davee and Guthman Foundations, the Bristol Myers Freedom to Discover Program and the Jane Goodall Institute.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the seven full ChiSCV genomes are GQ351272–GQ35127.
A supplementary figure showing recombination analyses is available with the online version of this paper.
References
- Allander, T., Emerson, S. U., Engle, R. E., Purcell, R. H. & Bukh, J. (2001). A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A 98, 11609–11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassami, M. R., Berryman, D., Wilcox, G. E. & Raidal, S. R. (1998). Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology 249, 453–459. [DOI] [PubMed] [Google Scholar]
- Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova, O., Kapoor, A., Victoria, J., Jones, M., Wolfe, N., Naeem, A., Shaukat, S., Sharif, S., Alam, M. M. & other authors (2009). Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J Virol 83, 4631–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M., Salamon, P., Andresen, B., Mahaffy, J. M., Segall, A. M., Mead, D., Azam, F. & Rohwer, F. (2002). Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci U S A 99, 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M., Hewson, I., Felts, B., Mahaffy, J. M., Nulton, J., Salamon, P. & Rohwer, F. (2003). Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185, 6220–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M., Haynes, M., Kelley, S., Angly, F., Edwards, R. A., Felts, B., Mahaffy, J. M., Mueller, J., Nulton, J. & other authors (2008). Viral diversity and dynamics in an infant gut. Res Microbiol 159, 367–373. [DOI] [PubMed] [Google Scholar]
- Cai, C. Z., Han, L. Y., Ji, Z. L., Chen, X. & Chen, Y. Z. (2003a). SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res 31, 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, C. Z., Wang, W. L., Sun, L. Z. & Chen, Y. Z. (2003b). Protein function classification via support vector machine approach. Math Biosci 185, 111–122. [DOI] [PubMed] [Google Scholar]
- Cheung, A. K. (2006). Rolling-circle replication of an animal circovirus genome in a theta-replicating bacterial plasmid in Escherichia coli. J Virol 80, 8686–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C. Y., Greninger, A. L., Kanada, K., Kwok, T., Fischer, K. F., Runckel, C., Louie, J. K., Glaser, C. A., Yagi, S. & other authors (2008). Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A 105, 14124–14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, P. W. & Helms, K. (1988). Novel virus-like particles containing circular single-stranded DNAs associated with subterranean clover stunt disease. Virology 167, 38–49. [DOI] [PubMed] [Google Scholar]
- Delwart, E. L. (2007). Viral metagenomics. Rev Med Virol 17, 115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R. A. & Rohwer, F. (2005). Viral metagenomics. Nat Rev Microbiol 3, 504–510. [DOI] [PubMed] [Google Scholar]
- Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A. (2005). Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Academic Press.
- Finkbeiner, S. R., Allred, A. F., Tarr, P. I., Klein, E. J., Kirkwood, C. D. & Wang, D. (2008). Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4, e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, M. J. & Weiller, G. F. (1999). Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci U S A 96, 8022–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, M. J., Smeianov, V. V., Steele, J. L., Upcroft, P. & Efimov, B. A. (2006). Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol Biol Evol 23, 1097–1100. [DOI] [PubMed] [Google Scholar]
- Harding, R. M., Burns, T. M. & Dale, J. L. (1991). Virus-like particles associated with banana bunchy top disease contain small single-stranded DNA. J Gen Virol 72, 225–230. [DOI] [PubMed] [Google Scholar]
- Hattermann, K., Schmitt, C., Soike, D. & Mankertz, A. (2003). Cloning and sequencing of Duck circovirus (DuCV). Arch Virol 148, 2471–2480. [DOI] [PubMed] [Google Scholar]
- Heath, L., Martin, D. P., Warburton, L., Perrin, M., Horsfield, W., Kingsley, C., Rybicki, E. P. & Williamson, A. L. (2004). Evidence of unique genotypes of beak and feather disease virus in southern Africa. J Virol 78, 9277–9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Victoria, J., Simmonds, P., Slikas, E., Chieochansin, T., Naeem, A., Shaukat, S., Sharif, S., Alam, M. M. & other authors (2008a). A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A 105, 20482–20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Victoria, J., Simmonds, P., Wang, C., Shafer, R. W., Nims, R., Nielsen, O. & Delwart, E. (2008b). A highly divergent picornavirus in a marine mammal. J Virol 82, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Slikas, E., Simmonds, P., Chieochansin, T., Naeem, A., Shaukat, S., Alam, M. M., Sharif, S., Angez, M. & other authors (2009). A newly identified bocavirus species in human stool. J Infect Dis 199, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katul, L., Maiss, E., Morozov, S. Y. & Vetten, H. J. (1997). Analysis of six DNA components of the faba bean necrotic yellows virus genome and their structural affinity to related plant virus genomes. Virology 233, 247–259. [DOI] [PubMed] [Google Scholar]
- Keele, B. F., Van Heuverswyn, F., Li, Y., Bailes, E., Takehisa, J., Santiago, M. L., Bibollet-Ruche, F., Chen, Y., Wain, L. V. & other authors (2006). Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele, B. F., Jones, J. H., Terio, K. A., Estes, J. D., Rudicell, R. S., Wilson, M. L., Learn, G. H., Beasley, T. M., Schumacher-Stankey, J. & other authors (2009). Increased mortality and AIDS-like immmunopathology in wild chimpanzees infected with SIVcpz. Nature 460, 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klute, K. A., Nadler, S. A. & Stenger, D. C. (1996). Horseradish curly top virus is a distinct subgroup II geminivirus species with rep and C4 genes derived from a subgroup III ancestor. J Gen Virol 77, 1369–1378. [DOI] [PubMed] [Google Scholar]
- Lefeuvre, P., Lett, J. M., Varsani, A. & Martin, D. P. (2009). Widely conserved recombination patterns among single-stranded DNA viruses. J Virol 83, 2697–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Victoria, J., Kapoor, A., Naeem, A., Shaukat, S., Sharif, S., Alam, M. M., Angez, M., Zaidi, S. Z. & Delwart, E. (2009). Genomic characterization of novel human parechovirus type. Emerg Infect Dis 15, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Worobey, M., Li, Y., Keele, B. F., Bibollet-Ruche, F., Guo, Y., Goepfert, P. A., Santiago, M. L., Ndjango, J. B. & other authors (2008). Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog 4, e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole, K. S., Bollinger, R. C., Paranjape, R. S., Gadkari, D., Kulkarni, S. S., Novak, N. G., Ingersoll, R., Sheppard, H. W. & Ray, S. C. (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. M., Hon, C. C., Lam, T. Y., Li, V. Y., Wong, C. K., de Oliveira, T. & Leung, F. C. (2007). Evidence for recombination in natural populations of porcine circovirus type 2 in Hong Kong and mainland China. J Gen Virol 88, 1733–1737. [DOI] [PubMed] [Google Scholar]
- Morilla, G., Janssen, D., García-Andrés, S., Moriones, E., Cuadrado, I. M. & Bejarano, E. R. (2005). Pepper (Capsicum annuum) is a dead-end host for Tomato yellow leaf curl virus. Phytopathology 95, 1089–1097. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Yang, C. S., Sakon, N., Ueda, M., Tougan, T., Yamashita, A., Goto, N., Takahashi, K., Yasunaga, T. & other authors (2009). Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One 4, e4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz-ul-Rehman, M. S. & Fauquet, C. M. (2009). Evolution of geminiviruses and their satellites. FEBS Lett 583, 1825–1832. [DOI] [PubMed] [Google Scholar]
- Neumann, E. J., Dobbinson, S. S., Welch, E. B. & Morris, R. S. (2007). Descriptive summary of an outbreak of porcine post-weaning multisystemic wasting syndrome (PMWS) in New Zealand. N Z Vet J 55, 346–352. [DOI] [PubMed] [Google Scholar]
- Ng, T. F., Manire, C., Borrowman, K., Langer, T., Ehrhart, L. & Breitbart, M. (2009). Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J Virol 83, 2500–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niagro, F. D., Forsthoefel, A. N., Lawther, R. P., Kamalanathan, L., Ritchie, B. W., Latimer, K. S. & Lukert, P. D. (1998). Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol 143, 1723–1744. [DOI] [PubMed] [Google Scholar]
- Okamoto, H. (2009). History of discoveries and pathogenicity of TT viruses. Curr Top Microbiol Immunol 331, 1–20. [DOI] [PubMed] [Google Scholar]
- Padidam, M., Beachy, R. N. & Fauquet, C. M. (1995). Classification and identification of geminiviruses using sequence comparisons. J Gen Virol 76, 249–263. [DOI] [PubMed] [Google Scholar]
- Padidam, M., Sawyer, S. & Fauquet, C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225. [DOI] [PubMed] [Google Scholar]
- Phenix, K. V., Weston, J. H., Ypelaar, I., Lavazza, A., Smyth, J. A., Todd, D., Wilcox, G. E. & Raidal, S. R. (2001). Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus circovirus of the family Circoviridae. J Gen Virol 82, 2805–2809. [DOI] [PubMed] [Google Scholar]
- Rohde, W., Randles, J. W., Langridge, P. & Hanold, D. (1990). Nucleotide sequence of a circular single-stranded DNA associated with coconut foliar decay virus. Virology 176, 648–651. [DOI] [PubMed] [Google Scholar]
- Santiago, M. L., Lukasik, M., Kamenya, S., Li, Y., Bibollet-Ruche, F., Bailes, E., Muller, M. N., Emery, M., Goldenberg, D. A. & other authors (2003). Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J Virol 77, 7545–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segales, J. & Domingo, M. (2002). Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet Q 24, 109–124. [DOI] [PubMed] [Google Scholar]
- Steinfeldt, T., Finsterbusch, T. & Mankertz, A. (2001). Rep and Rep′ protein of porcine circovirus type 1 bind to the origin of replication in vitro. Virology 291, 152–160. [DOI] [PubMed] [Google Scholar]
- Todd, D. (2004). Avian circovirus diseases: lessons for the study of PMWS. Vet Microbiol 98, 169–174. [DOI] [PubMed] [Google Scholar]
- Todd, D., McNulty, M. S., Adair, B. M. & Allan, G. M. (2001a). Animal circoviruses. Adv Virus Res 57, 1–70. [DOI] [PubMed] [Google Scholar]
- Todd, D., Weston, J. H., Soike, D. & Smyth, J. A. (2001b). Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286, 354–362. [DOI] [PubMed] [Google Scholar]
- Varsani, A., Shepherd, D. N., Monjane, A. L., Owor, B. E., Erdmann, J. B., Rybicki, E. P., Peterschmitt, M., Briddon, R. W., Markham, P. G. & other authors (2008). Recombination, decreased host specificity and increased mobility may have driven the emergence of maize streak virus as an agricultural pathogen. J Gen Virol 89, 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rocha, S., Byeon, I. J., Gronenborn, B., Gronenborn, A. M. & Campos-Olivas, R. (2007a). Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2. J Mol Biol 367, 473–487. [DOI] [PubMed] [Google Scholar]
- Vega-Rocha, S., Gronenborn, B., Gronenborn, A. M. & Campos-Olivas, R. (2007b). Solution structure of the endonuclease domain from the master replication initiator protein of the nanovirus faba bean necrotic yellows virus and comparison with the corresponding geminivirus and circovirus structures. Biochemistry 46, 6201–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria, J. G., Kapoor, A., Dupuis, K., Schnurr, D. P. & Delwart, E. L. (2008). Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog 4, e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria, J. G., Kapoor, A., Li, L., Blinkova, O., Slikas, B., Wang, C., Naeem, A., Zaidi, S. & Delwart, E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M. E., Mainprize, T. H., Friesen, P. D. & Miller, L. K. (1987). Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. J Virol 61, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey, M., Santiago, M. L., Keele, B. F., Ndjango, J. B., Joy, J. B., Labama, B. L., Dhed'A, B. D., Rambaut, A., Sharp, P. M. & other authors (2004). Origin of AIDS: contaminated polio vaccine theory refuted. Nature 428, 820. [DOI] [PubMed] [Google Scholar]
- Zhang, T., Breitbart, M., Lee, W. H., Run, J. Q., Wei, C. L., Soh, S. W., Hibberd, M. L., Liu, E. T., Rohwer, F. & Ruan, Y. (2006). RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.