Abstract
Having observed an m × n matrix X whose rows are possibly correlated, we wish to test the hypothesis that the columns are independent of each other. Our motivation comes from microarray studies, where the rows of X record expression levels for m different genes, often highly correlated, while the columns represent n individual microarrays, presumably obtained independently. The presumption of independence underlies all the familiar permutation, cross-validation, and bootstrap methods for microarray analysis, so it is important to know when independence fails. We develop nonparametric and normal-theory testing methods. The row and column correlations of X interact with each other in a way that complicates test procedures, essentially by reducing the accuracy of the relevant estimators.
Keywords: total correlation, effective sample size, permutation tests, matrix normal distribution, row and column correlations
1 Introduction
The formal statistical problem considered here can be stated simply: having observed an m × n data matrix X with possibly correlated rows, test the hypothesis that the columns are independent of each other. Relationships between the row correlations and column correlations of X complicate the problem’s solution.
Why are we interested in column-wise independence? The motivation in this paper comes from microarray studies, where X is a matrix of expression levels for m genes on n microarrays. In the “Cardio” study I will use for illustration there are m = 20426 genes each measured on n = 63 arrays, with the microarrays corresponding to 63 subjects, 44 healthy controls and 19 cardiovascular patients1. We expect the gene expressions to be correlated, inducing substantial correlations within each column (Owen, 2005; Efron, 2007a; Qiu, Brooks, Klebanov and Yakovlev, 2005a), but most of the standard analysis techniques begin with an assumption of independence across microarrays, that is, across the columns of X. This can be a risky assumption: all of the familiar permutation, cross-validation and bootstrap methods for microarray analysis, such as the popular SAM program of Tusher, Tibshirani and Chu (2001), depend on column-wise independence of X; dependence can invalidate the usual choice of a null hypothesis, as discussed next, leading to flawed assessments of significance.
An immediate purpose of the Cardio study is to identify genes involved in the disease process. For gene i we compute the two-sample t-statistic “ti” comparing sick versus healthy subjects. It will be convenient for discussion to convert these to z-scores,
| (1.1) |
with and Ф F61 the cumulative distribution functions (cdf) of standard normal and t61 distributions; under the usual assumptions, zi will have a standard N(0, 1) null distribution, called here the “theoretical null.” Unusually large values of zi or —zi are used to identify non-null genes, with the meaning of “unusual” depending heavily on column-wise independence.
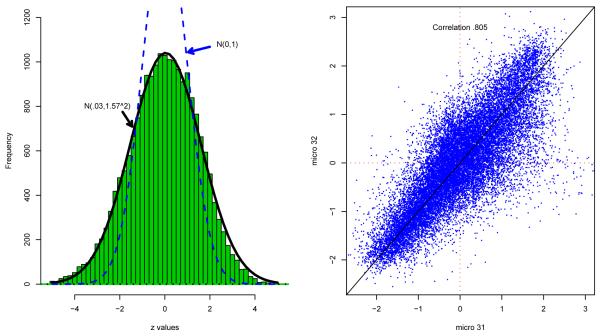
The left panel of Figure 1 shows the histogram of all 20426 zi values, which is seen to be much wider than N(0, 1) near its center. An “empirical null” fit to the center as in Efron (2007b) was estimated to be N(.03, 1.572). Null overdispersion has many possible causes (Efron, 2004, 2007a,b), one of which is positive correlation across the columns of X. Such correlations reduce the effective degrees of freedom for the t-statistic, causing (1.1) to yield overdispersed null zis, and of course changing our assessment of significance for outlying values.
Figure 1.
Left panel: histogram of m = 20426 z-values (1.1) for Cardio study; center of histogram is much wider than N(0, 1) theoretical null. Right panel: scatterplot of microarrays 31 and 32, (xi31, xi32) for i = 1, 2, …, m, after removal of row-wise gene means; the scattergram seems to indicate substantial correlation between the two arrays.
The right panel of Figure 1 seems to offer a “smoking gun” for correlation: the scattergram of expression levels for microarrays 31 and 32 looks strikingly correlated, with sample correlation coefficient .805. Here X has been standardized by subtraction of its row means, so the effect is not due to so-called ecological correlations. (X is actually “doubly standardized,” as defined in Section 2.) Nevertheless the question of whether or not correlation .805 is significantly positive turns out to be surprisingly close, as discussed in Section 4, because the row-wise correlations in X drastically reduce the degrees of freedom for the scatterplot. Despite the massive appearance of 20426 points, the scattergram’s accuracy is no more than would be given by 17 independent bivariate normal pairs.
Answering the title’s question, that is, testing for column-wise independence in the presence of row-wise dependence, has both easy and difficult aspects. Section 2 introduces a class of simple permutation tests which, in the case of the Cardio data, clearly discredit column-wise independence. However these tests depend on the ordering of the n columns, and can’t be used if the initial order is lost. It is natural and desirable to look for test statistics of column-wise independence that are invariant under permutation of the columns. Classical multivariate analysis, as in Anderson (2003), develops column independence tests in terms of the eigenvalues of an n by n Wishart matrix. However, this theory depends on the assumption of row-wise independence, disqualifying it for use here.
Sections 3 through 5 consider more general classes of independence tests, both from nonpara-metric and normal theory points of view. The theorem in Section 3 illustrates a key difficulty: correlation between the rows of X (ruled out in the classic theory) can give a misleading appearance of column-wise dependence. Similarly, row-wise dependence can greatly degrade the accuracy of the usual n × n sample covariance matrix of the columns, as shown by the theorem in Section 4. Various non-permutation normal-theory tests are discussed in Section 5, some promising, but with difficulties seen for all of them. The paper ends in Section 6 with a collection of remarks and details.
2 Permutation Tests of Column-Wise Independence
Simple permutation tests can provide strong evidence against column-wise independence, as we will see for the Cardio data. Our main example concerns the 44 healthy subjects, where X is now an m × n matrix with m = 20426 and n = 44. For convenience we assume that X has been “demeaned” by the subtraction of row and column means, giving
| (2.1) |
Our numerical results go further and assume “double standardization”: that in addition to (2.1),
| (2.2) |
i.e., that each row and column of X has mean 0 and variance 1; see Remark 6.4 in Section 6.
Let be the familiar estimate of the n × n covariance matrix between the columns of X,
| (2.3) |
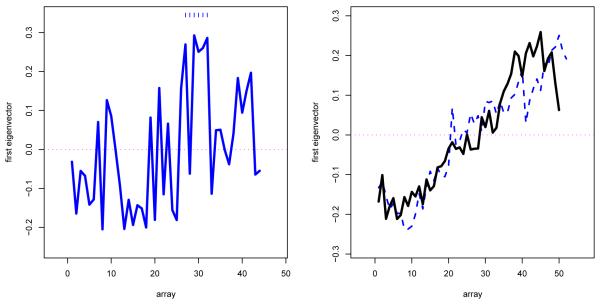
Under double standardization, is actually the sample correlation matrix, which we expect to be near the identity matrix In under column-wise independence. Also let v1 denote the first eigenvector of . The left panel of Figure 2 plots the components of v1 versus array number 1, 2, … , 44. Suppose that the columns of the original expression matrix, before standardization, are independent and identically distributed m-vectors (“i.i.d.”). Then it is easy to see, Remark 6.2 of Section 6, that all orderings of the components of v1 are equally likely. This is not what Figure 2 shows: the components seem to increase from left to right, with a noticeable block of large values for arrays 27-32.
Figure 2.
Left panel: Components of first eigenvector of row sample correlation matrix for the 44 healthy Cardio subjects, plotted versus array number 1, 2, … , 44; dashes emphasize the block of large components for arrays 27–32. Right panel: First eigenvectors for healthy (solid line) and cancer (dashed) subjects, prostate cancer study, Singh, Febbo, et al. (2002); there was a systematic drift in expression levels as the study progressed.
Let S(v1) be a statistic that measures structure, for instance a linear regression of v1 versus array index. Comparing S(v1) with a set of permuted values
| (2.4) |
v*l a random permutation of the components of v1, provides a quick test of the i.i.d. null hypothesis.
Permutation testing was applied to v1 for the Cardio data, using the “block” statistic
| (2.5) |
where B is the n × n matrix
| (2.6) |
The sum in (2.6) is over all vectors βh of the form
| (2.7) |
with the 1s forming blocks of length between 2 and 10 inclusive. A heuristic rationale for block testing appears below; intuitively, microarray experiments are prone to block disturbances because of the way they are developed and read; see Callow et al. (2000). After L = 5000 permutations, only three S* values exceeded the actual value S(v1), p-value .0006, yielding strong evidence against the i.i.d. null hypothesis.
The right panel of Figure 2 pertains to a microarray prostate cancer study (Singh et al., 2002) discussed in Efron (2008): m = 6033 genes were measured on each of n = 102 men, 50 healthy controls and 52 prostate cancer patients. The right panel plots first eigenvectors for , (2.3), computed separately for the healthy controls and the cancer patients (the two matrices being individually doubly standardized). Both vectors increase almost linearly from left to right. Taking S(v1) as the linear regression of v1 versus array number, permutation testing overwhelmingly rejected the i.i.d. null hypothesis, as it also did using the block test. The prostate study appears as a favorable example of microarray technology in Efron (2008). Nevertheless, Figure 2 indicates a systematic drift in the expression level readings as the study progressed. Some genes drift up, others down (the average drift equaling 0 because of standardization), inducing a small amount of column-wise correlation.
Section 5 discusses models for X where the n × n column covariance matrix is of the “single degree of freedom” form
| (2.8) |
for some known fixed vector β, the null hypothesis of column-wise independence being H0 : λ = 0. An obvious choice of test statistic in this situation is
| (2.9) |
a monotone increasing function of . If β is unknown we can replace Sβ with
| (2.10) |
where {β1, β2, … , βH, } is a catalog of “likely prospects” as in (2.7).
Permutation test statistics such as (2.5) can be motivated from the singular value decomposition (SVD) of X,
| (2.11) |
where K is the rank, d the diagonal matrix of ordered singular values, and U and V orthonormal matrices of sizes m × K and n × K,
| (2.12) |
IK the K × K identity. The squares of the diagonal elements, say
| (2.13) |
are the eigenvalues of X′X = V′d2V.
SB in (2.10) can now be written as
| (2.14) |
Model (2.8) suggests that most of the information against the null hypothesis H0 of independence lies in the first eigenvector v1, getting us back to test statistic as in (2.5).
What should the statistician do if column-wise independence is strongly rejected, as in the Cardio example? Use of an empirical null rather than a permutation or theoretical null, N (.03, 1.572) rather than N(0, 1) in Figure 1, removes the reliance on column-wise independence for hypothesis testing methods such as False Discovery Rates, at the expense of increased variability. Efron (2008) discusses these points.
Two objections can be raised to our permutation tests: (1) they are really testing i.i.d., not independence; (2) non-independence might not manifest itself in the order of v1 (particularly if the order of the microarrays has been shuffied in some unknown way).
Column-wise standardization makes the column distributions more similar, mitigating objection (1). Going further, “quantile standardization” — say replacing each column’s entries by normal scores (Bolstad, Irizarry, Åstrand and Speed, 2003) — makes the marginals exactly the same. The Cardio data was reanalyzed using normal scores, with almost identical results.
Objection (2) is more worrisome from the point of view of statistical power. The order in which the arrays were obtained should be available to the statistician, and should be analyzed to expose possible trends like those in Figure 22. It would be desirable, nevertheless, to have independence tests that do not depend on order — that is, test statistics invariant under column-wise permutations. The remainder of this paper concerns both the possibilities and difficulties in the development of “non-permutation” tests.
3 Row and Column Correlations
There is an interesting relationship between the row and column correlations of the matrix X, which complicates the question of column-wise independence. For the notation of this section define the n × n matrix of sample covariances between the columns of X as
| (3.1) |
called in Section 2, and likewise
| (3.2) |
for the m × m matrix of row-wise sample covariances (having more than 400, 000, 000 entries in the Cardio example!).
Theorem 1. If X has row and column means 0, (2.1), then the n2 entries of have empiricalmean 0 and variance c2,
| (3.3) |
with ek the eigenvalues (2.13), and so do the m2 entries of . Proof. The sum of ’s entries is
| (3.4) |
according to (2.1), while the mean of squared entries is
| (3.5) |
Replacing X′X with XX′ yields the same results for the row covariances .
Under double standardization (2.1)-(2.2), the covariances become sample correlations, say and for the columns and rows. Theorem 1 has a surprising consequence: whether or not the columns of X are independent, the column sample correlations will have the same mean and variance as the row correlations. In other words, substantial row-wise correlation can induce the appearance of column-wise correlation.
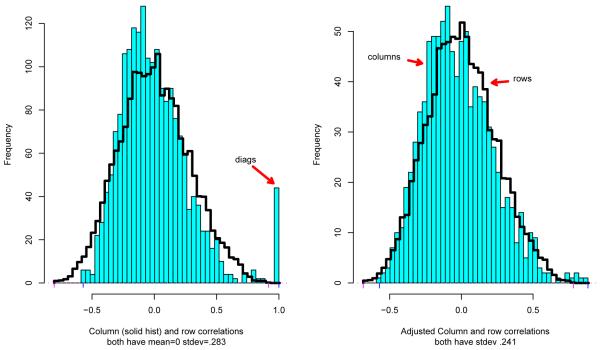
Figure 3 concerns the 44 healthy subjects in the Cardio study, with X an (m, n) = (20426, 44) doubly standardized matrix. All 442 column correlations are shown by the solid histogram, while the line histogram is a random sample of 10, 000 row correlations. Here c2 = .2832, so according to the Theorem both histograms have mean 0 and standard deviation .283.
Figure 3.
Left panel: solid histogram the 442 column sample correlations for X the doubly standardized matrix of healthy Cardio subjects; line histogram is sample of 10000 of the 204262 row correlations. Right panel: solid histogram the column correlations excluding diagonal 1s; line histogram the row correlations corrected for sampling overdispersion.
The 44 diagonal elements of protrude as a prominent spike at 1. (We can’t see the spike of 20426 diagonal elements for the row correlation matrix because they form such a small fraction of all 204262.) It is easy to remove the diagonal 1’s from consideration.
Corollary. In the doubly standardized situation, the off-diagonal elements of the column correlation matrix have empirical mean and variance
| (3.6) |
For n = 44 and c2 = .283 this gives
| (3.7) |
.
The corresponding diagonal-removing corrections for the row correlations (replacing n by m in (3.6)) are neglible for m = 20426. However c2 overestimates the variance of the row correlations for another reason: with only 44 points available to estimate each correlation, estimation error adds a considerable component of variance to the histogram in the left panel, as discussed next.
Suppose now that the columns of X are in fact independent, in which case the substantial column correlations seen in Figure 3 must actually be induced by row correlations, via Theorem 1. Let corii′ indicate the true correlation between rows i and i′ (that is, between Xij and Xi′j), and define α the total correlation to be the root mean square of the corii′ values,
| (3.8) |
.
Remark 6.5 of Section 6 shows that in (3.6) is an approximately unbiased estimate of α2, assuming column-wise independence. For the Cardio example , similar to the size of the microarray correlation estimates in Efron (2007a), Owen (2005), and Qiu et al. (2005a). Section 4 discusses the crucial role of α in determining the accuracy of estimates based on X.
The right panel of Figure 3 compares the histogram of the column correlations , now excluding cases j = j′, with the row correlation histogram corrected for sampling overdispersion via the shrinkage factor .241/.283. As predicted by Theorem 1, the similarity is striking. A possible difference lies in the long right tail of the distribution (including , the case illustrated in Figure 1), whose significance is examined in Section 4.
4 Normal Theory
The results of Sections 2 and 3 were developed nonparametrically. This section concerns multivariate normal theory, afterwards used in Section 5 to draw the connection with classical multivariate independence tests. We consider the matrix normal distribution for X,
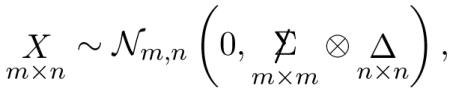 |
(4.1) |
where the Kronecker notation indicates covariance structure
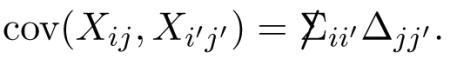 |
(4.2) |
Row xi of X has covariance matrix proportional to Δ,
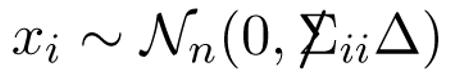 |
(4.3) |
(not independently across rows unless 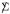 is diagonal), and likewise for column xj, xj ~ Nm(0, Δjj
is diagonal), and likewise for column xj, xj ~ Nm(0, Δjj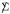 ). As in (2.1), we take all means equal 0.
). As in (2.1), we take all means equal 0.
Much of classical multivariate analysis focuses on the situation 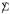 = I, where the rows xi are independent replicates3,
= I, where the rows xi are independent replicates3,
| (4.4) |
in which case the sample covariance matrix has a scaled Wishart distribution,
| (4.5) |
Distribution (4.5) has first and second moments
| (4.6) |
for j, k, l, h = 1, 2, … , n; see Mardia, Kent and Bibby (1979, p. 92).
Relation (4.6) says that when 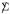 = I, that is when the rows of X are independent, unbiasedly estimates the row covariance matrix with accuracy proportional to m-1/2. Correlation between rows reduces the accuracy of , as shown next.
= I, that is when the rows of X are independent, unbiasedly estimates the row covariance matrix with accuracy proportional to m-1/2. Correlation between rows reduces the accuracy of , as shown next.
Returning to the general situation (4.1)-(4.3), define
| (4.7) |
where σ is the diagonal matrix with diagonal entries 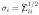 .
.
Theorem 2. Under model (4.1), has first and second moments
| (4.8) |
where α is the total correlation as in (3.8),
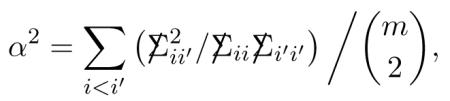 |
(4.9) |
and Δ(2) is the Wishart covariance (4.6).
Comparing (4.8) with (4.6), we see that correlation between the rows reduces “effective sample size” from m to : for α = .241 as in (3.7), the reduction is from m=20426 to (Notice that row standardization effectively makes (2.3), justifying the comparison.) The total correlation α shows up in other efficiency calculations; see Remark 6.7.
Proof. The row-standardized matrix X̃ = σ-1X has matrix normal distribution
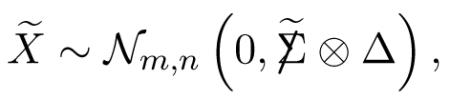 |
(4.10) |
where 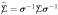 has diagonal elements
has diagonal elements  From (4.2) we see that
From (4.2) we see that 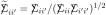 is the correlation between elements Xij and Xi′j in the same column of X; has entries and is unbiased for, Δ
is the correlation between elements Xij and Xi′j in the same column of X; has entries and is unbiased for, Δ
| (4.11) |
using (4.2).
The covariance calculation for involves expansion
| (4.12) |
| (4.13) |
Using the formula
| (4.14) |
for a normal vector (Z1Z2Z3Z4)′ with 0 means and covariances γij, (4.2) gives
| (4.15) |
and
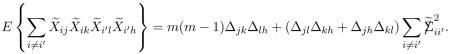 |
(4.16) |
Then (4.13) yields giving
| (4.17) |
giving
| (4.18) |
as in (4.8).
A corollary of Theorem 2, used in Section 5, concerns bilinear functions of and Δ and ,
| (4.19) |
where w is a given n-vector.
Corollary. Under model (4.1), has mean and variance
| (4.20) |
The proof follows that for Theorem 2; see Remark 6.9.
If 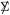 = I in (4.1), then and has a scaled chi-squared distribution,
= I in (4.1), then and has a scaled chi-squared distribution,
| (4.21) |
with mean and variance , so again the effect of correlation within 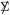 is to reduce the effective sample size from m to (4.8).
is to reduce the effective sample size from m to (4.8).
We can approximate (4.7), with
| (4.22) |
where is an estimate of 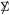 ii based on the observed variability in row i. If the rows of X have been standardized, then and returns to its original definition X′X/m.
ii based on the observed variability in row i. If the rows of X have been standardized, then and returns to its original definition X′X/m.
Both Theorem 2 and the Corollary encourage us to think of as, approximately, a scaled Wishart distribution based on an independent sample of size
| (4.23) |
The dangers of this approximation are discussed in Section 5, but it is, nevertheless, an evocative heuristic, as shown below.
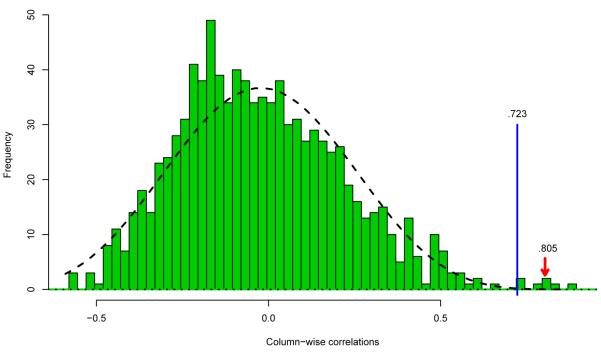
Figure 4 returns to the question of the seemingly overwhelming correlation .805 between arrays 31 and 32 seen in Figure 1. A one-sided p-value was calculated for each of the 946 column correlations, using as a null hypothesis the normal theory correlation coefficient distribution based on a sample size of = 17.2 pairs of N2(0, I) points (the correct null if Δ = I in (4.23)). Benjamini and Hochberg’s (1995) False Discovery Rate test, level q = .1, was applied to the 946 p-values. This yielded 7 significant cases, those with sample correlation .723; all 7 were from the block of arrays 27 to 32 indicated in Figure 2. Correlation .805 does turn out to be significant, but by a much closer margin than Figure 1’s scattergram suggests.
Figure 4.
Dashed curve is normal-theory null density for correlation coefficient from pairs of points; see Remark 6.6. Histogram is the 946 column correlations, right panel Figure 3. FDR test, q = .1, yielded 7 significant correlations, ≥.723, including .805 between arrays 31 and 32, Figure 1.
The Fdr procedure was also applied using the simpler null distribution N(—.023, .2412) (3.7). This raised the significance threshold from .723 to .780, removing two of the previously significant correlations.
Theorem 1 showed that the variance of the observed column correlations is useless for testing column-wise independence, since any value at all can be induced by row correlations. The test in Figure 4 avoids this trap by looking for unusual outliers among the column correlations. It does not depend on the order of the columns, objection (2) in Section 2 for permutation tests, but pays the price of increased modeling assumptions.
5 Other Test Statistics
Theorem 2 offers a normal-theory strategy for testing column-wise independence. We begin with X ~ Nm,n(0, 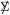 ⊗ Δ)(4.1), taking
⊗ Δ)(4.1), taking
| (5.1) |
as suggested by double standardization. The null hypothesis of column-wise independence is equivalent to the column correlation matrix equaling the identity,
| (5.2) |
since then (4.2) says that all pairs in different columns are independent.
To test (5.2), we estimate with Δ with , (4.22) or more simply after standardization, and compute a test statistic
| (5.3) |
where s(.) is some measure of distance between and 1. The accuracy approximation from (4.8), with Δ = 1, is used to assess the significance level of the observed S, maybe even employing the more daring approximation . Strategy (5.3) looks promising but, as the examples of this section will show, it suffers from serious difficulties that are absent under the classic assumption of independent rows.
One of the difficulties stems from Theorem 1. An obvious test statistic for H0 : Δ = I is
| (5.4) |
the average squared off-diagonal element of . But (3.1), so in the doubly standardized situation of (3.6), S is an increasing monotone function , the estimated total correlation. This disqualifies S as a test statistic for (5.2), since large values of can always be attributed to row-wise correlation alone.
Similarly, the variance of the eigenvalues (2.13),
| (5.5) |
looks appealing since the true eigenvalues all equal 1 when Δ = I. However (5.5) is also a monotonic function of ; see Remark 6.1.
The general difficulty here is “leakage,” the fact that row-wise correlations affect the observed pattern of column-wise correlations. This becomes clearer by comparison with classical multivariate methods, where row-wise correlations are assumed away by taking 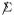 = I in (4.1). Johnson and Graybill (1972) consider a two-way ANOVA problem where, after subtraction of main effects, X has the form
= I in (4.1). Johnson and Graybill (1972) consider a two-way ANOVA problem where, after subtraction of main effects, X has the form
| (5.6) |
ai ~ N(0, λ) and εij ~ N(0, 1), all independently, with β = (β1, β2, …, βn) a fixed but unknown vector (representing “one degree of freedom for nonadditivity” in the two-way table X, Johnson and Graybill’s extension of Tukey’s procedure).
In the Kronecker notation (4.1), X ~ Nm,n(0,I ⊗ Δ) with
| (5.7) |
Now (5.2) becomes H0 : λ = 0. Johnson and Graybill show that, with β unknown, the likelihood ratio test rejects H0 for large values of the eigenvalue ratio (2.13),
| (5.8) |
Since the m rows of X are assumed independent, they can test H0 by comparison of S with values obtained from
| (5.9) |
as in (4.5).
Getting back to the correlated rows situation, Theorem 2 suggests comparing S with values S* from
| (5.10) |
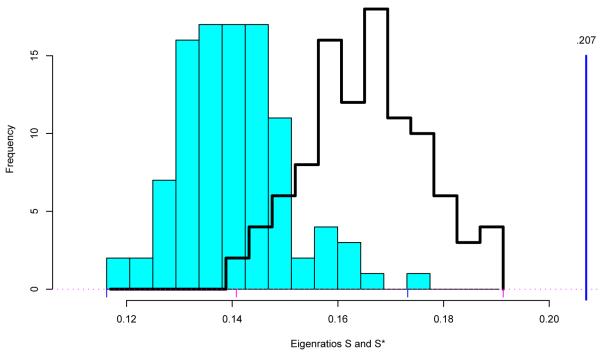
as in (4.8). The solid histogram in Figure 5 compares 100 S* values from (5.10), = 17.2 for the Cardio data, with the observed value S = .207 from the doubly standardized Cardio matrix for the healthy subjects used in Figure 3. All 100 S* values are much smaller than S, providing strong evidence against H0 : Δ = I.
Figure 5.
Eigenratio statistic (5.8) equals .207 for 20426 × 44 Cardio matrix X; solid histogram 100 simulations S* from Wishart (5.10), ; line histogram 100 simulations from correlated-row X* matrices (5.11), α = .241, Δ = I.
The evidence looks somewhat weaker, though, if we simulate S* values with obtained from random matrices
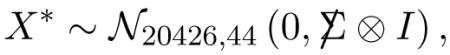 |
(5.11) |
doubly standardized, where 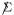 has total correlation α = .241, the estimated value for X, (4.9). The line histogram in Figure 5 shows 100 such S* values, all still smaller than S, but substantially less so. (Remark 6.8 describes the construction of X*.)
has total correlation α = .241, the estimated value for X, (4.9). The line histogram in Figure 5 shows 100 such S* values, all still smaller than S, but substantially less so. (Remark 6.8 describes the construction of X*.)
Why does (5.11) produce larger “null” S* values than (5.10)? The answer is simple: even though the first and second moments of match from (5.10), its eigenvalues do not. The non-zero eigenvalues of X*′X*/m equal those of 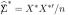 . This is another example of leakage, where the fact that
. This is another example of leakage, where the fact that 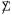 in (5.11) is not the identity Im distorts the estimated eigenvalue of even if Δ = In.
in (5.11) is not the identity Im distorts the estimated eigenvalue of even if Δ = In.
The eigenratio statistic S = e1/Σek is invariant under permutations of the columns of X, answering objection (2) to permutation testing of Section 2. Because of invariance, the eigenratio and permutation tests provide independent p-values for testing the null hypothesis of i.i.d. columns, and so can be employed together. Figure 5 is disturbing nonetheless, in suggesting that an appropriate null distribution for S depends considerably on the choice of the nuisance parameter 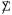 in (5.11).
in (5.11).
The bilinear form (4.19)-(4.20) yields another class of test statistics,
| (5.12) |
where w is a pre-chosen n-vector and . Delta-method arguments give for the coeffcient of variation of . Defining
| (5.13) |
yields the alternative form
| (5.14) |
.
In a two-sample situation like that for the Cardio study, sample sizes n1 and n2, we can choose
| (5.15) |
“1n” indicating a vector of n 1’s. This choice makes
| (5.16) |
the multiple of the mean response difference between the two samples that has variance 1 if Δ = I. In terms of (5.12), ∥w∥2 = 1 so τ2 = 1.
For the Cardio study, with n1 = 44, n2 = 19, and = 17.2, we obtain , coefficient of variation 0.17. This puts more than 2.8 standard errors above the null hypothesis value τ = 1, again providing evidence against column-wise independence. The Zi values from (5.16) are nearly indistinguishable from the zi values in Figure 1 — not surprisingly since with the rows of X standardized, Zi is an equivalent form of the two-sample t-statistic ti in (1.1).
Once again, however, there are difficulties with this as a test for column-wise independence. There is no question that the Zi’s are overdispersed compared to the theoretical value τ = 1. But problems other than column dependence can cause overdispersion, in particular unobserved covariate differences between subjects in the two samples (Efron, 2004, 2008).
The statistic in (5.15) does not depend upon the order of the columns of X within each of the two samples, answering objection (2) against permutation tests, but it is the only such choice for a two-sample situation. Other w’s might yield interesting results. The version of (5.15) comparing the first 22 healthy Cardio subjects with the second 22 provided the spectacular value , and here the “unobserved covariate” objection has less force.
Now, however, the test statistic depends on the order of the columns within the healthy subjects’ matrix, reviving objection (2). Again we might want to check a catalog of possible w vectors w1, w2, …, wH, leading back to test statistic
| (5.17) |
as in (2.10), the only difference being that the null distribution of now involves normal theory rather than permutations. Remark 6.9 shows that the null first and second moments of SB are similar to (5.12),
| (5.18) |
.
In summary, normal-theory methods are interesting and promising, but are not yet proven competitors for the permutation tests of Section 2.
6 Remarks
This section presents some brief remarks and details supplementing the previous material.
Remark 6.1. The constant c2 The variance constant c2 in Theorem 1 (3.3) can be expressed as
| (6.1) |
. so that c2 ≥ K(ē/mn)2, with equality only if the eigenvalues ek are equal. In the doubly standardized case ē = mn/K, giving
| (6.2) |
where K is the rank of X.
Remark 6.2. Permutation invariance If the columns of X are i.i.d. observations from a distribution on Rm, then the distribution of X is invariant under permutations: Xπ ~ X for any n × n permutation matrix π. Now suppose where L performs the same operation on each column of X, for example replacing each column by its normal scores vector. Then
| (6.3) |
showing that is permutation invariant.
Similarly, suppose , performing the same operation on each row of X, where now we require r(x)π = r(xπ) for all n-vectors x. The same argument as (6.3) demonstrates that is still permutation invariant. Iterating row and column standardizations as in Table 1 then shows that if the original data matrix X is permutation invariant, so is its doubly standardized version.
Table 1.
Successive row and column standardizations of the 20426 × 44 matrix of healthy Cardio subjects. “Col” empirical standard deviation of ; “Eig” from (3.6); “Row” from 1% sample of values, adjusted for overdispersion (6.6), sampling standard error :0034.
| Col | Row | Eig | Col | Row | Eig | ||
|---|---|---|---|---|---|---|---|
| demeaned | 0.252 | 0.286 | 0.000 | demeaned | 0.252 | 0.286 | 0.000 |
| col | 0.252 | 0.249 | 0.251 | row | 0.241 | 0.283 | 0.279 |
| row | 0.242 | 0.255 | 0.246 | col | 0.241 | 0.251 | 0.240 |
| col | 0.242 | 0.241 | 0.242 | row | 0.240 | 0.247 | 0.241 |
| row | 0.241 | 0.246 | 0.235 | col | 0.240 | 0.247 | 0.240 |
| col | 0.241 | 0.244 | 0.241 | row | 0.241 | 0.240 | 0.235 |
| row | 0.241 | 0.245 | 0.234 | col | 0.241 | 0.237 | 0.240 |
| col | 0.241 | 0.238 | 0.241 | row | 0.241 | 0.233 | 0.233 |
Remark 6.3. Covariances after demeaning Suppose that X is normally distributed, with covariances 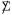 ⊗ Δ (4.2), all columns having the same expectation vector μ. Let be the demeaned matrix obtained by subtracting all the row and column means of X. Then
⊗ Δ (4.2), all columns having the same expectation vector μ. Let be the demeaned matrix obtained by subtracting all the row and column means of X. Then
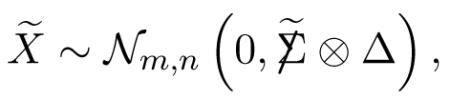 |
(6.4) |
where
| (6.5) |
dots indicating averaging over the missing subscripts, and similarly for 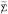 . This shows that de-meaning tends to reduce covariances by recentering them around 0.
. This shows that de-meaning tends to reduce covariances by recentering them around 0.
Remark 6.4. Standardization A matrix X is “column standardized” by individually subtracting the mean and dividing by the standard deviation of each column, and similarly for row standardization. Table 1 shows the effect of successive row and column standardizations on the 20426 × 44 demeaned matrix of healthy Cardio subjects. Here “Col” is the empirical standard deviation of the 946 column-wise correlations ; “Eig” is in (3.6); and “Row” is the empirical standard deviation of a 1% sample of the row correlations , but adjusted for overdispersion,
| (6.6) |
Sampling error of the Row entries is about ±.0034.
The doubly standardized matrix X used for Figure 3 was obtained after five successive column-row standardizations. This was excessive; the Figure looked almost the same after two iterations. Other microarray examples converged equally rapidly, though small counterexamples can be constructed where double standardization isn’t possible.
Microarray analyses usually begin with some form of column-wise standardization (Bolstad et al., 2003; Qiu, Klebanov and Yakovlev, 2005b), designed to negate “brightness” differences between the n microarrays. In the same spirit, row standardization helps prevent incidental gene differences (for example, very great or very small expression level variabilities) from obscuring the actual effects of interest. Standardization tends to reduce the apparent correlations as in Remark 6.3. Without standardization, the scatterplot in Figure 1 stretches out along the main diagonal, correlation .917, driven by genes with unusually large or small inherent expression levels.
Remark 6.5. Corrected estimates of the total correlation Suppose that the true row correlations corii′ have mean 0 and variance α2, as in (3.8) with , and that given corii’ , the usual estimate has mean and variance
| (6.7) |
(6.7) being a good normal-theory approximation (Johnson and Kotz, 1970, Chap. 32). Letting be the empirical variance of the values, a standard empirical Bayes derivation yields
| (6.8) |
as an approximately unbiased estimate of α2. (If is not assumed to equal 0, a slightly more complicated formula applies.) Of course if the right side of (6.8) is negative.
Theorem 1 implies that = 0 nearly equals c2, (3.3), in the doubly standardized situation. Formula (3.6), with say
| (6.9) |
is not identical to (6.8), but provides an excellent approximation for values of : with n = 44 and as in (3.6), while .
Remark 6.6. Column and row centerings The column correlation mean in (3.6) is forced by the row-wise demeaning Σj xij = 0, (2.1), centering the solid histogram in the right panel of Figure 3 at -.023. With m = 20426, the corresponding center for the line histogram is nearly 0, and the difference in the two centerings is noticeable. The dashed density curve in Figure 4, and the corresponding p-values for the FDR analysis, were shifted .023 units leftwards.
Remark 6.7. The total correlation α The total correlation α, which plays a key role in Theorem 2, (4.9), also is the central parameter of the theory developed in Efron (2007a). Equations (3.15)- (3.16) there are equivalent to (5.12) here. In both papers, α has the very convenient feature of summarizing the effects of an enormous m × m correlation matrix 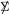 in a single number.
in a single number.
Remark 6.8. 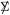 for simulation (5.11) The X* simulation used in Figure 5 began with m × n matrix Y = (yij),
for simulation (5.11) The X* simulation used in Figure 5 began with m × n matrix Y = (yij),
| (6.10) |
where I = 1, 2, 3, 4, 5 as i is in the first, second, …, last fifth of 1 through m; Y was then column standardized to give X*, so that 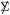 had a block form, with large positive correlations (about 0.61) in the (m/5) × (m/5) diagonal blocks. The choice λ = 1.23 was required to yield α = .241.
had a block form, with large positive correlations (about 0.61) in the (m/5) × (m/5) diagonal blocks. The choice λ = 1.23 was required to yield α = .241.
Remark 6.9. Bilinear statistics Since (4.8), it is clear that in Corollary (4.20). The variance calculation proceeds as in Theorem 2:
| (6.11) |
The verification of (5.18) is the same, except with element bjk of B replacing wjwk above, blh replacing wlwh, etc.
Footnotes
The entries of X are log(red/green) ratios obtained from oligonucleotide arrays.
The referee points out that when A ymetrix CEL files are available, array run dates will usually be found in the DatHeader lines.
Most multivariate texts reverse the situation, taking the columns as independent replicas of possibly correlated rows.
References
- Anderson TW. An Introduction to Multivariate Statistical Analysis. Third Wiley, New York: 2003. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M. Irizarry, Speed TP. Comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. http://web.mit.edu/biomicro/education/RMA.pdf Available at. [DOI] [PubMed]
- Callow M, Dudoit S, Gong E, Speed T, Rubin E. Microarray expression profiling identifies genes with altered expression in HDL-deficient mice. Genome Research. 2000;10:2022–2029. doi: 10.1101/gr.10.12.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: The choice of a null hypothesis. J. Amer. Statist. Assoc. 2004;99:96–104. [Google Scholar]
- Efron B. Correlation and large-scale simultaneous significance testing. J. Amer. Statist. Assoc. 2007a;102:93–103. [Google Scholar]
- Efron B. Size, power, and false discovery rates (2007) Ann. Statist. 2007b;35:1351–1377. [Google Scholar]
- Efron B. Microarrays, empirical Bayes, and the two-groups model. Statist. Sci. 2008;23:1–47. with discussion and Rejoinder. [Google Scholar]
- Johnson DE, Graybill FA. An analysis of a two-way model with interaction and no replication. J. Amer. Statist. Assoc. 1972;67:862–868. [Google Scholar]
- Johnson NL, Kotz S. Continuous Univariate Distributions-1. Houghton Mifflin Company; Boston: 1970. [Google Scholar]
- Mardia K, Kent J, Bibby J. Multivariate Analysis. Academic Press; London San Diego: 1979. [Google Scholar]
- Owen AB. Variance of the number of false discoveries. J. Roy. Statist. Soc. Ser. B. 2005;67:411–426. [Google Scholar]
- Qiu X, Brooks AI, Klebanov L, Yakovlev A. The effects of normalization on the correlation structure of microarray data. BMC Bioinformatics. 2005;6:120. doi: 10.1186/1471-2105-6-120. http://www.biomedcentral.com/1471-2105/6/120 Available at. [DOI] [PMC free article] [PubMed]
- Qiu X, Klebanov L, Yakovlev A. Correlation between gene expression levels and limitations of the empirical Bayes methodology for finding differentially expressed genes. Statist. Appl. Genet. Mol. Bio. 2005;4 doi: 10.2202/1544-6115.1157. http://www.bepress.com/sagmb/vol4/iss1/art34 article 34. Available at. [DOI] [PubMed]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Nat. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. http://www.pnas.org/cgi/content/full/98/9/5116 Available at. [DOI] [PMC free article] [PubMed]