Abstract
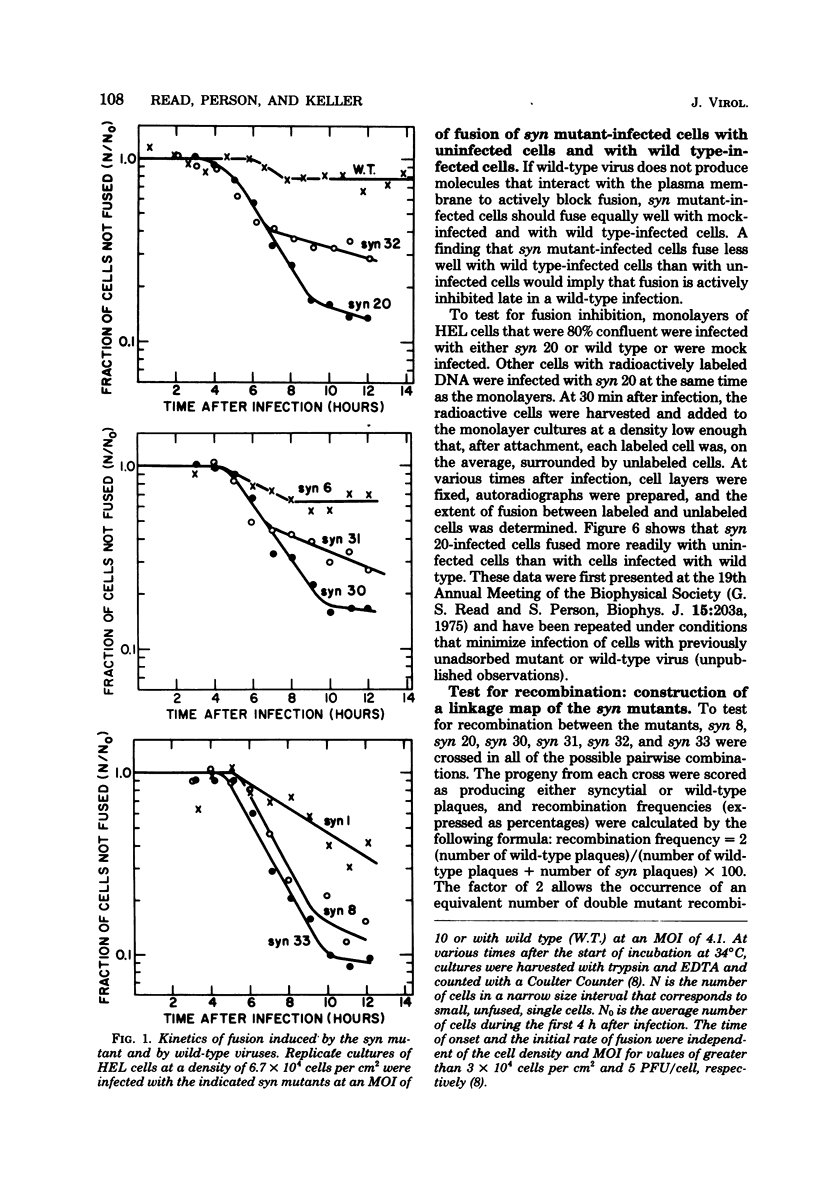
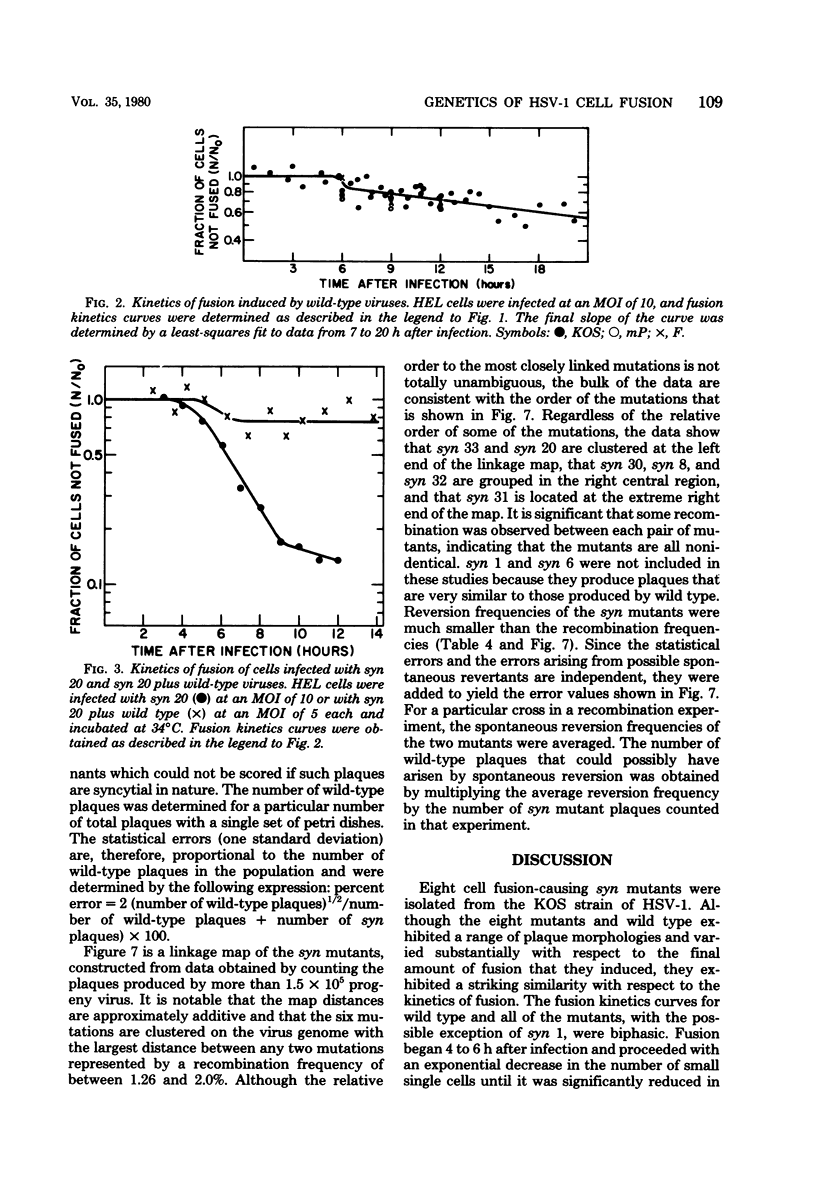
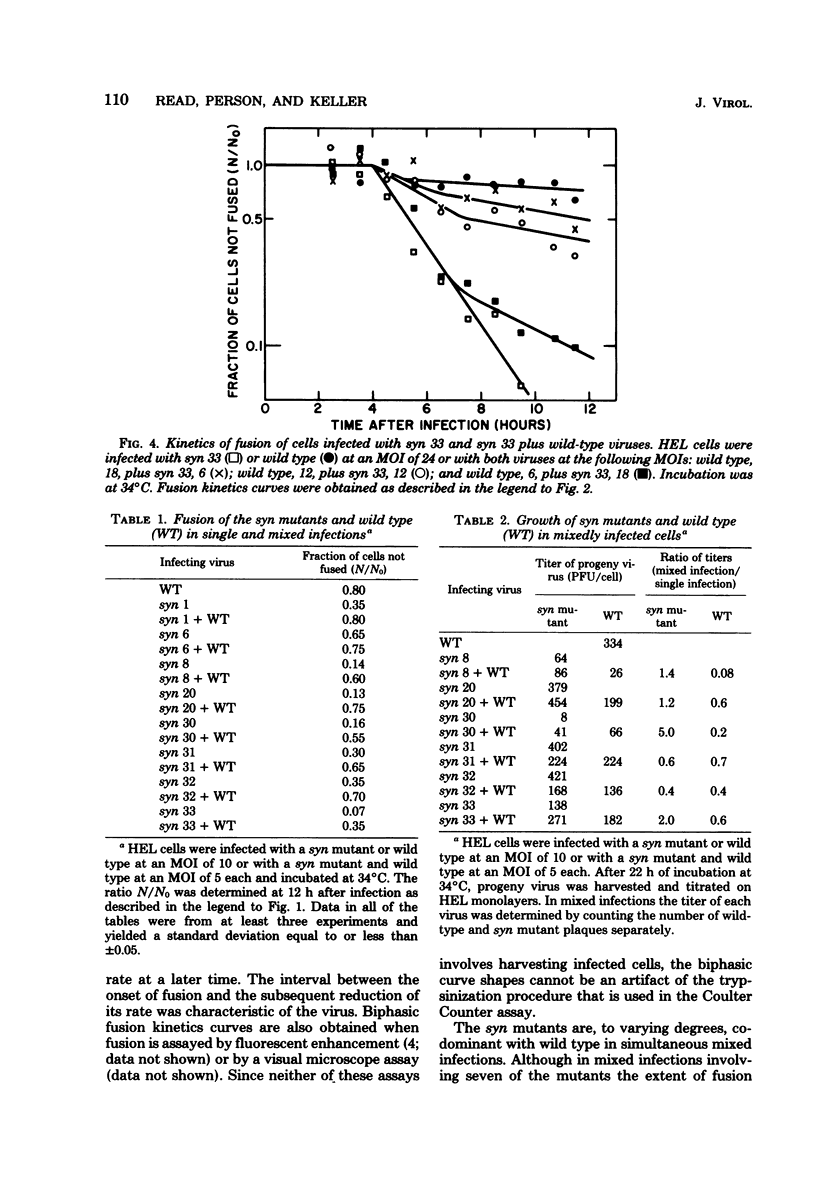
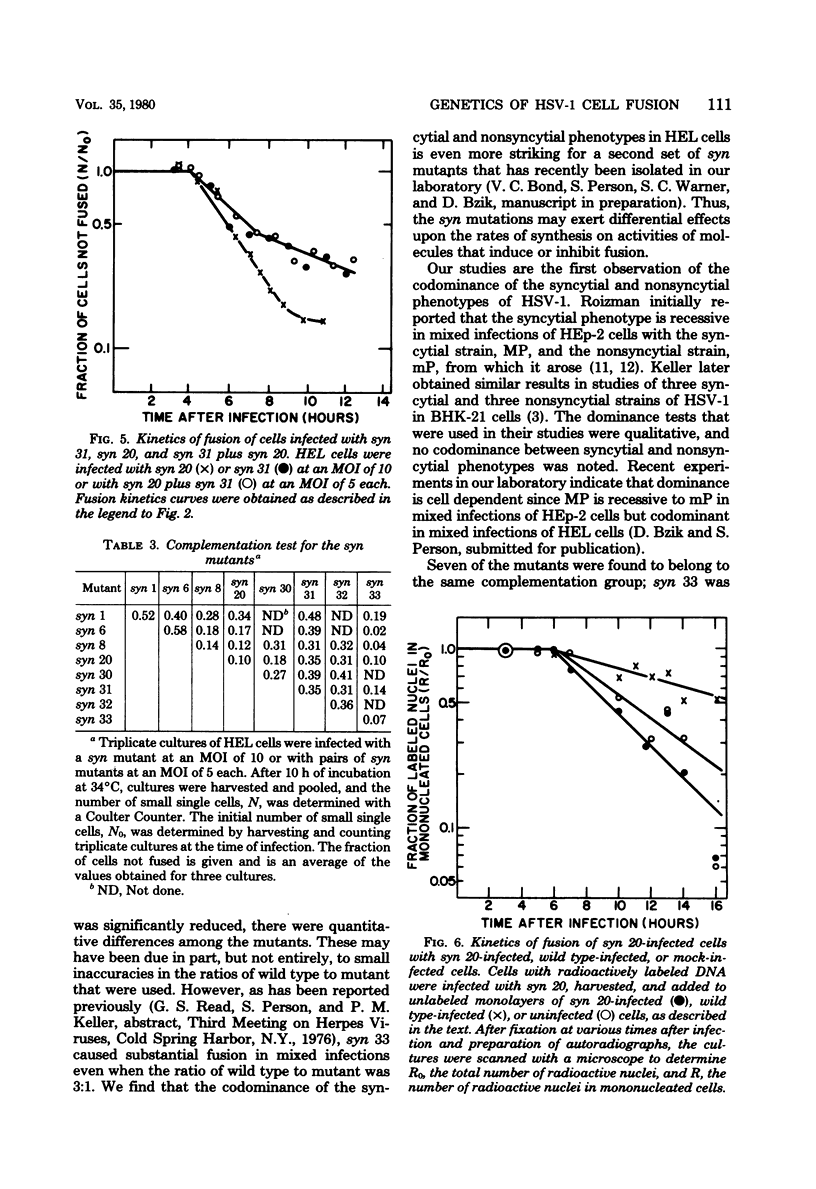
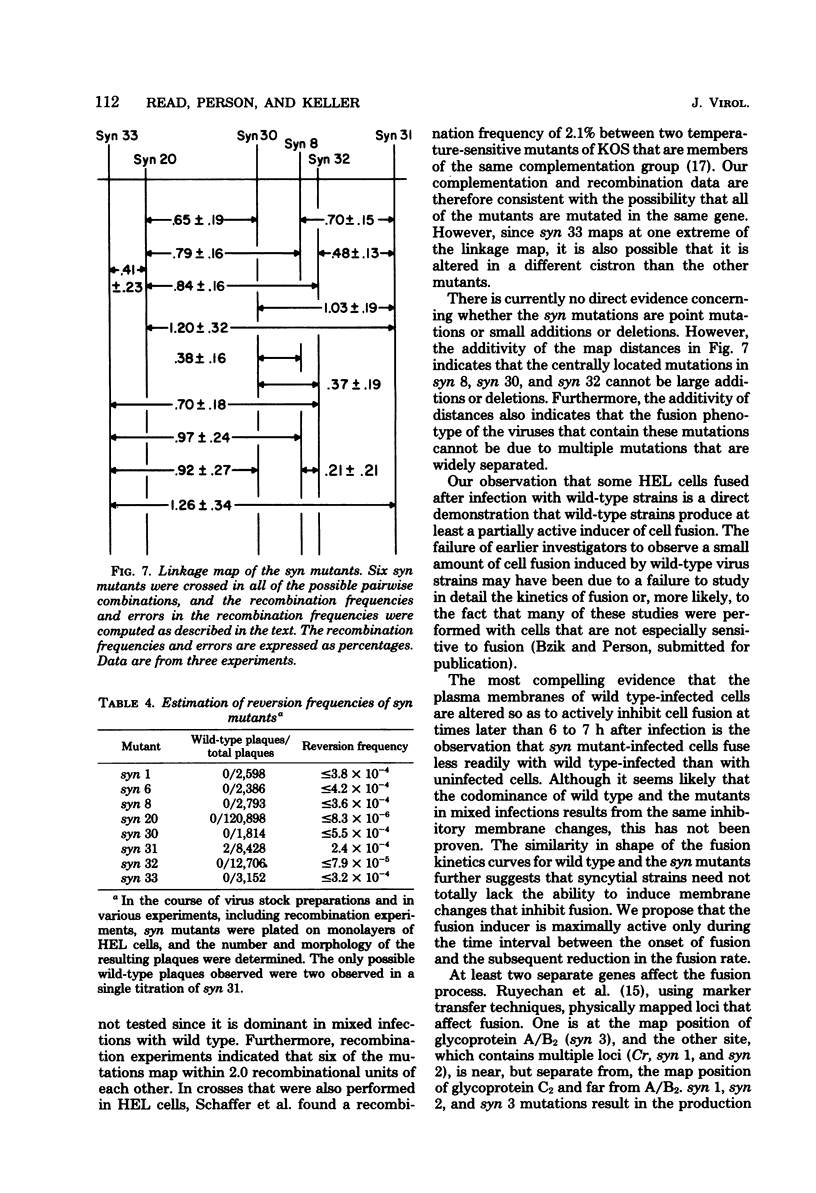
Eight cell fusion-causing syn mutants were isolated from the KOS strain of herpes simplex virus type 1. Unlike the wild-type virus, the mutants produced plaques containing multinucleated cells, or syncytia. Fusion kinetics curves were established with a Coulter Counter assay for the mutants and wild-type virus in single infections of human embryonic lung (HEL) cells, for the mutants and wild-type virus in mixed infections (dominance test), and for pairs of mutants in mixed infections (complementation test). In single infections, fusion began 4 to 6 h after infection and proceeded with an exponential decrease in the number of small single cells. At some later time that was characteristic of the mutant, there was a significant reduction in the rate of fusion for all but possibly one of the mutants. Although the wild-type virus did not produce syncytial plaques, it did induce a small amount of fusion that stopped abruptly about 2 h after it started. These data are consistent with the hypothesis that both mutants and wild type induce an active fusion inducer and that the activity of this inducer is subsequently inhibited. The extent of fusion is apparently determined by the length of the interval during which the fusion inducer is active. That fusion is actively inhibited in wild-type infections is indicated by the observation that syn mutant-infected cells fused more readily with uninfected cells than with wild-type infected cells. Fusion was decreased in mixed infections with the mutants and wild-type virus, but the mutants displayed a codominant fusion phenotype. Fusion was not decreased in mixed infection with pairs of mutants, indicating that the mutants, with one possible exception, are members of the same complementation group. A linkage map was established for six of the mutants by analysis of recombination frequencies.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973 Mar;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- HOGGAN M. D., ROIZMAN B. The isolation and properties of a variant of Herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959 Sep;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- Keller J. M. The expression of the syn- gene of herpes simplex virus type 1. I. Morphology of infected cells. Virology. 1976 Feb;69(2):490–499. doi: 10.1016/0042-6822(76)90479-7. [DOI] [PubMed] [Google Scholar]
- Keller P. M., Person S., Snipes W. A fluorescence enhancement assay of cell fusion. J Cell Sci. 1977 Dec;28:167–177. doi: 10.1242/jcs.28.1.167. [DOI] [PubMed] [Google Scholar]
- MUNK K., DONNER D. CYTOPATHISCHER EFFEKT UND PLAQUE-MORPHOLOGIE VERSCHIEDENER HERPES-SIMPLEX-VIRUSSTAEMME. Arch Gesamte Virusforsch. 1963 Aug 26;13:529–540. [PubMed] [Google Scholar]
- Manservigi R., Spear P. G., Buchan A. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NII S., KAMAHORA J. Cytopathic changes induced by herpes simplex virus. Biken J. 1961 Dec;4:255–270. [PubMed] [Google Scholar]
- Person S., Knowles R. W., Read G. S., Warner S. C., Bond V. C. Kinetics of cell fusion induced by a syncytia-producing mutant of herpes simplex virus type I. J Virol. 1975 Jan;17(1):183–190. doi: 10.1128/jvi.17.1.183-190.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poste G., Allison A. C. Membrane fusion. Biochim Biophys Acta. 1973 Dec 28;300(4):421–465. doi: 10.1016/0304-4157(73)90015-4. [DOI] [PubMed] [Google Scholar]
- Poste G. Mechanisms of virus-induced cell fusion. Int Rev Cytol. 1972;33:157–252. doi: 10.1016/s0074-7696(08)61451-5. [DOI] [PubMed] [Google Scholar]
- ROIZMAN B. Polykaryocytosis induced by viruses. Proc Natl Acad Sci U S A. 1962 Feb;48:228–234. doi: 10.1073/pnas.48.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROIZMAN B. Polykaryocytosis. Cold Spring Harb Symp Quant Biol. 1962;27:327–342. doi: 10.1101/sqb.1962.027.001.031. [DOI] [PubMed] [Google Scholar]
- Ruhlig M. A., Person S. Alterations of neutral glycolipids in cells infected with syncytium-producing mutants of herpes simplex virus type 1. J Virol. 1977 Nov;24(2):602–608. doi: 10.1128/jvi.24.2.602-608.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer P. A. Temperature-sensitive mutants of herpesviruses. Curr Top Microbiol Immunol. 1975;70:51–100. doi: 10.1007/978-3-642-66101-3_3. [DOI] [PubMed] [Google Scholar]
- Schaffer P. A., Tevethia M. J., Benyesh-Melnick M. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974 Mar;58(1):219–228. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- Tokumaru T. The nature of toxins of herpes simplex virus. I. Syncytial giant cell producing components in tissue culture. Arch Gesamte Virusforsch. 1968;24(1):104–122. doi: 10.1007/BF01242905. [DOI] [PubMed] [Google Scholar]


