Abstract
Non-natural amino acids have been genetically encoded in living cells, using aminoacyl-tRNA synthetase–tRNA pairs orthogonal to the host translation system. In the present study, we engineered Escherichia coli cells with a translation system orthogonal to the E. coli tyrosyl-tRNA synthetase (TyrRS)–tRNATyr pair, to use E. coli TyrRS variants for non-natural amino acids in the cells without interfering with tyrosine incorporation. We showed that the E. coli TyrRS–tRNATyr pair can be functionally replaced by the Methanocaldococcus jannaschii and Saccharomyces cerevisiae tyrosine pairs, which do not cross-react with E. coli TyrRS or tRNATyr. The endogenous TyrRS and tRNATyr genes were then removed from the chromosome of the E. coli cells expressing the archaeal TyrRS–tRNATyr pair. In this engineered strain, 3-iodo-l-tyrosine and 3-azido-l-tyrosine were each successfully encoded with the amber codon, using the E. coli amber suppressor tRNATyr and a TyrRS variant, which was previously developed for 3-iodo-l-tyrosine and was also found to recognize 3-azido-l-tyrosine. The structural basis for the 3-azido-l-tyrosine recognition was revealed by X-ray crystallography. The present engineering allows E. coli TyrRS variants for non-natural amino acids to be developed in E. coli, for use in both eukaryotic and bacterial cells for genetic code expansion.
INTRODUCTION
The repertoire of genetically encoded amino acids has been expanded in Escherichia coli, yeast, insect and mammalian cells, with the purpose of increasing the chemical and structural diversity in proteins (1–6). The genetically encoded ‘non-natural’ amino acids have unique chemical groups and functionalities not found among the 20 canonical amino acids, such as heavy atoms, photo-reactive linkers, fluorescent groups and chemical groups for site-specific labeling (7). The incorporation of these novel structures into proteins at desired sites promises to facilitate protein science and engineering.
The genetic encoding of a non-natural amino acid requires the development of an aminoacyl-tRNA synthetase (aaRS)–tRNA pair specific to the amino acid. To date, several aaRS species have been engineered to recognize useful non-natural amino acids, and among these, the majority are l-tyrosine or l-phenylalanine derivatives recognized by variants of tyrosyl-tRNA synthetase (TyrRS) (7). In living cells, non-natural amino acids are encoded with the amber codon in most cases, by using the amber suppressor tRNA–aaRS pair from a different organism that does not cross-react with the endogenous tRNA or aaRS (8). The bacterial TyrRS, for example, does not recognize the tRNATyr molecules from eukaryotes or archaea, while the TyrRS molecules from these two kingdoms do not recognize the bacterial tRNATyr. In fact, the bacterial TyrRS–tRNATyr pair does not cross-react with the endogenous tRNA and aaRS species in eukaryotic host cells, and has been used to achieve the site-specific incorporation of l-tyrosine derivatives into proteins only at the amber codon (2,9,10). On the other hand, the genetic encoding of l-tyrosine derivatives in E. coli cells was achieved using the TyrRS–tRNATyr pair from Methanocaldococcus jannaschii (1,7). Thus, the E. coli and archaeal TyrRS–tRNATyr pairs are used in eukaryotic and E. coli cells, respectively. Other aaRSs, such as tryptophanyl-tRNA synthetase (11), phenylalanyl-tRNA synthetase (5,12), leucyl-tRNA synthetase (13) and lysyl-tRNA synthetase (14), also take advantage of the orthogonal tRNA specificities across different kingdoms.
Pyrrolysyl-tRNA synthetase (PylRS) differs from these aaRS species, in that the PylRS–tRNAPyl pair from Methanosarcina mazei is reportedly orthogonal to both the E. coli and eukaryotic translation systems (15–17). Archaeal PylRS variants can be developed in E. coli cells, and are then used for incorporating l-lysine derivatives into proteins in E. coli, yeast and mammalian cells (16–20). With this advantage, the PylRS–tRNAPyl pair is a promising system for genetic code expansion. The usefulness of E. coli TyrRS variants would also be increased, if the E. coli translation system could be engineered to be orthogonal to the E. coli TyrRS–tRNATyr pair. This modification to E. coli cells may be achieved by substituting an archaeal or eukaryal tyrosine pair for the endogenous tyrosine pair.
In the present study, the genes encoding TyrRS and tRNATyr were disrupted in the chromosome of the E. coli cells expressing the M. jannaschii TyrRS–tRNATyr pair. The engineered cells then hosted the E. coli pair of a TyrRS variant and the amber suppressor tRNATyr, to genetically encode either 3-iodo-l-tyrosine or 3-azido-l-tyrosine, depending on the presence of the corresponding amino acid in the growth medium. This TyrRS variant, designated as iodoTyrRS-ec, was originally developed for 3-iodo-l-tyrosine (21), and has been used for the site-specific incorporation of this amino acid into proteins in mammalian cells (2,22). We found that iodoTyrRS-ec also recognizes 3-azido-l-tyrosine, and the structural basis for its recognition by this variant was elucidated by X-ray crystallography.
MATERIALS AND METHODS
Strains, growth conditions, growth rate, reagents and non-natural amino acids
Escherichia coli strains TOP10, BL21(DE3) and DH5α were purchased from Invitrogen, Novagen and Toyobo (Tokyo, Japan), respectively. Kanamycin (Km) (30 µg/ml), chloramphenicol (Cm) (34 µg/ml), ampicillin (Amp) (10 µg/ml), Zeocin (25 µg/ml), l-arabinose (0.2%, w/v) and d-glucose (0.4%, w/v) were added to LB media as indicated, except that Cm was added at a concentration of 25 µg/ml when non-natural amino acids were incorporated into proteins. The growth rate was monitored as the change in the natural logarithm of the optical density at 600 nm of the culture per hour. Zeocin was purchased from InvivoGen (San Diego, USA). 3-Iodo-l-tyrosine was purchased from Sigma-Aldrich. 3-Bromo-l-tyrosine, 3-azido-l-tyrosine and the chemical conjugate between triarylphosphine and fluorescein were commercially synthesized by Shinsei Chemical Company Ltd (Osaka, Japan).
Plasmids
The E. coli, M. jannaschii and Saccharomyces cerevisiae TyrRS genes, and the variant gene encoding iodoTyrRS-ec, were cloned with the tyrS promoter into pACYC184 (Nippon Gene Co. Ltd., Japan) to create pEcYS, pMjYS, pScYS and pEcIYS, respectively. The wild-type TyrRS genes with the promoter were also cloned in an ampicillin-resistant ColIb-P9 plasmid, pAp102 (23), to create pApEcYS, pApMjYS and pApScYS, respectively. The archaeal and yeast tRNATyr genes were cloned with the tyrT promoter into pACYC184, to create pMjYR and pScYR, respectively. These tRNATyr genes with the promoter were also cloned in pMjYS and pScYS, to create pMjYSYR and pScYSYR, respectively. Three copies of these tRNATyr genes were cloned after the tyrT promoter in pAp102, to create pApMjYR and pApScYR, respectively. The archaeal and yeast TyrRS genes with the tyrS promoter were cloned in these plasmids, pApMjYR and pApScYR, to create pApMjYSYR and pApScYSYR, respectively. The kanamycin resistance (kam) gene was cloned in the ‘amber suppressor control’ plasmid from the Interchange amber suppressor in vivo mutagenesis system (Promega), to create the plasmid pACamK. The resulting plasmid carries the replication origin of pACYC184 and an amber mutant chloramphenicol acetyltransferase (CAT) gene, in addition to the cloned kam gene. The E. coli amber suppressor tRNATyr gene was cloned with the tyrT promoter into pACamK, to create the plasmid pACamKsupF.
To create the plasmid pMJY, the coding sequence of the Zeocin resistance gene was amplified by polymerase chain reaction (PCR) from the vector pcDNA4/TO (Invitrogen). The sequence 5′-ACGACTCACTATAGGAGGGCC-3′, in which the ribosome-binding site (RBS) is underlined, was added at the 5′-end of the coding sequence. The amplified gene was cloned between the SspI–PstI sites of the pBR322 vector, downstream of the promoter of the ampicillin resistance gene and in place of this gene, to create the plasmid pBRZeo. The M. jannaschii TyrRS and tRNATyr genes with the tyrS and tyrT promoters, respectively, were cloned in pBRZeo to obtain pMJY.
For incorporating non-natural amino acids into proteins, the E. coli amber suppressor tRNATyr gene with the tyrT promoter was cloned in pAp102 to create pApsYR. The replication origin and the ampicillin resistance gene in the plasmid pGEX-4T-3 (GE Healthcare) were replaced with the origin and kam gene from the vector pRSF-1b (Novagen), to create the plasmid pRGexGST. The resulting plasmid retains the glutathione S-transferase (GST) gene under the control of the tac promoter. Then, the tyrosine codon at position 84 of GST was mutated to the amber codon, to create pRGexGST(Am84). The wild-type gst gene and its mutant with the amber codon at position 25 (24) were cloned after the T7 promoter in pRSF-1b to create pRSFGST and pRSFGST(Am25), respectively. The rat calmodulin gene and its mutant with the amber codon at position 80 (25) were also cloned after the T7 promoter in pRSF-1b, to create pRSFCAM and pRSFCAM(Am80), respectively.
Construction of the conditional tyrS mutant by chromosome engineering
All of the modifications to the E. coli chromosome were made using the RT/ET kit (Gene Bridges GmbH, Germany), which utilizes the bacteriophage λ recombination system to promote homologous recombination (26,27). The DNA encoding the RBS, the araBAD promoter and the araC gene—designated as the ara block—was introduced into the chromosome in place of the tyrS promoter. For this engineering, the ara block was amplified by PCR from the pBAD/His vector (Invitrogen) with the primers 5′-GGTTAATTCCTCCTGTTAGC-3′ and 5′-CCAATTATGACAACTTGACG-3′. The right-hand arm (rhm) corresponding to the first 50 nt of the tyrS coding sequence was then added at the RBS end of the ara block. The kam gene with the promoter was amplified from the plasmid pHSG299 (Takara Bio Inc., Japan), and the rrnC terminator was added downstream of the kam gene. The left-hand arm (lhm) with the sequence from 160- to 110-nt upstream the tyrS coding region (5′-ATGCGTGGAAGATTGATCGTCTTGCACCCTGAAAAGATGCAAAAATCTTG-3′) was added to the kam gene after the terminator. Then, the kam gene and the ara block were linked with each other at the ends opposite to lhm or rhm. The resulting double-stranded DNA fragment consists of lhm, kam, araC, the araBAD promoter, RBS and rhm in this order, and was used to replace the promoter and RBS of tyrS in the chromosome. The substitution was confirmed by a sequence analysis of the tyrS locus.
Engineering of the tyrS, tyrT, tyrU and tyrV loci in the E. coli chromosome
The cat gene with the promoter was amplified from pACYA184, using the primers A (5′-ACCCGACGCACTTTGCGCCG-3′) and B (5′-TTACGCCCCGCCCTGCCACTC-3′), while the gene without a promoter was amplified using primer B and another primer (5′-GATTTTCAGGAGCTAAGGAAGC-3′). In the following experiments, the cat gene was introduced into the chromosome in the same direction as the genes to be replaced. To disrupt tyrS, the sequences lhm-S (5′-ATGCGTGGAAGATTGATCGTCTTGCACCCTGAAAAGATGCAAAAATCTTG-3′) and rhm-S (5′-ACAGGGAACATGATGAAAAATATTCTCGCTATCCAGTCTCACGTTGTTTA-3′) were added at the 5′- and 3′-ends, respectively, of the cat gene with the promoter. To disrupt tyrT and tyrV, lhm-T1 (5′-AAAATAACTGGTTACCTTTAATCCGTTACGGATGAAAATTACGCAACCAG-3′) and rhm-T (5′-AGTCCCTGAACTTCCCAACGAATCCGCAATTAAATATTCTGCCCATGCGG-3′) were added at the 5′- and 3′-ends, respectively, of the cat gene with the promoter. To disrupt tyrU, lhm-U (5′- GTAATCAGTAGGTCACCAGTTCGATTCCGGTAGTCGGCACCATCAAGTCC-3′) and rhm-U (5′-GGCCACGCGATGGCGTAGCCCGAGACGATAAGTTCGCTTACCGGCTCGAA-3′) were added at the 5′- and 3′-ends, respectively, of the cat gene without the promoter.
The knocked-in cat gene was removed using a linear DNA fragment consisting of the lhm and rhm sequences linked directly to each other. Cells deprived of cat were selected in the growth media supplemented with Cm (10 µg/ml) and ampicillin (200 µg/ml). The cells that are resistant to Cm and are able to grow in this medium should undergo lysis in the presence of ampicillin, while only sensitive cells should survive. The colonies formed by the surviving cells were replicated on an LB plate containing Cm (10 µg/ml), to check for the sensitivity to Cm.
The E. coli amber suppressor tRNATyr was introduced into the chromosome in place of the knocked-in cat in the tyrU locus, using a linear DNA consisting of lhm-U, the tRNA coding sequence and rhm-U in this order. For the introduction of the suppressor tRNATyr in the tyrT and tyrV loci, the cat gene was first introduced into these loci, with lhm-T2 (5′-ACTTTACAGCGGCGCGTCATTTGATATGATGCGCCCCGCTTCCCGATAAG-3′) and rhm-T at the 5′- and 3′-ends, respectively. Then, the knocked-in cat gene was replaced by the sequence encoding the suppressor tRNA, amplified using primers (5′-TAATTCACCACAGCGATGTG-3′ and 5′-TTTGAAAGTGATGGTGGTGG-3) from the genomic DNA of a supF strain, in which the tyrV gene was mutated to the amber suppressor tRNATyr.
Site-specific incorporation of non-natural amino acids into proteins
FT3 cells were transformed with pEcIYS, pApsYR and pRGexGST(Am84), and were grown in medium containing Cm, Amp and Km. The expression of the gst gene was induced by the addition of IPTG (1 mM, final), and at the same time, 3-iodo-l-tyrosine (0.3 mg/ml, final) was added to the medium. FB3 cells were transformed with pEcIYS and either pRSFGST(Am25) or pRSFCAM(Am80), and were grown in medium containing Kam and Cm. The expression of the recombinant proteins was induced with IPTG (1 mM, final), and at the same time, 3-iodo-l-tyrosine or 3-azido-l-tyrosine (0.3 mg/ml, final) was added to the medium. Labeling with the triarylphosphine-fluorescein conjugate was performed as described previously (20,25). Fluorescence from an SDS-gel was detected using an image analyzer, LAS-1000 (Fuji Film, Japan). Mass scpectrometric analysis was commercially performed by Shimadzu Biotech (Japan).
Structure determination
The iodoTyrRS-ec catalytic domain (residues 1–322) was prepared as described previously (28). The samples were dialyzed against 20 mM Tris–Cl (pH 7.5) containing 50 mM NaCl, 10 mM 2-mercaptoethanol and 1 mM 3-azido-l-tyrosine, and then they were concentrated to 4–6 mg/ml with an Amicon Ultra 4 (Millipore). Crystallization of the complex between iodoTyrRS-ec and 3-azido-l-tyrosine was performed as described (28). The samples and crystals were shaded to avoid the photolysis of 3-azido-l-tyrosine. The X-ray diffraction experiment of the crystals was performed at the beamline BL26B1 at SPring-8 (Harima, Japan). The data were processed with HKL2000 (29). The iodoTyrRS-ec–3-azide-l-tyrosine structure was determined by molecular replacement with the program Molrep (30), using the structure of the complex between iodoTyrRS-ec and 3-iodo-l-tyrosine (28) as a search model. The refinement and model building were performed as described (28). The model of 3-azido-l-tyrosine was generated by the Dundee PRODRG2 server (31).
RESULTS AND DISCUSSION
Complementation of a conditional E. coli tyrS mutant by the archaeal and eukaryal TyrRS–tRNATyr pairs
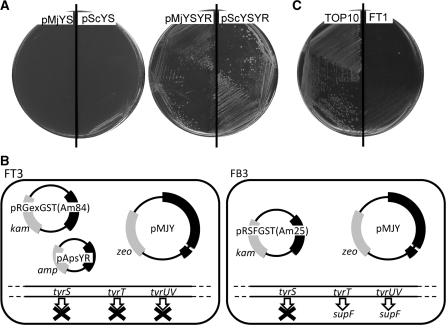
We examined whether the translational function of E. coli TyrRS can be replaced with that of a eukaryal or archaeal TyrRS–tRNATyr pair in E. coli cells. The expression system for the arabinose operon was used to control the expression of the chromosomal tyrS gene encoding E. coli TyrRS. The araBAD promoter, together with the araC gene, was introduced into the chromosome in place of the tyrS promoter, to allow the induction of TyrRS expression with l-arabinose. Transcription from the araBAD promoter is tightly repressed in the presence of d-glucose (32); the created conditional tyrS mutant of E. coli grew in the presence of l-arabinose, but did not grow in the presence of d-glucose (Supplementary Figure S1A). Then, the mutant was transformed with the multicopy vector pACYA184, and then reproduced the conditional growth (Supplementary Figure S1B). This conditional growth was complemented by the tyrS gene carried on the multicopy plasmid pEcYS, derived from pACYC184 (Supplementary Figure S1B). On the other hand, no complementation was observed with either the multicopy plasmid pMjYS carrying the M. jannaschii TyrRS gene or the multicopy plasmid pScYS carrying the S. cerevisiae TyrRS gene; these genes were expressed from the tyrS promoter (Figure 1A). This observation is consistent with the reports that these archaeal and eukaryal TyrRS species do not recognize E. coli tRNATyr (8,33).
Figure 1.
The tyrS complementation tests with pMjYS, pScYS, pMjYSYR and pScYSYR (A). The mutant cells transformed with these plasmids were inoculated on the LB plates containing 1% d-glucose and 34 µg/ml Cm. The three sectors of each half plate represent dilutions of the cells. An illustration of the genetic modifications and plasmid systems in FT3 and FB3 cells (B). The absence of the E. coli TyrRS activity in the strain FT1 (C). TOP10 and FT1 cells were each transformed with pACamKsupF and were then inoculated on the LB plate with 10 µg/ml Cm.
Next, the archaeal and eukaryal TyrRS species were expressed, together with their cognate tRNATyr species, from the multicopy plasmids pMjYSYR and pScYSYR, respectively. In these plasmids, the tRNATyr genes were placed after the promoter of the tyrT gene, which is one of three genes encoding E. coli tRNATyr. These plasmids were found to complement the tyrS mutant (Figure 1A), indicating that the function of the endogenous TyrRS in E. coli can be substituted by the archaeal and eukaryal TyrRS–tRNATyr pairs, which are each orthogonal to the host translation system.
To determine whether the overproduction of these TyrRS–tRNATyr pairs from multicopy plasmids is necessary for the complementation, we used the low-copy plasmid pAp102 (23) to introduce the tyrosine pairs into the tyrS mutant. This plasmid has the ColIb-P9 replication origin and its copy number is 1.7 per cell (34). In a control experiment, the vector pAp102 did not complement the tyrS mutant, while a pAp102-derived plasmid carrying the tyrS gene (pApEcYS) displayed the complementing activity (Supplementary Figure S1B). The M. jannaschii and S. cerevisiae TyrRS genes were then each cloned in this low-copy vector, together with three copies of their cognate tRNATyr genes, to create the plasmids pApMjYSYR and pApScYSYR, respectively. Neither of these plasmids showed the complementing activity (Supplementary Figure S1C), indicating that these exogenous TyrRS or tRNATyr molecules must be expressed from a multicopy plasmid.
We then determined which molecule, TyrRS or tRNATyr, needed to be overproduced. First, for the M. jannaschii tyrosine pair, the enzyme was expressed from the multicopy plasmid pMjYS, with the tRNATyr being expressed from the low-copy plasmid pApMjYS. No complementing activity was observed for this pair of plasmids (Supplementary Figure S1D). Next, TyrRS was expressed from the low-copy plasmid pApMjYS, with the tRNATyr expressed from the multicopy plasmid pMjYR, but again, no complementation was detected (Supplementary Figure S1D). These observations indicated that the complementation requires the overproduction of both the archaeal TyrRS and tRNATyr molecules.
We also expressed S. cerevisiae TyrRS from either the multicopy plasmid pScYS, with the yeast tRNATyr being expressed from the low-copy plasmid pApScYR, or the low-copy plasmid pApScYS, with the tRNATyr being expressed from the multicopy plasmid pScYR. Neither combination achieved the complementation (Supplementary Figure S1D), indicating that both the yeast TyrRS and tRNATyr need to be overproduced. Similar complementation tests were performed for the yeast TyrRS–tRNATyr pair at 30°C, the normal growth temperature for yeast. However, no complementation was observed (data not shown), indicating that the optimal temperature for these molecules is probably not relevant to the requirement of the overproduction of the TyrRS–tRNATyr pair. Although the reason why the exogenic tyrosine pairs must be overproduced is not clear, this requirement is consistent with a recent report showing that the overproduction of the M. jannaschii TyrRS–tRNATyr pair in E. coli cells is prerequisite for the efficient incorporation of non-natural amino acids into proteins (35).
Replacement of the E. coli TyrRS–tRNATyr pair by the M. jannaschii pair in an E. coli strain
The TyrRS and tRNATyr genes were removed from the chromosome in an E. coli K12 strain expressing the M. jannaschii tyrosine pair from the multicopy plasmid pMJY, constructed as described in the ‘Materials and Methods’ section (Figure 1B). First, the CAT gene was introduced into the chromosome in place of the tyrS gene by homologous recombination. Then, the cat gene was removed from the chromosome to create the strain FT1 [TOP10 ΔtyrS pMJY]. A DNA fragment including the tyrS locus was amplified from this strain and analyzed to confirm the disruption of the tyrS gene. The absence of the E. coli TyrRS activity in this strain was also confirmed by expressing the E. coli amber suppressor tRNATyr, together with a cat amber mutant gene, from the plasmid pACamKsupF. This plasmid conferred chloramphenicol resistance to the parent strain TOP10, but not to FT1 cells (Figure 1C), showing that the E. coli TyrRS activity was absent in FT1 cells, and that the E. coli suppressor tRNATyr is orthogonal to the translation system of the cell.
Next, we removed all three of the E. coli tRNATyr genes from the chromosome of the FT1 cells. Two of them, tyrT and tyrV, are located close to each other on the E. coli chromosome, and share the tyrT promoter for expression. The cat gene was introduced into the chromosome of FT1 in place of these genes and their promoter, and the knocked-in cat gene was then removed to create the strain FT2 [TOP10 ΔtyrS ΔtyrT ΔtyrV pMJY]. A DNA fragment encompassing the tyrT and tyrV loci was amplified from this strain and analyzed to confirm the disruption of these tRNATyr genes and the tyrT promoter. Then, to remove the tyrU gene, which is the third copy of the tRNATyr gene and is co-transcribed with two tRNAThr species and a tRNAGly, the cat gene without a promoter was introduced into the chromosome of FT2 in place of tyrU, for transcription together with the other tRNAs. The cat gene was then removed to create the strain FT3 [TOP10 ΔtyrS ΔtyrT ΔtyrU ΔtyrV pMJY]. The absence of the tRNATyr gene in the tyrU locus was confirmed by a sequence analysis.
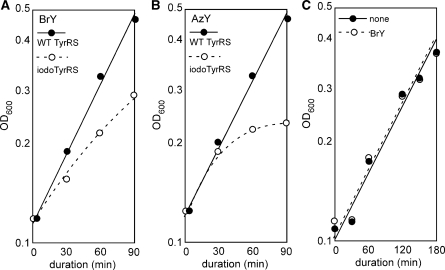
In order to show that the strain FT3 lacks E. coli tRNATyr activity, we expressed iodoTyrRS-ec, an E. coli TyrRS variant that recognizes 3-iodo-l-tyrosine, and investigated growth inhibition. When 3-bromo-l-tyrosine was added to the growth media, the growth rate of DH5α cells, an E. coli K-12 stain, overproducing the variant was significantly reduced, as compared to that of the cells overproducing the wild-type E. coli TyrRS (Figure 2A). This observation suggested that iodoTyrRS-ec is able to efficiently attach 3-bromo-l-tyrosine to the E. coli tRNATyr, and to inhibit the growth by randomly incorporating this amino acid into proteins at tyrosine positions. On the other hand, strain FT3 is immune to this growth inhibition (Figure 2C), which requires the interaction between iodoTyrRS-ec and E. coli tRNATyr, and this observation indicates the absence of the E. coli tRNATyr in the cell. Thus, the E. coli translation system was successfully engineered to be orthogonal to the E. coli TyrRS–tRNATyr pair.
Figure 2.
E. coli DH5α cells, an E. coli K-12 strain, expressing the wild-type E. coli TyrRS from plasmid pEcYS (filled circles) and those expressing iodoTyrRS-ec from plasmid pEcIYS (open circles) were grown in the presence of 3-bromo-l-tyrosine (0.5 mg/ml) (A) and 3-azido-l-tyrosine (0.3 mg/ml) (B). FT3 cells expressing iodoTyrRS-ec were grown in the absence (filled circles) and presence (open circles) of 3-bromo-l-tyrosine (C). The growth was monitored upon the addition of the non-natural amino acid to the growth media.
The iodoTyrRS-ec–E. coli suppressor tRNATyr pair site-specifically incorporates 3-iodo-l-tyrosine into proteins in FT3 cells
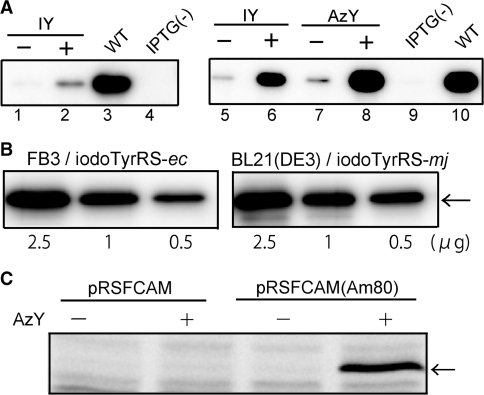
We used iodoTyrRS-ec to incorporate 3-iodo-l-tyrosine into proteins at the amber position in the strain FT3. This TyrRS variant was expressed from the tyrS promoter on the multicopy plasmid pEcIYS, which is compatible with pMJY expressing the archaeal tyrosine pair. Since the expression of the E. coli amber suppressor tRNATyr from a multicopy plasmid negatively affected the growth of the E. coli cells, the tRNA gene was expressed from the low-copy plasmid pApsYR. The gene encoding GST with an interrupting amber codon was carried on the third plasmid pRGexGST(Am84). Thus, it was necessary to maintain four different plasmids in FT3 cells for the site-specific incorporation of non-natural amino acids into proteins. The gst amber mutant gene produced the full-length product only when 3-iodo-l-tyrosine was supplemented in the growth media (Figure 3A, lanes 1 and 2), indicating that 3-iodo-l-tyrosine was incorporated into GST at the amber position, although the suppression efficiency was low.
Figure 3.
(A) Site-specific incorporation of 3-iodo-l-tyrosine (IY) and 3-azido-l-tyrosine (AzY) into GST in FT3 cells (lanes 1–4) and FB3 cells (lanes 5–10). The full-length GST was detected by western blotting. The growth media were supplemented with IY (lanes 2 and 6) and AzY (lane 8). The wild-type GST (WT) was expressed from pRGexGST (lane 3) or pRSFGST (lane 10). IPTG(–) means no induction of the gst expression (lanes 4 and 9). (B) The full-length GST product, indicated by the arrow, was detected by western blotting in extracts from the FB3 and BL21(DE3) cells. The applied quantity of total protein from the extract is indicated at the bottom of each lane. (C) Fluorescent labeling of rat calmodulin containing 3-azido-l-tyrosine. Extracts from the cells grown in the absence and presence of AzY were mixed with the fluorescein-triarylphosphine conjugate and then applied to SDS–PAGE. The band of calmodulin was indicated by the arrow.
The growth rate of FT3 was found to be just half that of its parent strain TOP10; the growth rates were determined as 0.7 and 1.4 h–1 for FT3 and TOP10, respectively, in LB medium with no antibiotics. Due to this slow growth rate, the maintenance of four different plasmids, and the low yield of suppression products, FT3 cells were apparently not useful for yielding large amounts of proteins containing non-natural amino acids. We tried to circumvent this problem by further engineering, starting with another E. coli strain.
Replacement of the E. coli TyrRS–tRNATyr pair by the M. jannaschii tyrosine pair in strain BL21(DE3)
The E. coli TyrRS and tRNATyr genes were replaced by the M. jannaschii pair in strain BL21(DE3), an E. coli B strain. This strain carries the gene encoding the bacteriophage T7 RNA polymerase in its chromosome, for the high-level expressions of recombinant proteins. The order of gene disruptions was not the same as that with TOP10. In addition, the E. coli amber suppressor tRNATyr gene was introduced into the chromosome of BL21(DE3), instead of on a plasmid, to reduce the number of required plasmids (Figure 1B). First, the cat gene was substituted for the tyrU gene, and was then replaced by the sequence encoding the E. coli suppressor tRNATyr. This step effectively mutated tyrU to the suppressor tRNATyr gene, thus creating the strain FB1 [BL21 λDE3 tyrU]. This strain was transformed with pMJY, and the tyrS gene was then removed from the chromosome to create FB2 [BL21 λDE3 tyrU ΔtyrS pMJY]. The tyrT and tyrV genes in FB2 were replaced with one copy of the suppressor tRNATyr gene. This step created the strain FB3 [BL21 λDE3 tyrU tyrV ΔtyrS ΔtyrT pMJY], in which tyrT was removed and tyrV was mutated to another copy of the suppressor tRNATyr gene, expressed from the tyrT promoter. Thus, the created FB3 strain expresses the archaeal TyrRS–tRNATyr pair and the E. coli suppressor tRNATyr, while it lacks E. coli TyrRS and wild-type tRNATyr. The growth rates of the FB3 and BL21(DE3) strains were determined as 1.1 and 1.3 h–1, respectively; the FB3 cells grow almost as fast as the parent strain, and much faster than FT3. The modifications to the BL21(DE3) chromosome had no serious effects on the viability of the cells.
To demonstrate the site-specific incorporation of 3-iodo-l-tyrosine into proteins in the strain FB3, iodoTyrRS-ec was expressed from pEcIYS, and the gst gene with the amber codon at position 25, gst(Am25), was expressed from a separate multicopy plasmid, pRSFGST(Am25), under the control of the T7 promoter. The FB3 strain bearing these plasmids produced the full-length GST only in the presence of 3-iodo-l-tyrosine, and its yield was significantly higher than that in FT3 cells (Figure 3A, lanes 5 and 6). We previously used the amber suppressor tRNATyr and the 3-iodo-l-tyrosine-specific variant of TyrRS (iodoTyrRS-mj) from M. jannaschii, in order to site-specifically incorporate 3-iodo-l-tyrosine into proteins in E. coli cells (36). The yield of the full-length product of gst(Am25) in the presence of 3-iodo-l-tyrosine was then compared between FB3 cells expressing the iodoTyrRS-ec–E. coli suppressor tRNATyr pair and the parent BL21(DE3) cells expressing the iodoTyrRS-mj–archaeal suppressor tRNATyr pair. Comparable amounts of the full-length product were detected in the extracts from these cells (Figure 3B).
Finally, the full-length GST from FB3 cells was analyzed by mass spectrometry. This amber suppression product was subjected to a trypsin digestion, for comparison with the peptide data previously obtained for the wild-type GST (36). The wild-type protein generates a peptide (Tyr25 peptide) with an average mass corresponding to the theoretical value (m/z = 1327.7) for residues 23–33, NSYSPILGYWK (tyrosine at position 25 is underlined). This peptide was not detected from the amber suppression product from FB3 cells; instead, a peptide with an average mass (1453.6) was detected (Supplementary Figure S2). This value is larger than that of the Tyr25 peptide by a value (125.9) corresponding to the mass of an iodine atom, less the mass of a hydrogen atom, indicating the incorporation of 3-iodo-l-tyrosine at the amber position.
Genetic encoding of 3-azido-l-tyrosine in the engineered E. coli cells
A variant of yeast TyrRS, engineered to recognize 3-iodo-l-tyrosine, reportedly also recognizes 3-azido-l-tyrosine (37). Using this variant, 3-azido-l-tyrosine was site-specifically incorporated into proteins in E. coli cell-free translation, and was shown to be useful for site-specific protein modification (25). However, since this yeast TyrRS variant still recognizes l-tyrosine efficiently, it has not been used for genetically encoding l-tyrosine derivatives in E. coli cells. We found that iodoTyrRS-ec also recognizes 3-azido-l-tyrosine efficiently; this amino acid supplemented in the growth media inhibits the growth of normal E. coli cells expressing iodoTyrRS-ec, as observed for 3-bromo-l-tyrosine (Figure 2B). Therefore, iodoTyrRS-ec was used to incorporate 3-azido-l-tyrosine into proteins in FB3 cells. The full-length product of gst(Am25) was obtained only in the presence of 3-azido-l-tyrosine, and its yield was comparable to that of the product with 3-iodo-l-tyrosine (Figure 3A, lanes 7 and 8). Thus, FB3 cells expressing iodoTyrRS-ec and the E. coli suppressor tRNATyr produced the proteins site-specifically containing either 3-azido-l-tyrosine or 3-iodo-l-tyrosine, depending on which amino acid is supplemented in the growth media.
To demonstrate the site-specific protein modification, rat calmodulin containing 3-azido-l-tyrosine was produced in FB3 cells expressing the iodoTyrRS-ec–suppressor tRNATyr pair. The full-length product was expressed from the amber mutant gene, carried on plasmid pRSFCAM(Am80), only in the presence of 3-azido-l-tyrosine, and was modified with a fluorescent probe by the Staudinger–Bertozzi reaction, while the wild-type protein expressed from plasmid pRSFCAM was not labeled (Figure 3C). p-Azido-l-phenylalanine and a l-lysine derivative with the azido group have already been genetically encoded in E. coli cells (20,38). Tyrosines tend to reside on the protein surface (39), and such tyrosines on the surface could be safely replaced with 3-azido-l-tyrosine for site-specific protein labeling.
Structural basis for 3-azido-l-tyrosine recognition by iodoTyrRS-ec
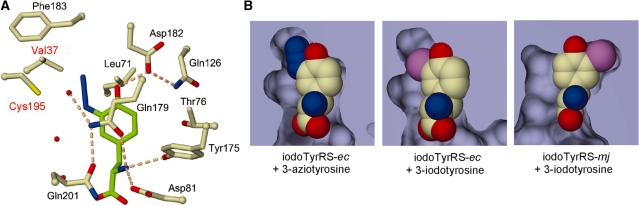
E. coli TyrRS was previously engineered into iodoTyrRS-ec by introducing a double substitution, Tyr37Val and Gln195Cys (Figure 4A) (21,28). To reveal the structural basis for the additional recognition of 3-azido-l-tyrosine by iodoTyrRS-ec, the crystal structure of a complex between these molecules was determined at 1.8-Å resolution (PDB ID: 2YXN). The statistics of the data set are listed in Table 1. The overall structure of the variant superposed well on the reported crystal structure of iodoTyrRS-ec complexed with 3-iodo-l-tyrosine (28). The azido group of 3-azido-l-tyrosine is accommodated in the space created by the double substitution at the amino-acid binding pocket (Figure 4B). Although no enzyme residues appear to make firm contacts or interactions with the azido group, the shape of the space snugly fits the substituent. By contrast, in the reported structure, iodoTyrRS-ec recognizes 3-iodo-l-tyrosine by making van der Waals contacts between the mutated residues, Val37 and Cys195, and the iodine atom (Figure 4B). The binding of l-tyrosine to iodoTyrRS-ec, on the other hand, is weakened by the Tyr37Val substitution, which removes the hydrogen bond between the phenolic hydroxyl groups of l-tyrosine and Tyr37 (28) (Figure 4A).
Figure 4.
(A) The amino-acid binding pocket in the crystal structure of iodoTyrRS-ec bound with 3-azido-l-tyrosine. The mutated residues are denoted in red. The carbon atoms of the ligand are shown in green. The nitrogen, oxygen and sulfur atoms are in blue, red and yellow, respectively. The hydrogen bonds are shown as pink broken lines. (B) The surface structures of the binding pockets of iodoTyrRS-ec and iodoTyrRS-mj accommodating 3-aido-l-tyrosine or 3-iodo-l-tyrosine, as indicated below each panel. The enzyme surface and section are shown in light blue. The iodine atom is in purple.
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P3121 |
| Cell dimensions | |
| a, b, c (Å) | 83.16, 83.16, 93.67 |
| α, β, γ(°) | 90, 90, 120 |
| Wavelength (Å) | 1.0000 |
| Resolution (Å) | 50–1.8 |
| Measured reflections | 382 492 |
| Unique reflections | 35 502 |
| Redundancy | 10.8 |
| Completeness (%) | 100.0 |
| I/σ (I) | 23.0 |
| Rsym (%)a | 11.3 |
| Refinement statistics | |
| Resolution (Å) | 39.26–1.8 |
| Protein atoms | 2536 |
| Substrates (per subunit) | 1 |
| Water oxygens | 295 |
| Mean B value (Å2) | 21.3 |
| Rwork (%)b | 19.9 |
| Rfree (%)c | 24.7 |
| R.m.s. deviations | |
| Bond length (Å) | 0.011 |
| Bond angle (°) | 1.40 |
aRsym = Σhkl|Iavg − Ii| / ΣhklIi.
bRwork = Σhkl||FO| − |FC|| / Σhkl|FO|.
cRfree = Rwork using 10% of FO sequestered before refinement.
The M. jannaschii iodoTyrRS-mj–suppressor tRNATyr pair does not incorporate 3-azido-l-tyrosine into proteins (data not shown), indicating that this archaeal TyrRS variant does not recognize 3-azido-l-tyrosine. In the reported crystal structure of iodoTyrRS-mj complexed with 3-iodo-l-tyrosine (36), the space created by the mutations just fits the size and shape of the iodine atom, with five enzyme residues making van der Waals contacts with this atom (Figure 4B). The azido substitutent would collide with Thr158, one of these five residues, in the binding pocket of iodoTyrRS-mj, because an azido group has a more extended shape and is longer than an iodine atom. This explains the inability of this variant to recognize 3-azido-l-tyrosine.
CONCLUSION
A variety of tyrosine or phenylalanine derivatives have been genetically encoded in mammalian cells and E. coli to facilitate protein science and technology. In mammalian cells, for example, photo-reactive cross-linking amino acid was used for analyzing protein–protein interactions (9), and the incorporated 4-azido-l-phenylalanine allowed the probing of conformational changes in a membrane protein (40). E. coli cells have been useful for producing large amounts of proteins containing non-natural amino acids. The incorporation of 4-azido-l-phenylalanine allowed the site-specific protein modification with polyethylene glycol (41), and iodine-atom-containing amino acids have been utilized in X-ray crystallography (36,42). For each of these non-natural amino acids, specific archaeal and bacterial TyrRS variants both needed to be developed, when the amino acid was used in the bacterial and eukaryotic cells. Our engineered E. coli strains promise to reduce the labor for variant development, because they can be engineered in the E. coli cells, and then used in both eukaryotic cells and bacteria. Our strategy could be applied to other aaRS species, and would facilitate genetic code expansion in living cells.
ACCESSION NUMBER
2YXN.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Targeted Proteins Research Program (TPRP); RIKEN Structural Genomics/Proteomics Initiative (RSGI) in the National Project on Protein Structural and Functional Analyses, and in part, by a Grant-in-Aid for Scientific Research (B) 19380195 to K.S. from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Special Postdoctoral Researchers Program at RIKEN to T.K. Funding for open access charge: RIKEN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Nobuhiro Hayashi for kindly providing the rat calmodulin gene, Dr Takamitsu Hosoya and Dr Masaaki Suzuki for kindly providing the first lot of the triarylphosphine-fluorescein conjugate used in this study, and Ms Azusa Ishii and Ms Tomoko Nakayama for clerical assistance.
REFERENCES
- 1.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 4.Ye S, Köhrer C, Huber T, Kazmi M, Sachdev P, Yan EC, Bhagat A, RajBhandary UL, Sakmar TP. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 5.Kwon I, Tirrell DA. Site-specific incorporation of tryptophan analogues into recombinant proteins in bacterial cells. J. Am. Chem. Soc. 2007;129:10431–10437. doi: 10.1021/ja071773r. [DOI] [PubMed] [Google Scholar]
- 6.Mukai T, Wakiyama M, Sakamoto K, Yokoyama S. Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells. Protein Sci. 2010 doi: 10.1002/pro.322. 10.1002/pro.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 8.Kowal AK, Köhrer C, RajBhandary UL. Twenty-first aminoacyl-tRNA synthetase––suppressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc. Natl Acad. Sci. USA. 2001;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat. Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Alfonta L, Tian F, Bursulaya B, Uryu S, King DS, Schultz PG. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furter R. Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci. 1998;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson JC, Schultz PG. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- 14.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc. Natl Acad. Sci. USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 16.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 17.Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, Schultz PG. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Ed. Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polycarpo CR, Herring S, Bérubé A, Wood JL, Söll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 20.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multi-step engineering of pyrrolysyl-tRNA synthetase to genetically encode Nε-(o-azidobenzyloxycarbonyl)lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Naklayama H, Takio K, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA. 2002;99:9715–9723. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hino N, Hayashi A, Sakamoto K, Yokoyama S. Site-specific incorporation of non-natural amino acids into proteins in the mammalian cells with an expanded genetic code. Nat. Protoc. 2007;1:2957–2962. doi: 10.1038/nprot.2006.424. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto K, Ishimaru S, Kobayashi T, Walker JR, Yokoyama S. The Escherichia coli argU10(Ts) phenotype is caused by a reduction in the cellular level of the argU tRNA for the rare codons AGA and AGG. J. Bacteriol. 2004;186:5899–5905. doi: 10.1128/JB.186.17.5899-5905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chumpolkulwong N, Sakamoto K, Hayashi A, Iraha F, Shinya N, Matsuda N, Kiga D, Urushibata A, Shirouzu M, Oki K, et al. Translation of ‘rare’ codons in a cell-free protein synthesis system from Escherichia coli. J. Struct. Funct. Genomics. 2006;7:31–36. doi: 10.1007/s10969-006-9007-y. [DOI] [PubMed] [Google Scholar]
- 25.Ohno S, Matsui M, Yokogawa T, Nakamura M, Hosoya T, Hiramatsu T, Suzuki M, Hayashi N, Nishikawa K. Site-selective post-translational modification of proteins using an unnatural amino acid, 3-azidotyrosine. J. Biochem. 2007;141:335–343. doi: 10.1093/jb/mvm036. [DOI] [PubMed] [Google Scholar]
- 26.Murphy KC. Use of bacteriophage λ recombination function to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyrers J.PP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Sakamoto K, Takimura T, Sekine R, Vincent K, Kamata K, Nishimura S, Yokoyama S. Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion. Proc. Natl Acad. Sci. USA. 2005;102:1366–1371. doi: 10.1073/pnas.0407039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr, Sweet RM, editors. Methods in Enzymology, Macromolecular Crystallography, Part A, New York: 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 31.Schuettelkopf AW, van Aalten D.MF. PRODRG—a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 32.Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Washington DC: ASM Press; 1996. [Google Scholar]
- 33.Wang L, Magliery TJ, Liu DR, Schultz PG. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc. 2000;122:5010–5011. [Google Scholar]
- 34.Clewell DB, Helinski DR. Existence of the colicinogenic factor-sex factor ColIb -P9 as a supercoiled circular DNA-protein relaxation complex. Biochem. Biophys. Res. Commun. 1970;41:150–156. doi: 10.1016/0006-291x(70)90481-x. [DOI] [PubMed] [Google Scholar]
- 35.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto K, Murayama K, Oki K, Iraha F, Kato-Murayama M, Takahashi M, Ohtake K, Kobayashi T, Kuramitsu S, Shirouzu M, et al. Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure. 2009;17:335–344. doi: 10.1016/j.str.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Ohno S, Yokogawa T, Nisihikawa K. Changing the amino acid specificity of yeast tyrosyl-tRNA synthetase by genetic engineering. J. Biohem. 2001;130:417–423. doi: 10.1093/oxfordjournals.jbchem.a003001. [DOI] [PubMed] [Google Scholar]
- 38.Tsao M.-L, Tian F, Schultz PG. Selective staudinger modification of proteins containing p-azidephenylalanine. ChemBioChem. 2005;6:2147–2149. doi: 10.1002/cbic.200500314. [DOI] [PubMed] [Google Scholar]
- 39.Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 1991;217:721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- 40.Ye S, Huber T, Vogel R, Sakmar TP. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG. Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg. Med. Chem. Lett. 2004;14:5743–5745. doi: 10.1016/j.bmcl.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 42.Xie J, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. The site-specific incorporation of p-iodo-l-phenylalanine into proteins for structure determination. Nat. Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.