Abstract
Recently, modularity has emerged as a general attribute of complex biological systems. This is probably because modular systems lend themselves readily to optimization via random mutation followed by natural selection. Although they are not traditionally considered to evolve by this process, biological ligands are also modular, being composed of recurring chemical fragments, and moreover they exhibit similarities reminiscent of mutations (e.g. the few atoms differentiating adenine and guanine). Many ligands are also promiscuous in the sense that they bind to many different protein folds. Here, we investigated whether ligand chemical modularity is reflected in an underlying modularity of binding sites across unrelated proteins. We chose nucleotides as paradigmatic ligands, because they can be described as composed of well-defined fragments (nucleobase, ribose and phosphates) and are quite abundant both in nature and in protein structure databases. We found that nucleotide-binding sites do indeed show a modular organization and are composed of fragment-specific protein structural motifs, which parallel the modular structure of their ligands. Through an analysis of the distribution of these motifs in different proteins and in different folds, we discuss the evolutionary implications of these findings and argue that the structural features we observed can arise both as a result of divergence from a common ancestor or convergent evolution.
INTRODUCTION
Modularity is emerging as a general attribute of complex biological systems (1). A system is defined as modular when it is composed of discrete units that can be combined in different ways to give rise to a diversity of functions. The ubiquity of modular systems in nature is due to modularity being a fundamental requirement for the possibility to evolve by random mutation followed by natural selection. Perhaps, the most often considered types of modularity in biological systems are those related to the shuffling of exons, that can lead to rearrangements of genes or gene fragments and of protein domains (2). This is particularly prevalent in eukaryotic signaling, where it is now established that rearrangements of particular catalytic (e.g. phosphatases, kinases) and recognition (e.g. SH2, SH3, PDZ, etc.) domains, together with point mutations, have led to a great diversity of proteins that have evolved to form signaling pathways (e.g. Wnt, Akt, G-protein signaling, etc.) (3,4).
Indeed, the study of genetic algorithms has shown that improvement by random mutations is possible when function is encoded in independent units therefore reducing the likelihood that a mutation will have a pleiotropic effect. In this way, one component can be improved by mutation/selection without disrupting units already optimized (5).
Modularity extends to a higher order of proteome organization. Protein complexes show a modularity in that sets of proteins, or sub-complexes, appear to be involved in more than one complex. For instance, RNA polymerases I, II and III share five subunits that can be considered a modular sub-complex (6), and analysis of the repertoire of yeast molecular machines suggests dozens of additional examples (7).
There is also an evidence of modularity at a scale smaller than that of domains. Sturniolo et al. (8) hypothesized that Human Leukocyte Antigen-group DR (HLA-DR)-binding grooves can be described as a juxtaposition of binding pockets characterized by distinct preferences for specific residues. The modular structure of the binding groove allowed them to predict promiscuous HLA class II ligands by composing the residue preferences as experimentally determined. A similar approach was used by Brannetti et al. (9) to predict preferred ligands to different members of the SH3 gene family. In this case, the interaction between the protein and the binding peptide was described as the sum of independent interactions between their contacting residues. Similarly, Petsalaki et al. (10) showed that it is possible to predict peptide-binding sites by combining 3D scoring matrices describing the binding preferences of each amino acid in the peptide. This is akin to linking predicted single residue-binding sites to form a complete peptide-binding patch. Moreover, Reichmann et al. (11) extended the concept of modularity to protein–protein interfaces. They demonstrated that the surface of the interface can be divided in groups of highly interconnected residues, which make a few interactions outside their cluster. Interestingly, they also experimentally showed that these modules can be removed by mutation with limited effects on binding affinity.
Here, we present evidence for modularity in the molecular features of protein-binding sites. The rationale for this is that ligands themselves are composed of well-defined fragments whose combination produces an infinite variety of molecules. Indeed, this property has important practical applications as demonstrated by fragment-based drug design (12). To search for candidate modular binding sites, we first identified structural motifs that were associated with a common ligand in at least two distinct protein folds (13) (a requirement that excludes motifs due to obvious homology), and then sought instances where these motifs were used in combination.
We focused on nucleotides as they are well represented in the structural database binding to many different folds, and they are highly prominent in all major biological functions, from signaling to metabolism. Moreover, as they are one of the earliest cofactors bound to proteins (14), they are excellent candidates for studying binding site evolution. These binding sites have been extensively probed for commonalities across non-homologous proteins, and several structural motifs or principles have been described previously (15–25), though to our knowledge there has been no investigation into their possible modularity.
We found that nucleotide-binding sites are very often composed of small 3D motifs that are common to a number of different folds (between two and nine depending on the motif) and are associated with the same nucleotide fragment (e.g. nucleobase, ribose or phosphate). The resulting network of protein folds and spatial binding motifs questions and complements current views about convergent or divergent evolution.
METHODS
In this work, we define motifs as sets of three or more protein residues that occur in more than one protein fold and interact with a common fragment of a nucleotide molecule (i.e. adenine, ribose or phosphate). Adenine- and ribose-binding motifs were derived from a previous work (13) dealing with the identification of structural motifs associated with specific ligand fragments. The main steps of the procedure are recapitulated in the following paragraphs. We used 24 402 Protein Data Bank (PDB) entries classified in Structural Classification Of Proteins (SCOP). Binding pockets were defined by selecting all the protein residues that had an atom whose distance from any atom of the ligand was <3.5 Å. This dataset comprises 65 467 binding pockets, mapping to 4050 different ligands, which were used to perform an all-against-all structural comparison with the program Query3d (26). The Root Mean Square Deviation (RMSD) threshold was set to 0.4, 0.8, 1.3 and 1.5 Angstroms for matches comprising three, four, five and six or more residues, respectively; only residues with a similarity score of at least 1 in a BLOSUM62 matrix were allowed to match. Because we were not interested in similarities readily explained by homology, we did not compare with each other binding pockets belonging to proteins assigned to the same sequence cluster (at the 30% sequence identity level; as downloaded from the PDB website). We also discarded all matches between proteins belonging either to the same Class, Architecture, Topology, Homologous superfamily architecture or SCOP fold. The FunClust multiple structural comparison algorithm (27) was then used in order to identify structural similarities common to more than two folds starting from pairwise matches. We then developed an automated procedure to analyze the coordinates of the ligands bound by these motifs and identified the largest common ligand fragment. Note that our motif definition also requires the common portion of the ligands to align spatially. The 330 fragment-associated motifs, obtained in this way, were visually inspected and only those binding the base or sugar portion of a nucleotide were retained. Finally, nucleotide-binding motifs were manually analyzed in order to merge together motifs that the automatic procedure failed to identify as similar because their RMSD after superimposition was higher than the threshold used.
Phosphate-binding motifs were kindly provided by Prof. Richard M. Jackson and are as described previously (23).
In order to identify all the instances of modularity in a systematic way, we first extracted from the network in Figure 1 all the instances pertaining to each of the four cases (Figure 2). We then used Query3d to search the 14 motifs in the dataset of binding pockets in order to identify the complement of motifs possessed by each structure. This enabled the systematic identification of all the proteins possessing the combination of motifs corresponding to each instance of modularity. Motifs 13 and 14 often appear together in proteins of the Flavin Adenine Dinucleotide (FAD)/nicotinamide adenine dinucleotide phosphate [NAD(P)] fold. Indeed, Brakoulias and Jackson (23) describe motif 13 as analogous to motif 14, but more sparsely conserved. For the purpose of this analysis, we have assigned these structures to motif 14. The analysis was subsequently repeated by pooling together all motifs in the same family and superfamily, i.e. considering a protein as sharing all the motifs found in any protein of its family/superfamily.
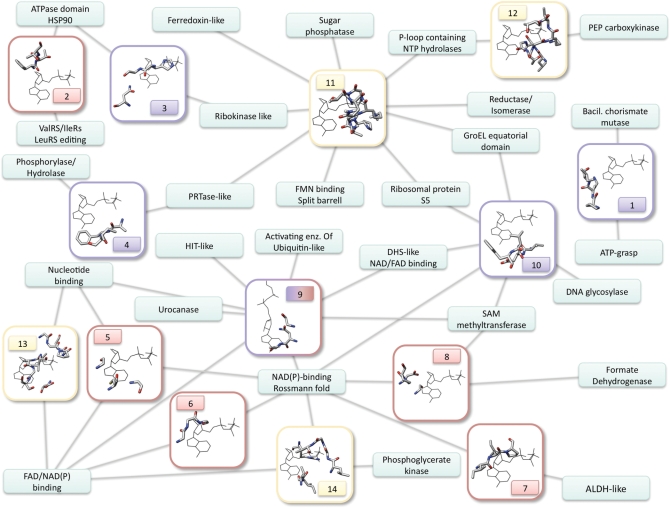
Figure 1.
Network representing the distribution of nucleotide-binding motifs across different protein folds. Each small rectangle is a fold, with the name written inside. The bigger rectangles represent structural motifs, numbered as in Supplementary Table S1, and with the frame and number colored according to the bound ligand fragment (lila: nitrogen base; red: ribose; yellow: phosphate). The edges connect each motif to the folds on which it was found. The network comprises 14 motifs and 27 folds. The pictures of the motifs show one representative structure belonging to one of the folds involved. All the ligands were aligned to a reference ligand by superimposing the relevant molecular fragments (i.e. for ribose-binding motifs, the ribose of the structure was aligned to the ribose of the reference ligand, etc.). The pictures, therefore, show the protein residues together with the reference ligand, which has always the same position. This was not possible for motif number 9, which binds both ribose and adenine in a conformation that cannot be aligned to the reference ligand. Accordingly, this is the only case in which the ligand in the picture is different.
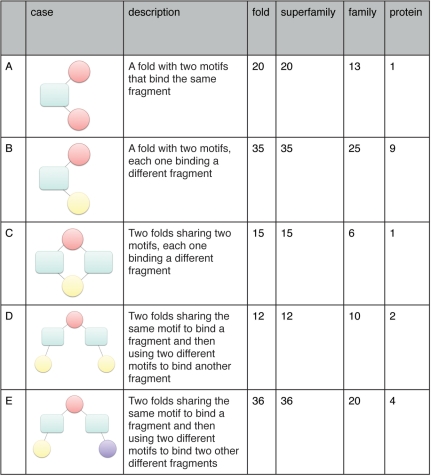
Figure 2.
Total number of occurrences for each one of the five examples of modularity we considered (see text for details). The ‘case’ column gives a schematic representation of each case with folds depicted as turquoise rectangles. Each different motif is represented as a circle; motifs of the same color bind the same ligand fragment. The ‘description’ column contains an explanation of each case of modularity. The ‘fold’ column represents the combinations extracted from the network, i.e. pooling together all the motifs belonging to the same fold. This is the maximum number of instances, only a fraction of which will be verified in single structures. The ‘protein’, ‘family’ and ‘superfamily’ columns contain the number of instances of each case that were identified in single proteins and pooling together all the motifs in the same family and superfamily, respectively. The single case where a protein shares two motifs for the same fragment (A) involves motifs 9 and 10. Motif 9 binds both adenine and ribose (i.e. the ‘sugar’ edge of the nucleotide). This mode of binding leaves adenine free to interact with a different motif.
RESULTS
Overview of the methodology
We created a curated, non-redundant, well-defined set of nucleobase, ribose and phosphate-binding spatial motifs, only considering those shared by at least two distinct folds. This restricted the set to those motifs that have either evolved multiple times or been conserved across great evolutionary distances, and thus allowed the key common, and functionally significant elements to be readily identified.
For the identification of adenine- and ribose-binding motifs, we used a previously developed method for the identication of structural motifs associated with specific ligand fragments (13). For phosphate-binding motifs, we used the comprehensive classification of Brakoulias and Jackson (23), keeping those common to two or more distinct folds (as defined in SCOP). We were left with a total of 14 motifs mapping to 27 protein folds. The conservation in the respective superfamilies and families is shown in Supplementary Table S2.
Among nucleotide-binding sites, the acceptor–donor–acceptor (ADA) motif, which can interact with the three edges of the nucleotide molecule (‘Watson–Crick’: N6 + N1, ‘Sugar’: N3 + O2′, ‘Hoogsten’: N6 + N7) (15), is the most common motif (motifs 1, 4, 9 and 10 in our set). Phosphate is most often bound by glycine-rich loops via hydrogen bonds to main chain nitrogen atoms, including the P-loop (28) (motifs 11 and 12), the dinucleotide-binding motif of Rossmann-type folds (29) (motifs 13 and 14), or other non-canonical P-loops (22) (motif 11). The binding mode of ribose is more diverse and mainly involves either main chain atoms or the side chains of arginine/lysine and aspartate/glutamate, as described previously (17).
A network of structural motifs
The network in Figure 1 shows the distribution of the 14 motifs across fold space and reveals substantial motif modularity and promiscuity; for instance, six motifs are shared by three or more folds, and 11 folds contain two or more motifs. The three most highly connected motifs involve the ADA motif, interacting with the sugar edge, the Watson–Crick edge and the P-loop, respectively. No ribose-binding motif was found in more than three folds reflecting its previously described diversity (see above).
The two largest clusters correspond to Rossmann or classical P-loop folds, which normally bind di- or mono-nucleotides, respectively, and are believed to be among the most ancient topologies (30,31). The Rossmann folds contain a greater number of motifs, though this is probably as it is one of the most common folds in the nature (and the database) (32).
We defined five different cases of modularity and quantitatively assessed their prevalence in the network. We first considered all the examples where a single fold had more than one motif binding to the same nucleotide fragment (Figure 2A). We then looked at all the instances where a single fold (B) or two different folds (C) have two motifs binding different portions of the ligand. Subsequently, we looked at cases where two folds share a motif for a fragment but possess two different motifs binding the same (D) or different fragments (E).
We used the network to extract all the combinations pertaining to each case (‘fold’ column in Figure 2). It should be noted that Figure 1 describes only the motifs possessed by each fold, but does not give any information about whether two motifs associated with the same fold are indeed located in the same protein. Therefore, the number of combinations extracted from the network represents the maximum number of examples of modularity in our dataset, only a fraction of which will be effectively verified in single structures. To address this issue, we searched for the 14 motifs in all the structures classified in SCOP (the distribution of each motif in the SCOP hierarchy is reported in Supplementary Table S2). This analysis was also repeated by pooling together the motifs at the family and superfamily levels (i.e. considering cases where the motifs occurred in the same family/superfamily but not necessarily in the same structure). In all cases but one (C), more than half of the combinations are effectively verified in a single family. Therefore, the examples of modularity appear to be quite widespread at the family and superfamily levels even if the number of cases where the motifs occur in the same structure is limited.
Modular composition of specific binding pockets
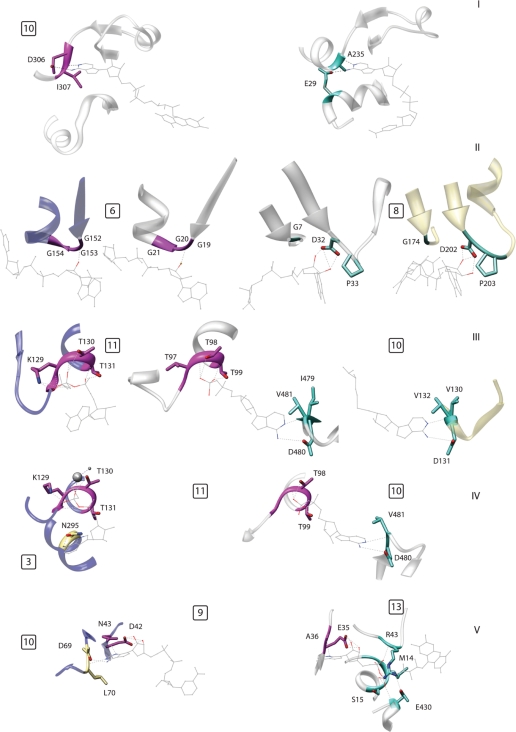
The panel in Figure 3 depicts several examples of modularity in the composition of nucleotide-binding sites. It is important to note that, as previously stated, the network does not contain binding modes, which are encoded in a single protein fold. Indeed, the same overall architecture can bind the same ligand fragment in different ways. One such example is depicted in panel 3(I). The proteins involved are an electron transferring flavoprotein (ETF) from Methylophilus methylotrophus and a lysine deacetylase of the Sir2 family (Sir2Af2). In Sir2Af2, the residue interacting with the N6 of adenine is located in an α-helix, which is much farther away from the ligand in ETF, resulting in a different binding mode in the latter protein.
Figure 3.
Examples of modular composition of specific binding pockets; the numbers in the small squares indicate the motifs, numbered as in Figure 1 and Supplementary Table S1. (I) Two proteins belonging to the same fold (DHS-like NAD/FAD-binding domain) that bind the same ligand fragment, namely adenine, in different ways. Left: ETF from M. methylotrophus (PDB: 1o96); right: lysine deacetylase of the Sir2 family (1s7g). (II) An example of alternative motifs in a single fold (Rossmann fold). The two central proteins are the Siroheme synthase CysG from Salmonella typhimurium (1pjs, inner left) and glyceraldehyde-3-phosphate dehydrogenase (1ihx, inner right). These proteins belong to the same fold but use different ribose-binding motifs shared with another fold. Outer left: thioredoxin reductase (1f6m, FAD/NAD(P)-binding domain), outer right: formate dehydrogenase H (1fdi, formate dehydrogenase/Dimethyl Sulfoxide reductase fold). (III) The center protein shares a phosphate binding motif with the one on the left and an adenine motif with the one on the right. All three proteins belong to different folds. Left: MurC (1gqy, ribokinase-like fold); center: group II chaperonin from Thermococcus strain KS-1 (1q3s, GroEL equatorial domain-like); right: methyltransferase from Dengue virus (1l9k, SAM-dependent methyltransferases). (IV) Two proteins sharing a phosphate-binding motif but using two different motifs to bind adenine. Left: MurC (1gqy, ribokinase-like fold); right: group II chaperonin from Thermococcus (1q3s, GroEL equatorial domain like). (V) The same motif, binding the sugar edge of adenine, is associated with a motif binding the ‘Watson–Crick’ edge (left) and with a phosphate-binding motif (right) in two different structures. Left: Gal10p (1z45, NAD(P)-binding Rossmann fold); right: polyamine oxidase (1b37, FAD/NAD(P)-binding domain).
Another possibility of variation in a single fold is where two alternative motifs for the same fragment exist and these are in turn shared with two other folds. Figure 3(II) shows that the siroheme synthase CysG, from Salmonella typhimurium, and glyceraldehyde-3-phosphate dehydrogenase have different ribose-binding motifs (6 and 8, respectively), common to different folds, even though both proteins adopt a Rossmann fold.
The analysis of the relationships between binding pockets belonging to two or more different folds reveals more complicated examples of modularity. Figure 3(III) depicts the binding site of a group II chaperonin from Thermococcus strain, KS-1. This pocket shares its adenine- and phosphate-binding motifs (10 and 11, respectively) with the proteins MurC (ribokinase-like fold) and methyltransferase from Dengue virus [S-Adenosyl methionine (SAM)-dependent methyltransferases]. Interestingly, in these two folds, the motifs do not occur together. Extending this relationship along the network reveals another intriguing example of modularity. Indeed, MurC itself has a distinct adenine-binding motif. Therefore, the chaperonin and MurC share the same phosphate-binding element but have alternative ways of binding adenine [Figure 3(IV)].
This example shows the key point that a single motif can be paired with multiple and distinct additional motifs in two binding pockets. These alternative partners may represent different modes of binding the same fragment, as in this case. However, they can also interact with another fragment of the ligand. Polyamine oxidase and Gal10p [Figure 3(V)] display exactly this phenomenon. They share a motif that simultaneously binds adenine and ribose (motif 9). This motif is paired with another one binding the ‘Watson–Crick’ edge of adenine in Gal10p (10), and with a phosphate-binding element characteristic of the FAD/NAD(P)-binding domain in polyamine oxidase (13). However, the latter two elements are never paired together in any fold.
Evolutionary origin of the binding motifs
The prevalence of this modularity raises several interesting questions. Perhaps, the first is whether the similarities we have detected are the result of evolutionary divergence from a common, albeit ancient ancestor, or convergence to a common molecular solution. This question, as ever, is difficult if not impossible to answer, though additional analysis of the dataset reveals that certainly some of the similarities could be the result of divergence.
We used DALI (33) to check for examples where the global superimposition of the structures coincides with the superimposition corresponding to the structural motif. For such examples, it is reasonable to hypothesize that the presence of the motif is the result of divergence. Clearly, because a single motif can be shared by more than two folds, there are cases where the motif can be considered divergent or convergent depending on the pair of folds considered. Overall, eight out of 14 motifs are found in at least one pair of folds whose global structural similarity suggests remote homology.
More specifically, a well-defined cluster of similar protein architectures is formed by the Rossmann, SAM-dependent methyltransferases, nucleotide-binding domains, FAD/NAD(P) binding, DHS-like NAD/FAD binding, formate dehydrogenase, urocanase and activating enzymes of the ubiquitin-like proteins folds. These similarities have already been well documented (34). These folds adopt a scaffold of five or more strands in a parallel β-sheet with helices on either side whose prototype is the Rossmann fold itself. P-loop ATPases also share elements of this fold, though the question of whether they are homologous is still open (35–37). In addition to that our analysis shows the ribokinase-like fold to be related to the P-loop ATPases fold.
DISCUSSION
We have shown that nucleotide-binding pockets often have a modular arrangement of structural elements responsible for binding different regions of their ligands. Modularity is advantageous as discretizing functional units is ultimately more resilient to mutation (5), and their exchange can allow for efficient emergence of new functions (1,2,38). Several of the binding sites discussed here have the first property, but it is unlikely that such small units would ever be shuffled in evolution. Akin to linear motifs (39), these binding modules are probably more easily mutable because of their small size. Thus, the modularity is likely the result of a mixture of ancient divergence and convergences of very small protein motifs and is probably more the reflection of a functional need than an evolutionary divergence. The same motifs are found in various combinations in different binding sites, but this does not imply that they have a common evolutionary origin. Convergence seems very likely due to the constraints imposed by the structure of the ligand, which is akin to the proposition that reaction chemistry acts as a constraint in convergently evolved enzyme active site similarities (40). This is in contrast to other modularities in nature, such as modular protein domain architecture or modularity in complexes, where the units are almost certainly divergent from a common ancestor.
As a concept, modularity has both evolutionary and functional implications. The binding motifs we describe do not necessarily have the same evolutionary origin. However, we demonstrate that they can be functionally interchangeable: that one can, in principle, be used in place of another. This observation has interesting functional implications because it shows that binding pockets can be decomposed in small modules instead of being treated as whole functional units. Our findings might also shed light on the general observation that, with the exception of the protease catalytic triad, convergences of entire functional sites are rare events. If one allows for modularity, many more convergently evolved binding sites of the same type become evident.
As many other ligands show a modular arrangement of chemical fragments (e.g. carbohydrates, peptides), we expect that this principle will reveal additional binding site modularities once sufficient structural data become available. The identification of these protein motifs and their modular positioning suggests interesting possibilities to detect new binding sites based on spatial proximity of the motifs on protein surfaces, which will be of great promise for assigning ligands to protein structures and ultimately for the design of chemicals that could modulate their function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was funded by the European Commission under FP7, LEISHDRUG Project (to M.H.C.), and under FP6, contract LSHG-CT-2005-512028 (to R.B.R.). Funding for open access charge: LEISHDRUG EC FP7.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Profs Alfonso Valencia, Anna Tramontano and Gianni Cesareni for interesting discussion and for critically reading the manuscript.
REFERENCES
- 1.Carroll SB. Chance and necessity: the evolution of morphological complexity and diversity. Nature. 2001;409:1102–1109. doi: 10.1038/35059227. [DOI] [PubMed] [Google Scholar]
- 2.Patthy L. Genome evolution and the evolution of exon-shuffling–a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 5.Wagner GP, Altenberg L. Perspective: complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 6.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl Acad. Sci. USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 8.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 9.Brannetti B, Via A, Cestra G, Cesareni G, Helmer-Citterich M. SH3-SPOT: an algorithm to predict preferred ligands to different members of the SH3 gene family. J. Mol. Biol. 2000;298:313–328. doi: 10.1006/jmbi.2000.3670. [DOI] [PubMed] [Google Scholar]
- 10.Petsalaki E, Stark A, García-Urdiales E, Russell RB. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput. Biol. 2009;5:e1000335. doi: 10.1371/journal.pcbi.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc. Natl Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 13.Ausiello G, Gherardini PF, Gatti E, Incani O, Helmer-Citterich M. Structural motifs recurring in different folds recognize the same ligand fragments. BMC Bioinformatics. 2009;10:182. doi: 10.1186/1471-2105-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji HF, Kong DX, Shen L, Chen LL, Ma BG, Zhang HY. Distribution patterns of small-molecule ligands in the protein universe and implications for origin of life and drug discovery. Genome Biol. 2007;8:R176. doi: 10.1186/gb-2007-8-8-r176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denessiouk KA, Johnson MS. “Acceptor-donor-acceptor” motifs recognize the Watson-Crick, Hoogsteen and Sugar “donor-acceptor-donor” edges of adenine and adenosine-containing ligands. J. Mol. Biol. 2003;333:1025–1043. doi: 10.1016/j.jmb.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Traut TW. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 17.Vetter IR, Wittinghofer A. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys. 1999;32:1–56. doi: 10.1017/s0033583599003480. [DOI] [PubMed] [Google Scholar]
- 18.Cappello V, Tramontano A, Koch U. Classification of proteins based on the properties of the ligand-binding site: the case of adenine-binding proteins. Proteins. 2002;47:106–115. doi: 10.1002/prot.10070. [DOI] [PubMed] [Google Scholar]
- 19.Mao L, Wang Y, Liu Y, Hu X. Molecular determinants for ATP-binding in proteins: a data mining and quantum chemical analysis. J. Mol. Biol. 2004;336:787–807. doi: 10.1016/j.jmb.2003.12.056. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi N, Go N. A method to search for similar protein local structures at ligand binding sites and its application to adenine recognition. Eur. Biophys. J. 1997;26:135–144. doi: 10.1007/s002490050065. [DOI] [PubMed] [Google Scholar]
- 21.Nobeli I, Laskowski RA, Valdar WS, Thornton JM. On the molecular discrimination between adenine and guanine by proteins. Nucleic Acids Res. 2001;29:4294–4309. doi: 10.1093/nar/29.21.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinoshita K, Sadanami K, Kidera A, Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 23.Brakoulias A, Jackson RM. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 24.Watson JD, Milner-White EJ. A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2002;315:171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- 25.Via A, Ferre F, Brannetti B, Valencia A, Helmer-Citterich M. Three-dimensional view of the surface motif associated with the P-loop structure: cis and trans cases of convergent evolution. J. Mol. Biol. 2000;303:455–465. doi: 10.1006/jmbi.2000.4151. [DOI] [PubMed] [Google Scholar]
- 26.Ausiello G, Via A, Helmer-Citterich M. Query3d: a new method for high-throughput analysis of functional residues in protein structures. BMC Bioinformatics. 2005;6(Suppl. 4):S5. doi: 10.1186/1471-2105-6-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausiello G, Gherardini PF, Marcatili P, Tramontano A, Via A, Helmer-Citterich M. FunClust: a web server for the identification of structural motifs in a set of non-homologous protein structures. BMC Bioinformatics. 2008;9(Suppl. 2):S2. doi: 10.1186/1471-2105-9-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saraste M, Sibbald PR, Wittinghofer A. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 29.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 30.Ma B, Chen L, Ji H, Chen Z, Yang F, et al. Characters of very ancient proteins. Biochem. Biophys. Res. Commun. 2008;366:607–611. doi: 10.1016/j.bbrc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Boca SM, Kalelkar R, Mittenthal J, Caetano-Anolles G. A phylogenomic reconstruction of the protein world based on a genomic census of protein fold architecture. Complexity. 2006;12:27–40. [Google Scholar]
- 32.Day R, Beck D.AC, Armen RS, Daggett V. A consensus view of fold space: combining SCOP, CATH, and the Dali domain dictionary. Protein Sci. 2003;12:2150–2160. doi: 10.1110/ps.0306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 34.Aravind L, Mazumder R, Vasudevan S, Koonin EV. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 2002;12:392–399. doi: 10.1016/s0959-440x(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 35.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheek S, Zhang H, Grishin NV. Sequence and structure classification of kinases. J. Mol. Biol. 2002;320:855–881. doi: 10.1016/s0022-2836(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 37.Kull FJ, Vale RD, Fletterick RJ. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J. Muscle Res. Cell Motil. 1998;19:877–886. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- 38.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol. Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg M.JE. Convergent evolution of enzyme active sites is not a rare phenomenon. J. Mol. Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.