Abstract
Escherichia coli can survive extreme acid stress for several hours. The most efficient acid resistance system is based on glutamate decarboxylation by the GadA and GadB decarboxylases and the import of glutamate via the GadC membrane protein. The expression of the corresponding genes is controlled by GadE, the central activator of glutamate-dependent acid resistance (GDAR). We have previously shown by genetic approaches that as well as GadE, the response regulator of the Rcs system, RcsB is absolutely required for control of gadA/BC transcription. In the presence of GadE, basal activity of RcsB stimulates the expression of gadA/BC, whereas activation of RcsB leads to general repression of the gad genes. We report here the results of various in vitro assays that show RcsB to regulate by direct binding to the gadA promoter region. Furthermore, activation of gadA transcription requires a GAD box and binding of an RcsB/GadE heterodimer. In addition, we have identified an RcsB box, which lies just upstream of the −10 element of gadA promoter and is involved in repression of this operon.
INTRODUCTION
Escherichia coli is a neutrophilic bacteria that can survive for a long time at very low pH (<2.5). This property is due to three clearly defined acid resistance (AR) systems: glutamate-dependent, arginine-dependent and oxidative systems (1). The glutamate-dependent (Gad) system is the most effective. Two glutamate decarboxylase isoforms, GadA and GadB, consume intracellular protons in decarboxylating glutamate, and GadC, an integral membrane protein, exchanges extracellular glutamate for the decarboxylation product, γ-amino butyric acid (GABA) (2,3). The gadB and gadC genes are cotranscribed, while gadA forms an operon with gadX, which encodes an activator of the gad system (4). gadA and gadX belong to the acid fitness island (AFI), a 15-kb region unique to E. coli and Shigella species, which specifies 12 proteins and a small RNA that contribute to E. coli acid resistance in different ways (5). One of these genes, gadE, codes for a master regulator of glutamate-dependent acid resistance (GDAR) belonging to the LuxR-like family (6,7). GadE is the key activator of gadA and gadBC genes. It binds to a GAD box, a 20-bp consensus sequence centered 63 bp upstream of the transcriptional start sites of these genes (8). Regulation of gadE expression is extremely complex. At least nine regulators appear to converge on the gadE promoter region to either activate or repress its transcription in response to different environmental conditions (1).
Previously, we demonstrated by genetic approaches that RcsB, a response regulator, is another essential regulator of gadA/BC transcription (9). RcsB is part of the RcsCDB phosphorelay, a signal transduction system conserved in members of the Enterobacteriaceae. This system was first identified as a regulator of capsule synthesis and is involved in pathogenesis, development of biofilms, and the regulation of envelope composition or trafficking (10). RcsB can also be activated independently of the phosphorelay, by binding of different co-regulators: RcsA, the main one, and RmpA, TviA and PhoP (11–14). Presumably, this allows multiple inputs into RcsB-mediated regulation of specific promoters.
However, the ability of RcsB to stimulate gadA/B expression is independent of its Rcs partners, both that involved in the phosphorelay and the RcsA activator. In addition, although the activity of RcsB requires a functional gadE allele, its effect is not mediated by activation of gadE transcription, as is the case for the majority of other regulators of GDAR. In contrast, activation of RcsB, either through the phosphorylation pathway or by RcsA, leads to general repression of gad gene, including gadA, gadB and gadE itself, and a corresponding reduction in acid resistance (9).
The goal of the present study was to investigate the mechanism of regulation of the gad genes by GadE and RcsB. We show that RcsB exerts direct control over gadA/BC expression. RcsB is a critical partner of GadE and the binding of both regulators as a heterodimer to the GAD box activates gadA transcription. In addition, we identified an RcsB box located directly upstream from the −10 element of the gadA promoter, which contributes to the repression of gadA transcription upon Rcs activation.
MATERIALS AND METHODS
Strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. The rcsA and rcsCDB deletions were constructed as described by Datsenko and Wanner (15). The whole ORFs were deleted and replaced by the chloramphenicol resistance (cat) gene. To construct MPC398 strain, we first transduced ΔrcsCDB::cat into MC4100 lon− strain (UGM2). The resistance gene was eliminated by using a FLP helper plasmid (15). Then, we transduced ΔrcsA::cat into the previous strain and finally a ΔgadE::kan (8).
Table 1.
Bacterial strains and plasmids in this study
| Genotype | Source or reference | |
|---|---|---|
| Strains | ||
| UGM2 | MC4100 lon- (ΔPstI) | Lab. collection |
| MPC398 | MC4100 lon- ΔrcsCDB ΔrcsA::cat ΔgadE::kn | This study |
| MPC295 | trpDC::putPA1303-Km-gadA::lacZ (–165 to +788) | (9) |
| MPC297 | trpDC::putPA1303-Km-gadB::lacZ (–203 to + 788) | (9) |
| MPC299 | trpDC::putPA1303-Km-gadE::lacZ (–804 to +331) | (9) |
| CF6343 | MG1655 ΔlacIZ (MluI) | Lab. Collection |
| Plasmids | ||
| pHRcsB | pFab1, p15A-derived vector carrying ϕ(lacPUV5-his6-rcsB) | (20) |
| pHGadE | p15A-derived vector carrying ϕ(lacPUV5-his6-gadE) | This study |
| pMF533 | pMALc2E (New England Biolabs) containing malE-gadE fusion | (8) |
| pMRcsB | pMALc2E (New England Biolabs) containing malE-rcsB fusion | This study |
| pRS550 | ColE1 derived cloning vector, ApR, KnR | (17) |
| pRSgadA-lacZ | pRS550 containing gadAp fused to lacZ | This study |
pHGadE plasmid was constructed by amplifying gadE orf from MG1655 strain (gi:948023) using gadEup and gadEdown primers. The polymerase chain reaction (PCR) product was digested with NdeI and BamHI and cloned in the corresponding sites of pAPT156 (16).
pMRcsB plasmid was constructed by amplifying rcsB orf from MG1655 strain (gi:947441) using rcsBupM and rcsBdownM primers. The PCR product and the pMALc2E plasmid were digested by BamHI and HindIII enzymes, and ligated.
pRSgadA-lacZ plasmids were constructed as follows. PCR-generated fragments were cloned between the BamHI and EcoRI sites of pRS550 (17). The gadA promoter from −101 to +24 relative to the transcription start of the gadA gene was amplified with the primers gadAup and gadAdown. The introduction of the mutations M1, M2 and M3 in the gadAp promoter region was done by PCR using primers carrying the corresponding mutations.
Oligonucleotide sequences (5′ to 3′)
gadAup (GTTTATATTATAAAAAGTCG), gadAdown (GAACTCCTTAAATTTATTTG), gadEup (GGAATTCCATGCATCACCATCACCATCACATGATTTTTCTCATGACG), gadEdown (CGCGGAT CCCTAAAAATAAGATGTG), lacZ24 (CGCCAGGGTTTTCCCAGTCACGAC), rcsBupM (CGCGGA TCCAACAATATGAACGTAATTATTGCC), rcsBdownM (CGCAAGCTTTTAGTCTTTATCTGCCG GACT).
Purification of MBP-GadE and MBP-RcsB fusion proteins
Escherichia coli MPC398 containing the pMF533 or the pMRcsB plasmid expressing MBP (Maltose Binding Protein product of the malE gene) fused to GadE (Uniprot: P63204) or RcsB (Uniprot: P69407) protein respectively (8; our work), was grown at 37°C with aeration in 1 l of LB broth supplemented with 0.2% glucose and 100 µg/ml ampicillin. The purification procedure is described in Ma et al. (8).
Purification of (His)6-RcsB and (His)6-GadE fusion proteins
Escherichia coli MPC398 containing pHRcsB or pHGadE was grown at 37°C with aeration in 1 l of LB broth supplemented with 0.2% glucose and 75 µg/ml of spectinomycin. At OD600 ∼ 0.3, IPTG was added to a final concentration of 0.5 mM. The culture was incubated for an additional 3 h. Cells were then treated with lysozyme (0.75 mg/ml) for 30 min and disrupted by sonication in 10 ml of column buffer (50 mM NaP buffer pH 7, 300 mM NaCl). Lysates were centrifuged at 12 000g for 30 min. The supernatants were loaded onto 1 ml TALON columns (Ozyme). The loaded columns were washed with 20 ml of column buffer and 5 ml of column buffer plus 10 mM imidazole. The proteins were eluted with 5 ml of column buffer containing 150 mM imidazole. Samples with highest protein contents were combined and dialyzed against column buffer containing 40% glycerol. Protein concentrations were determined by Bio-Rad protein assays.
Electrophoretic mobility shift assay
Probes were PCR-generated with 32P 5′ end-labeled primers and purified using S300 columns (GE Healthcare). With proteins, 5000 c.p.m. per reaction were incubated at room temperature for 30 min in 10 µl of binding buffer (110 mM NaP pH 7, 10 mM EDTA, 20 mM NaCl, 1 mM DTT, 50 mM potassium glutamate, 1 µg poly-(dI-dC) (Sigma), 7 µM bovine serum albumin). Samples were loaded onto 5% TBE non denaturing gels and electrophoresed at room temperature in 0.5 X TBE. After migration, gels were dried, exposed to Fujifilm sensitive screen. Quantifications were made using Multi Gauge Software.
DNase I footprinting
Experiments were performed using protocol described by Francez-Charlot et al. (18). Reactions contained 5−10 nM of purified gadA promoter-containing templates which have been generated by PCR using gadAup and gadAdown primers. gadAup primer was end-labeled with [γ-32P] ATP by polynucleotide kinase.
In vitro transcription assays
Reactions (14 µl) were performed in buffer containing 20 mM Tris−HCl pH 8, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 50 µg/ml bovine serum albumin (BSA), 5% glycerol, 0.1 mg/ml heparin, 200 µM ATP, 200 µM CTP, 200 µM GTP, 10 µM UTP and 5 µCi of [α-32P] UTP (Promega). Linear DNA templates were generated by PCR using gadAup and lacZ24 primers from pRSgadAlacZ plasmid, gel purified using a Qiagen gel extraction kit; 20 nM of templates were used per reaction. When required GadE and RcsB proteins were added, then the reaction was initiated by the addition of 0.3 U of RNAP (Tebu). After 10 min at 30°C, samples were analyzed by denaturing gel electrophoresis. Transcript products were quantified using a Phosphorimager (Fujifilm) and Multi-Gauge software.
Beta-galactosidase assay
Cells were grown in LBG (LB plus 0.4% glucose) at 37°C. Overnight cultures were diluted 7000-fold, grown for five/10 generations, prior to addition of 500 µM IPTG. The cultures were sampled at different intervals for assay of β-galactosidase activities (19).
RESULTS
Both RcsB and GadE directly regulate gadA transcription
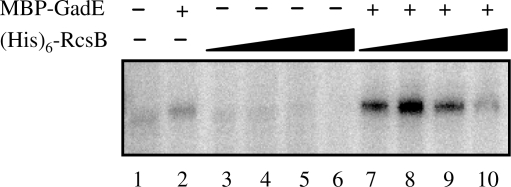
To test whether RcsB acts directly at gad promoters, we carried out an in vitro transcription assay using linear DNA fragment containing the gadA promoter region fused to lacZ as template. This assay was based on His-tagged version of RcsB and a MBP-tagged version of GadE (8,20). With RNA polymerase alone, a transcript of the expected size was visible (Figure 1, lane 1). The signal was weak, in agreement with the observation that in vivo the activity of the gadA promoter is low in the absence of either GadE or RcsB (9). Addition of MBP-GadE to the reaction did not appreciably change the amount of transcript (Figure 1, lane 2). In contrast, in the presence of MBP-GadE, the addition of 3 or 30 nM of (His)6-RcsB (Figure 1, lanes 7 and 8, respectively) strongly stimulated gadA transcription (up to 7-fold). This result therefore indicates that GadE and RcsB act synergistically to stimulate transcription of gadA. At higher concentrations of (His)6-RcsB, stimulation of gadA transcription by RcsB in the presence of GadE declined, essentially disappearing at 3 µM RcsB (Figure 1, lane 10). This behavior implies that the ratio of the two regulators is important for the balance activation/repression. Furthermore, (His)6-RcsB addition alone (Figure 1, lanes 3–6) had a concentration dependent negative effect on gadA transcription, indicating that RcsB alone directly represses the basal level of gadA transcription.
Figure 1.
Synergistic regulation of in vitro transcription initiation at the gadA promoter by GadE and RcsB. The gadA promoter-containing linear template was pre-incubated as indicated with RNA polymerase alone (lane 1), plus 760 nM MBP-GadE (lane 2), plus 3, 30, 300 or 3000 nM (His)6-RcsB (lanes 3–6, respectively), or plus MBP-GadE and (His)6-RcsB at the same concentrations (lanes 7–10, respectively). The figure shows a representative autoradiogram.
RcsB and GadE protection of the gadA regulatory region
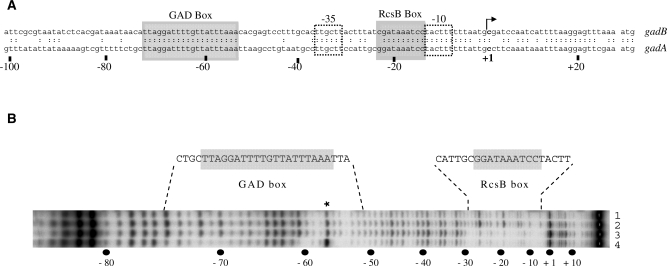
A GAD box, located 16 bp upstream of the −35 element of the gadA promoter, is required for the activity of GadE regulator (Figure 2A; Ref. 8). We had previously noticed that the left part of the GAD box was similar to the half site of the consensus RcsB box, and suggested that both GadE and RcsB might bind to it in order to stimulate gadA transcription (9). In addition, we identified a putative RcsB box located immediately upstream of the −10 element that could be required for the repression of gadA transcription. These two boxes are strictly conserved in the gadB promoter region (Figure 2A), except for a C instead of a G at the left end of the RcsB box. The reality of the predicted RcsB box in the gadA regulatory region was demonstrated using a DNase I protection assay. As shown in Figure 2B, (His)6-RcsB protected a 23-bp region containing the putative RcsB box from DNase I digestion (Figure 2B, lane 3). MBP-GadE alone did not protect either the GAD box or the RcsB box (Figure 2B, lane 2). MBP-GadE and (His)6-RcsB together protected a region extending from −50 to −76, including the GAD box (Figure 2B, lane 4). In these conditions [20 µM of MBP-GadE and 40 µM of (His)6-RcsB] the binding of (His)6-RcsB to its box was unaffected (Figure 2B: compare lane 4 to lane 3). Protection of the GAD box was still apparent with 1 µM of MBP-GadE and 0.1 µM of (His)6-RcsB (data not shown). However, in this case RcsB no longer protected the RcsB box, suggesting that the affinity of RcsB for the GAD box when GadE is present was higher than for the RcsB box. Therefore, there are two binding sites in the gadA regulatory region, the RcsB box next to the −10 element, where RcsB at high concentration binds independently of GadE, and the GAD box, where RcsB acts at lower concentrations provided GadE is present, suggesting a functional interaction between these two regulators.
Figure 2.
Identification of MBP-GadE and (His)6-RcsB binding sites in the gadA regulatory region. (A) Nucleotide sequences of the gadA and gadB promoter regions. Putative –35 and –10 elements of the gad promoters are boxed. The GAD and putative RcsB boxes are shaded. +1 denotes the transcription start point and numbers below indicate distance relative to this point. (B) DNase I footprinting assays were performed with 20 µM of MBP-GadE and/or 40 µM of (His)6-RcsB on the non-template strand. Protected regions and corresponding sequences are indicated above the autoradiogram. A DNase I hypersensitive site is indicated by an asterisk. The positions relative to the +1 start of transcription are shown below. No protein (lane 1), 20 µM MBP-GadE (lane 2), 40 µM (His)6-RcsB (lane 3), 20 µM MBP-GadE and 40 µM (His)6-RcsB (lane 4).
Binding of RcsB and GadE to the gadA regulatory region
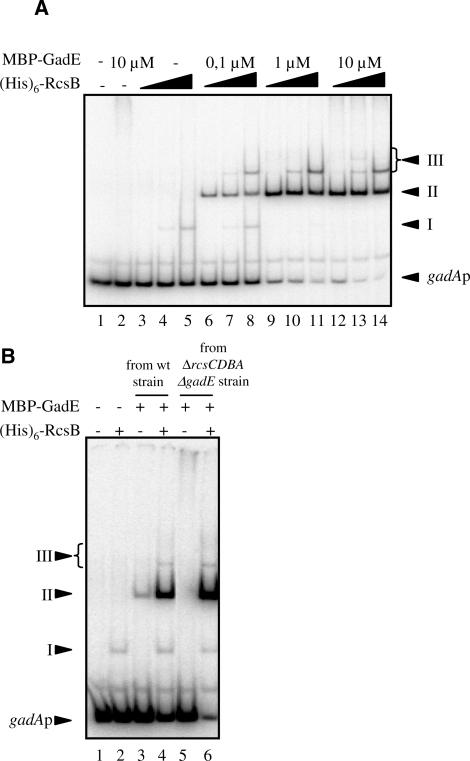
To probe more deeply into the relationship between RcsB, GadE and the two binding sites in the gadA promoter, we used electrophoretic mobility shift assay (EMSA) to analyze binding to a fragment extending from −101 to +24, containing both the GAD and the RcsB boxes. When the DNA was incubated with (His)6-RcsB protein alone at concentrations 1 µM and above, a shifted complex appeared (Figure 3A; complex I, lanes 4 and 5). This result indicated that RcsB alone could bind stably to the gadA regulatory region, probably to the RcsB box. On the other hand, we detected no retarded complex with MBP-GadE alone, even at 10 µM (Figure 3A, lane 2). With concentrations of RcsB and GadE at or above 0.01 µM (Figure 3A and data not shown), a major retarded complex, named complex II, appeared; furthermore, the limiting factor for complex II formation seems to be GadE since increasing quantities of complex II were obtained only by raising the concentration of MBP-GadE (Figure 3A). This corroborated the DNase I protection result. Other shifted complexes, called complexes III, that migrated more slowly than complex II appeared as the concentration of RcsB rose [Figure 3A: 10 µM of (His)6-RcsB in lanes 8, 11 and 14]. Hence, RcsB seems to bind pre-formed complex II, either at the RcsB box or at the GAD box already covered by RcsB and/or GadE.
Figure 3.
Binding of MalE-GadE and (His)6-RcsB to the gadA promoter region. (A) EMSA with gadA promoter region-templates (from –101 to +24 relative to the +1start of transcription). No protein (lane 1), 10 µM MBP-GadE (lane 2), 0.1, 1 and 10 µM (His)6-RcsB plus 0.1 µM MBP-GadE (lanes 6–8) or plus 1 µM MBP-GadE (lanes 9–11), plus 10 µM MBP-GadE (lanes 12–14). (B) EMSA using protein from wt or rcs mutants with gadA promoter region templates. Proteins were purified from either a wild-type strain (lanes 3 and 4) or a ΔrcsA ΔrcsCDB ΔgadE strain (lanes 2, 5 and 6). No protein (lane 1), 1 µM (His)6-RcsB (lane 2), 3 µM MBP-GadE (lanes 3 and 5), 3 µM MBP-GadE and 1 µM (His)6-RcsB (lanes 4 and 6).
These results contradict published reports that GadE regulator alone could bind to the GAD box (8,21,22). A likely explanation for this discrepancy was obtained when MBP-GadE purified from a wild-type strain was used in the EMSA instead of that purified from a ΔrcsCDBA ΔgadE strain used until now in this work. With 3 µM of this MBP-GadE preparation, we detected a retarded complex migrating at the same position as the RcsB and GadE-dependent complex II (Figure 3B, compare lane 3 with lanes 4 and 6). This result strongly suggests that the binding of MBP-GadE prepared from the wild-type strain is due to the copurification of RcsB with GadE. Finally, the presence of RcsB in the preparation of MBP-GadE purified from wild-type strain was confirmed by western-blot analysis using antibody raised against RcsB (kind provided by Anne Vianney; data not shown). These results suggested that GadE and RcsB act synergistically in binding to the gadA regulatory region and that both proteins interact directly.
RcsB/GadE heterodimer binds to the GAD box
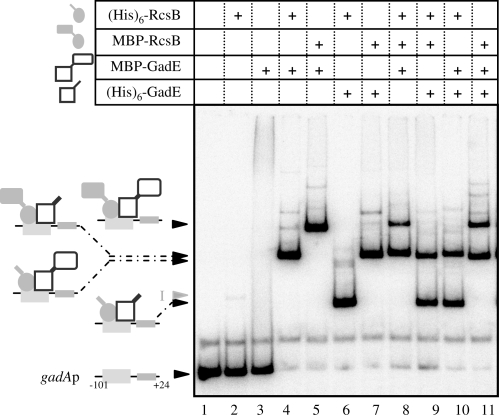
The formation of complex II could result either to binding of both proteins or to binding of only one after modification by the other. The presence of GadE and RcsB in complexes II and III was tested by using combinations of His-tagged and MBP-tagged versions of GadE and RcsB. Owing to the differences in fusion protein molecular weights [∼20 kDa for (His)6-GadE versus ∼62 kDa for MBP-GadE, and ∼23.6 kDa for (His)6-RcsB versus ∼66 kDa for MBP-RcsB], migration patterns were expected to depend on the combination of proteins present in the different complexes. As shown in Figure 4, this was the case. Complex II exhibited lower mobility when an MBP-tagged version of each protein was used instead of the corresponding His-tagged form (Figure 4, compare complex II migrations in lanes 4 and 5, or 4 and 6, or 5 and 7 or 6 and 7), as expected if complex II consisted of a RcsB/GadE oligomer bound to the gadA regulatory region. Combinations of three proteins, both tagged versions of one and one tagged version of the other, gave only two major retarded complexes II in this assay (Figure 4, lanes 8–11). The lack of additional shifted bands indicated that equivalent amounts of RcsB and GadE bound to the gadA promoter region. Taking into account that both regulators belong to protein families known to bind DNA as a dimer (23,24) and the size of the protected region at the GAD box, we could reasonably conclude that complex II is the result of an RcsB/GadE heterodimer sitting on the gadA regulatory region, at the GAD site.
Figure 4.
RcsB and GadE bind GAD box as a heterodimer. EMSA was performed as described in Figure 3 using the following proteins at 1 µM: (His)6-RcsB, MBP-RcsB, MBP-GadE and (His)6-GadE. Schematic representations of complexes II obtained are presented on the left-hand side.
Binding activities of RcsB and GadE at the gadA promoter
The affinity of the RcsB/GadE heterodimer for GAD box was estimated by EMSA with increasing concentrations of RcsB in reactions containing a saturating concentration of GadE (2.5 µM). With this approach, the apparent binding constant of the RcsB/GadE heterodimer was calculated to be ∼10 nM. We noticed that a large proportion of MBP-GadE formed homodimers in solution (data not shown). Hence, to obtain enough GadE monomers and to favor the RcsB/GadE heterodimer formation, we used a saturating concentration of GadE. We could not estimate the apparent binding constant of the RcsB homodimer for the RcsB box because even with 10 µM RcsB, only 6% of the probe was retarded (Figure 3A; lane 5). This indicates that the activation of gadA transcription could take place at low concentrations of both proteins, whereas repression of gadA transcription would require higher level of RcsB.
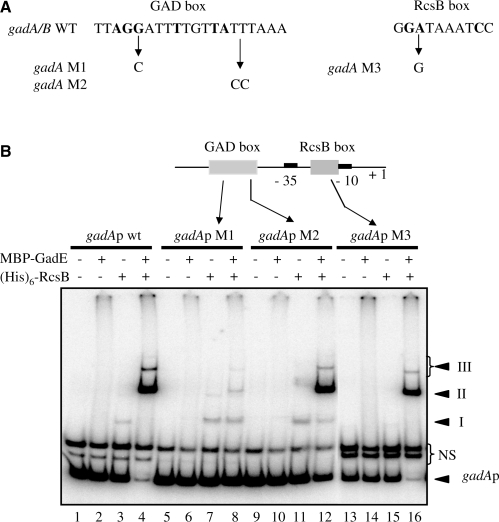
Independent binding on the RcsB and the GAD boxes
In an effort to dissect protein–protein and DNA–protein interactions, various mutations were introduced into the GAD box or in the RcsB box (Figure 5A) and their effects were analyzed by EMSA (Figure 5B). The M1 mutation changes a highly conserved G to a C. This G is also highly conserved in the RcsB box. As shown by the EMSA in Figure 5B (lanes 5–8), this mutation caused a dramatic diminution of complexes II and III, indicating that this G is critical for the binding of the RcsB/GadE heterodimer to the GAD box. In contrast, the formation of complex I was not compromised, in agreement with the idea that this complex was formed by RcsB bound to the RcsB box. The M2 mutation also introduced a modification TT to CC in the right-hand part of the GAD box, but in a less conserved region. This mutation caused a small decrease in the affinity of the RcsB/GadE proteins (Figure 5B, compare lanes 4 and 12), indicating that these nucleotides are not critical for binding RcsB/GadE. Again, no effect on the binding of RcsB to the RcsB box was observed. Finally, the M3 mutation replaced the conserved A in the RcsB box with a G. This mutation completely inhibited complex I formation by RcsB (Figure 5B, lane 15), but had no impact on the formation of complexes II and III by RcsB and GadE (Figure 5B, lane 16). Taken together, these results indicate that binding to one box was largely independent of binding to the other box. It should be pointed out that, as for the major complex II, the formation of minor complexes III was mainly independent of the RcsB box, indicating that these slow migrating complexes III do not result principally to simultaneous occupation of the Rcs and GAD boxes. This was confirmed by EMSA with a shorter DNA fragment containing only the GAD box. This fragment was still able to form two retarded complexes whose migrations were equivalent to that of complexes II and III (Supplementary Figure S1).
Figure 5.
Effects of GAD or RcsB box mutations on RcsB and GadE binding. (A) Point mutations introduced into the GAD and RcsB boxes. Conserved bases are shown in bold. (B) EMSA of gadA promoter fragment mutants in the presence of (His)6-RcsB (5 µM), and/or MBP-GadE (4 µM). NS: non-specific DNA.
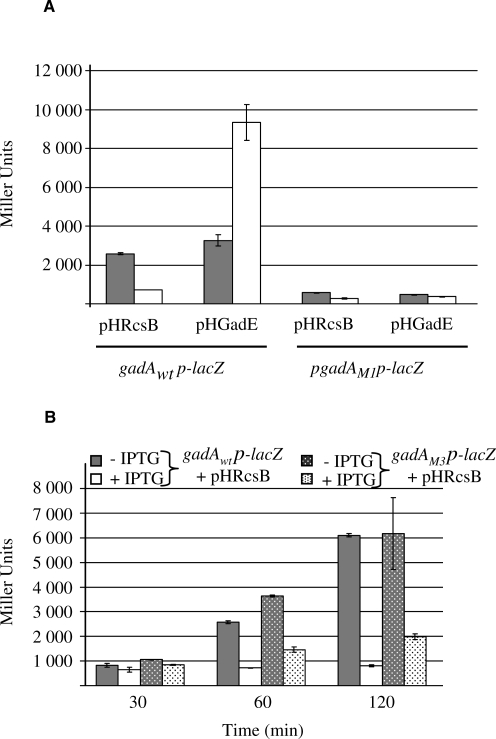
Effects of point mutations on transcriptional activity of gadA in vivo
The in vivo transcriptional activity of the gadA promoter was monitored using plasmid-borne transcriptional fusions between gadA regulatory region (−101 to +24) and lacZ (Figure 6). As previously reported (9), in a wild-type background, overproduction of (His)6-GadE activates the transcription of gadA, whereas overproduction of (His)6-RcsB represses. In contrast, the construction with the inactivated GAD box (mutation M1) displayed a lower intrinsic activity that was no longer stimulated by GadE (Figure 6A). Repression of the β-galactosidase activity was still observed after 1 h of overproduction of RcsB, even if the promoter was no more activated. This repression indicated that the GAD box is not involved in repression of gadA expression by RcsB.
Figure 6.
Effects of RcsB or GAD box mutations on the transcriptional regulation of gadA in vivo. (A) β-Galactosidase activities of transcriptional gadA-lacZ fusions (either wild-type or M1 mutant promoter) with or without overexpressed (His)6-GadE or (His)6-RcsB. Strains carrying gadAp-lacZ and either pHGadE or pHRcsB plasmids with (white bars) or without (gray bars) 0.5 mM IPTG added for 1 h. (B) Same experiment as in A for gadApwt-lacZ and gadApM3-lacZ fusions with or without induction of RcsB synthesis. Activities were measured 30, 60 and 120 min after IPTG addition.
With the M3 fusion (mutation in the RcsB box), the basal activity of the gadA promoter was slightly higher and RcsB repression little affected (Figure 6B). In the case of the mutant promoter, the transcription level was more than twice that of a wild-type promoter after 120 min of RcsB over expression (2000 versus 800 Miller units). However, a repression was still observed with this mutant promoter indicating that at least part of the RcsB-dependent repression of gadA is observed without normal binding of RcsB to the RcsB box. This was, however, consistent with our previous finding that overproduction of RcsB leads to general repression of the gad genes, including those coding for the activators of gadA expression (9). Therefore, in the case of the M3 fusion, the repression observed after RcsB overproduction was probably due to a lower level of GadE and/or GadX(W).
DISCUSSION
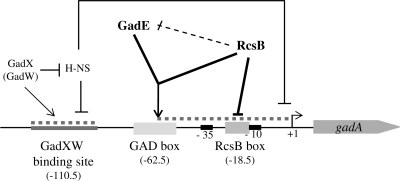
In this study, we report novel findings that shed light on the highly intricate regulation of glutamate-dependent acid resistance in E. coli (Figure 7). In particular, we have provided evidence that RcsB is a critical co-activator along with GadE of the gadA transcription. Genetic approaches had shown that in addition to the central regulator GadE, the response regulator RcsB is strictly required to activate the expression of the three genes gadA/BC and to develop resistance to low pH (9). Here, we demonstrate through in vitro assays that RcsB and GadE act directly and synergistically to activate gadA transcription (Figure 1). Localization of the binding site responsible for this activation by genetic (Figure 6) and footprinting (Figure 2) experiments confirmed the identity of the GAD box previously identified (8). However, our data demonstrate that the GadE protein alone is not able to bind this GAD box, and that GadE requires the formation of hetero-oligomers with the co-regulator RcsB to bind this site in the gadA regulatory region (Figures 2–5). These data contradict previous reports that GadE alone could bind to the GAD box (8,21,22). As shown in Figure 3B, this discrepancy can be explained by contamination of MBP-GadE overproduced in wild-type strains by RcsB protein.
Figure 7.
Regulatory model of gadA transcription. Schematic representation of gadA regulatory region with the GadXW (33), GAD and RcsB boxes (gray boxes). The H-NS binding sites responsible for gadA repression are represented as dotted line (37). Direct regulation is shown as a continuous arrow. Repression of gadE transcription by RcsB, in which a direct effect has not been confirmed, is shown as a dotted arrow. This putative feed-forward loop could coordinate gadA expression.
Importantly, we demonstrate here that RcsB and GadE both bind to the GAD box. Indeed, retarded complexes II and III, formed in EMSA with gadA promoter region in presence of both RcsB and GadE contain both proteins (Figure 4) and are abolished by mutation of a highly conserved nucleotide in the GAD box (Figure 5). Although our experiments do not define the stoichiometry of the proteins in the protein/DNA complexes, it is likely by analogy with other response regulators and LuxR-like activators (23,24) that the GAD box can accommodate one RcsB/GadE heterodimer. At present, the difference between complexes II and III is not clear, but the GAD box appeared sufficient to generate these complexes (Figures S1 and 5). We note that formation of complex III depended on RcsB concentration (Figure 3), which could be consistent with secondary binding of RcsB to the already bound RcsB/GadE or alternatively to the RcsB box, this latter corroborates the DNase I protection of the two boxes (Figure 2). The RcsB/GadE heterodimer binds the GAD box with an apparent binding constant of 10 nM, which is 10-fold lower than that measured for the RcsA/RcsB heterodimer (25). The point mutations introduced into the GAD box, on either the right or the left side, did not enable definition of the half-binding site for each partner. Although we note the left half-site is very similar to an RcsB site, further studies will be needed to settle this issue.
It is still not known whether both RcsB and GadE are directly involved in the DNA recognition process or whether one protein stimulates the binding of the other through conformational change triggered by protein–protein interaction. Heterodimerization of transcriptional regulators is prevalent in eukaryotic regulation mechanisms, where it allows the recognition of additional DNA targets (26). However, it is quite uncommon in prokaryotic systems. An in vitro study raised the possibility of interaction between hetero-pairs of response regulators from the OmpR/PhoB subfamily (27), and Knusten et al. (28) suggested that a ComE/BlpR heterodimer (these proteins belong to the LytR subfamily of response regulator) recognizes a hybrid DNA motif for convergent gene regulation. A RcsB/GadE heterodimer would be the first example of heterodimer formation between a LuxR-like transcription regulator (GadE) and a NarL/FixJ response regulator (RcsB). This noncanonical interaction is sustained by data obtained in the interactome analysis of E. coli (29), in which SPA-RcsB was purified with GadE (previously named YhiE). In agreement with this in vivo interaction, we found that RcsB co-purified with MBP-GadE (data not shown). All these strongly support that there is a specific interaction between GadE and RcsB. Both regulators displayed in their C-terminal region a motif related to the LuxR-type DNA-binding HTH (Prosite PS50043 HTH_LUXR_2). It is conceivable that these regulators interact through this motif since structural studies indicated that the fourth helix in this motif facilitates dimerization (30,24).The question arises as to whether GadE ever binds efficiently to DNA on its own or always needs a co-regulator or alternatively does it need a signal to become active. As shown by Sayed and Foster (31), this regulator collaborates with a still unknown co-activator to stimulate its own transcription. However, it remains possible that GadE regulates some target genes independently of co-regulators. For example, we noticed that GadE tends to form dimers during purification. Further studies are needed to understand how GadE controls transcription of ∼40 genes, including the ones in the AFI (21).
RcsB can function as an activator of gadA/B, although we had previously shown that the overproduction of RcsB or its activation through the Rcs phosphorelay or RcsA pathways represses gadA/BC transcription and results in GDAR reduction (9). In the present work, we identified an RcsB box displaying a sequence characteristic of canonical RcsB binding sites, located just upstream of the −10 element of the gadA promoter (Figures 2 and 7). This box is probably involved in the repression of gadA transcription by obstructing RNA polymerase binding to the promoter. However, the affinity of RcsB for the RcsB box is very low, several orders of magnitude below that of the RcsB/GadE heterodimer for the GAD box, suggesting that the basal level of RcsB is more efficiently oriented to the activation of the gadA/BC expression. This is coherent with the results of our in vitro transcription assay where in the presence of the two regulators RcsB behaved as an activator at lower concentration and as a repressor at higher concentration (Figure 1). It is however conceivable that either a cofactor or the phosphorylation of RcsB increases the affinity of RcsB for the RcsB box, thus making the direct repression more efficient. A potential co-regulator could be RcsA, since heterodimerization with RcsB was shown to considerably enhance DNA binding efficiency (32,24). On the other hand, our genetic data indicated that the effect of RcsA is minor for gad repression: its overproduction decreased GDAR 2-fold compared with a 20-fold decrease upon RcsB overexpression (9). Furthermore, repression of gadA/BC transcription seemed to combine a direct effect, probably resulting from binding of RcsB to the RcsB box, and an indirect one, since overexpression of RcsB also repressed transcription of gadE itself, as well as of gadX and gadW (Figure 7;Castanié-Cornet,M.P. and Cam,K. unpublished data) which encode AraC-like regulators involved in gadE and gadA/BC transcription activation (33,34,35). Sequence analysis revealed a putative RcsB box upstream the −10 element of the gadE promoter that could allow RcsB to control a feed-forward loop over gadA transcription (36). This putative RcsB-GadE-gadA network belongs to the minority of the so-called Coherent-type 2, in which RcsB (X node) will prevent the activation of gadA (Z node) directly and indirectly by repressing gadE expression (Y node). However, we have so far no evidence that this site is important for gadE repression.
Finally, what could be the physiological rationale of the dual effect of RcsB on expression of the Gad system? RcsB has been shown to activate gene expression upon the formation of biofilms and it may control a feed-forward loop involved in gad transcription, allowing repression of GDAR upon Rcs activation following interactions of cells with surfaces, and more generally after envelope perturbation. The purpose of this negative regulation could be to avoid costly runaway expression of gad genes in conditions where cell membrane properties or membrane proteins are modified or damaged, conditions known to activate Rcs system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Université Paul Sabatier and the Centre National de la Recherche Scientifique. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Anne Vianney for gift of anti-RcsB antibodies. We thank Jean-Yves Bouet, Béatrice Clouet-D’Orval for helpful discussions. Many thanks to Dave Lane and Philippe Rousseau for critical reading of the manuscript.
REFERENCES
- 1.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 2.Castanié-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J. Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 4.Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002;184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 2007;189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 2002;189:6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 8.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 2003;49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 9.Castanié-Cornet M.-P, Treffandier H, Francez-Charlot A, Gutierrez C, Cam K. The glutamate-dependent acid resistance system in Escherichia coli: essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology. 2007;153:238–246. doi: 10.1099/mic.0.29278-0. [DOI] [PubMed] [Google Scholar]
- 10.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 11.Brill JA, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 1998;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassif X, Honoré N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 1998;3:1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 13.Virlogeux I, Waxin H, Ecobichon C, Lee JO, Popoff MY. Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J. Bacteriol. 1996;178:1691–1698. doi: 10.1128/jb.178.6.1691-1698.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouslim C, Latifi T, Groisman EA. Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 2003;278:50588–50595. doi: 10.1074/jbc.M309433200. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polard P, Chandler M. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 1995;9:2846–2858. doi: 10.1101/gad.9.22.2846. [DOI] [PubMed] [Google Scholar]
- 17.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and gene fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 18.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanié-Cornet MP, Gutierrez C, Cam K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 2003;49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller JH. A Short Course in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 20.Carballès F, Bertrand C, Bouché JP, Cam K. Regulation of E. coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 1999;34:442–450. doi: 10.1046/j.1365-2958.1999.01605.x. [DOI] [PubMed] [Google Scholar]
- 21.Hommais F, Krin E, Coppee JY, Lacroix C, Yeramian E, Danchin A, Bertin P. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004;150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 22.Itou J, Eguchi Y, Utsumi R. Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 2009;73:870–878. doi: 10.1271/bbb.80795. [DOI] [PubMed] [Google Scholar]
- 23.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pristovsek P, Sengupta K, Löhr F, Schäfer B, von Trebra MW, Rüterjans H, Bernhard F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 2003;278:17752–17759. doi: 10.1074/jbc.M301328200. [DOI] [PubMed] [Google Scholar]
- 25.Wehland M, Kiecker C, Coplin DL, Kelm O, Saenger W, Bernhard F. Identification of an RcsA/RcsB recognition motif in the promoters of exopolysaccharide biosynthetic operons from Erwinia amylovora and Pantoea stewartii subspecies stewartii. J. Biol. Chem. 1999;274:3300–3307. doi: 10.1074/jbc.274.6.3300. [DOI] [PubMed] [Google Scholar]
- 26.Reményi A, Schöler HR, Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 27.Gao R, Tao Y, Stock AM. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol. Microbiol. 2008;69:1358–1372. doi: 10.1111/j.1365-2958.2008.06355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutsen E, Ween O, Håvarstein LS. Two separate quorum-sensing systems upregulate transcription of the same ABC transporter in Streptococcus pneumoniae. J. Bacteriol. 2004;186:3078–3085. doi: 10.1128/JB.186.10.3078-3085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 30.Vannini A, Volpari C, Cargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayed AK, Foster JW. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 2009;71:1435–1450. doi: 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 32.Kelm O, Kiecker C, Geider K, Bernhard F. Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol. Gen. Genet. 1997;256:72–83. doi: 10.1007/s004380050547. [DOI] [PubMed] [Google Scholar]
- 33.Tramonti A, De Canio M, Delanyl I, Scarlato V, De Biase D. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 2006;188:8118–8127. doi: 10.1128/JB.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 35.Richard H, Foster JW. Sodium regulates Escherichia coli acid resistance, and influences GadX- and GadW-dependent activation of gadE. Microbiology. 2007;153:3154–3161. doi: 10.1099/mic.0.2007/007575-0. [DOI] [PubMed] [Google Scholar]
- 36.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giangrossi M, Zattoni S, Tramonti A, De Biase D, Falconi M. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2005;280:21498–21505. doi: 10.1074/jbc.M413255200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.