Abstract
Expression of genes involved in the pathogenesis of Staphylococcus aureus is controlled by global regulatory loci, including two-component regulatory systems and transcriptional regulators. The staphylococcal-specific SarA family of transcription regulators control large numbers of target genes involved in virulence, autolysis, biofilm formation, stress responses and metabolic processes, and are recognized as potential therapeutic targets. Expression of some of these important regulators has been examined, mostly in laboratory strains, while the pattern of expression of these genes in other strains, especially clinical isolates, is largely unknown. In this report, a comparative analysis of 10 sarA-family genes was conducted in six different S. aureus strains, including two laboratory (RN6390, SH1000) and four clinical (MW2, Newman, COL and UAMS-1) strains, by Northern and Western blot analyses. Transcription of most of the sarA-family genes showed a strong growth phase-dependence in all strains tested. Among these genes, no difference was observed in expression of the sarA, sarV, sarT and sarU genes, while a major difference was observed in expression of the sarX gene only in strain RN6390. Expression of mgrA, rot, sarZ, sarR and sarS was observed in all strains, but the level of expression varied from strain to strain.
INTRODUCTION
Staphylococcus aureus is an opportunistic commensal pathogen and among the most common causes of bacterial infections in the community and the hospital. It can cause acute food poisoning, pneumonia, meningitis, skin conditions (e.g. acne, boils and cellulitis), arthritis, osteomyelitis, endocarditis and toxic shock syndrome (Gordon & Lowy, 2008; Klevens et al., 2007; Lowy, 1998). Many S. aureus strains are resistant to a wide spectrum of antibiotics, including meticillin and vancomycin, which often makes treatment of S. aureus infections extremely difficult. Therefore, there is considerable interest in identifying novel drug targets based on an analysis and a better understanding of staphylococcal molecular genetics. Most S. aureus infections begin as minor colonization of skin or soft tissue, from which the organism can spread to the bloodstream and subsequently disseminate into various tissues (Gordon & Lowy, 2008; Klevens et al., 2007; Lowy, 1998). Once inside the host or tissues, S. aureus produces a large number of factors that include adhesins, enzymes, toxins, capsular polysaccharides and other gene products that facilitate tissue colonization, tissue destruction and immune evasion. The expression of these factors is coordinately controlled by regulatory systems, such as two-component regulatory systems (e.g. agr, arlRS, saeRS, srrAB) and transcriptional regulatory systems (e.g. sarA family, sigB, tcaR) (Bronner et al., 2004; Cheung et al., 2004, 2008; Kornblum et al., 1990; Novick et al., 1993; Novick, 2003).
Among transcription regulators, the SarA family of proteins belongs to the winged-helix family of transcription factors (Alekshun et al., 2001; Cheung et al., 2008). The SarA protein family share sequence (22–35 % identity and 48–65 % similarity) and structural homology with each other as well as with the MarR (multidrug resistance) family of proteins in other bacteria (Alekshun et al., 2001; Bronner et al., 2004; Cheung et al., 2004, 2008; Novick, 2003). Analysis of sequenced staphylococcal genomes reveals that at least nine major SarA paralogues are present in most of these genomes (Kuroda et al., 2001). SarA (124 residues) is the first characterized member of the SarA protein family (Cheung et al., 1992) and others that have been subsequently characterized are SarR (115 residues; Manna & Cheung, 2001), SarS (250 residues; Cheung et al., 2001; Tegmark et al., 2000), Rot (133 residues; McNamara et al., 2000; Manna & Ray, 2007), SarT (119 residues; Schmidt et al., 2001), SarU (247 residues; Manna & Cheung, 2003), SarV (116 residues; Manna et al., 2004), MgrA (147 residues; Ingavale et al., 2003; Luong et al., 2003; Truong-Bolduc et al., 2003), SarX (119 residues; Manna & Cheung, 2006a) and SarZ (148 residues; Ballal et al., 2009; Kaito et al., 2006). Among these 10 SarA-family proteins, SarT and SarU, which are located in a position adjacent to the genome and transcribed divergently, are missing in many sequenced clinical S. aureus isolates, including UAMS-1, MRSA252 and RF122, and in other Staphylococcus species (Cassat et al., 2005; Lindsay et al., 2006). The SarA protein family can be divided into two subgroups based on their length: single-domain proteins 115–148 residues long and two-domain proteins of 247–250 residues long. The most interesting aspect of the two-domain proteins SarS and SarU is that each gene encodes a polypeptide with two homologous domains, where the two domains have significant homology not only to each other but also to smaller SarA-family proteins. Structural analysis of four SarA paralogues suggests that smaller SarA proteins exist as dimers (e.g. SarA, SarR, MgrA), while the two-domain proteins are monomers (e.g. SarS) (Chen et al., 2006; Li et al., 2003; Liu et al., 2001, 2006). Structurally these proteins have similar folding and DNA recognition regions (Li et al., 2003; Liu et al., 2001, 2006). In fact, mutational analysis of SarA, SarR and SarZ proteins suggest the existence of several conserved residues within the winged-helix region that are involved in target DNA recognition (Kaito et al., 2006; Liu et al., 2006; Manna & Cheung, 2006b). Despite evidence of conserved DNA-binding regions among the SarA proteins, the DNA-binding sites mapped by DNase I footprint analysis for four SarA paralogues, SarA (Chien et al., 1999; Manna et al., 2004; Manna & Cheung, 2006b; Rechtin et al., 1999), SarR (Manna & Cheung, 2001), MgrA (Manna et al., 2004) and SarT (Schmidt et al., 2003), are different.
Transcriptional analysis and physical mapping of various sar-family genes has been reported (Ballal et al., 2009; Bayer et al., 1996; Ingavale et al., 2003; Manna & Cheung, 2001, 2003, 2006a; Manna et al., 1998, 2004; Manna & Ray, 2007; Tegmark et al., 2000).
Phenotypic characterization of the sarA family of genes, mostly in laboratory strains derived from 8325-4, suggests that many of these genes have distinct phenotypes, which arise due to regulation of the expression of virulence factors, regulatory genes, and genes involved in biofilm formation, oxidative stress and metabolic processes. Several members of the sarA family are involved in regulation of the agr locus, a pivotal regulator for virulence factors expression and a potential drug target in S. aureus; specifically, SarZ and MgrA are involved in activation, SarX plays a role in repression, and SarA and SarR are involved in activation in the early phases of growth (Ballal et al., 2009; Cheung et al., 2008; Kaito et al., 2006; Manna & Cheung, 2006a, b; Tamber & Cheung, 2009). A few members of the sarA family, such as sarU, sarT and sarV, are either not transcribed or are transcribed at very low levels under normal conditions of growth (Manna & Cheung, 2003; Manna et al., 2004; Schmidt et al., 2001). An undetectable or low level of transcription is attributed to repression of normally expressed sarA-family genes. For instance, a very low level of sarV or undetectable sarT transcription is associated with repression mediated by MgrA and SarA, while undetectable sarU transcription is enhanced in a sarT mutant (Manna & Cheung, 2003; Manna et al., 2004; Schmidt et al., 2001). Transcriptional profiling studies have shown that individual mutation of sarA, rot, mgrA and sarZ genes leads to altered expression of large numbers of genes in S. aureus, thus demonstrating their important role in gene regulation (Cassat et al., 2006; Chen et al., 2009; Dunman et al., 2001; Luong et al., 2006; Said-Salim et al., 2003). In fact, the effects of the selected sarA-family genes (sarA, rot, sarU, mgrA and sarZ) on virulence have been examined in several vertebrate and invertebrate infection models (Bae et al., 2004; Chen et al., 2006, 2009; Cheung et al., 2004; Kaito et al., 2006; McNamara & Bayer, 2005; Wang et al., 2008). Overall, experimental findings have demonstrated that the sarA family of genes is important and contributes to reductions in both virulence and bacterial counts in different organs.
Our knowledge of global regulatory systems is predominantly based on studies with strains derived from the easily manipulated laboratory strain 8325-4, which has a mutation in the rsbU gene that results in reduced production of the stress-responsive alternative SigB factor (Giachino et al., 2001). Therefore, the gene expression profiles and regulatory pathways of laboratory strains may not be the same as those of clinical isolates. It is worth mentioning that strain-to-strain variation as well as growth conditions also play an important role in target gene expression. Although transcriptional analysis and physical mapping of various sar-family genes have been reported, comparative expression profiles, both transcriptional and translational, in different clinical strains have not been demonstrated. Therefore, to understand the expression pattern of the sarA-family genes, comparative expression profiling was performed in both laboratory and clinical strains.
METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are: RN6390 (three-prophage-cured derivative from the human isolate 8325; Novick et al., 1993), COL (first hospital-associated MRSA isolate; www.jcvi.org), Newman (a human clinical isolate; McDevitt et al., 1994), SH1000 (a single-copy rsbU-complemented derivative of 8325-4; Horsburgh et al., 2002), MW2 (a highly virulent community-associated MRSA human isolate; Baba et al., 2002) and UAMS-1 (an osteomyelitis isolate; Blevins et al., 2002). S. aureus strains were routinely grown in tryptic soy broth (TSB) without antibiotic selection. The cells were grown overnight in TSB medium and then inoculated at 0.5 % (v/v) (OD600 ∼0.05) into 10 ml fresh TSB and grown with vigorous aeration at 230 r.p.m. in a 45 degree angle attached stand in a New Brunswick air shaker at 37 °C in 18 mm diameter borosilicate glass tubes.
Extraction of RNA and Northern blot hybridization.
Total RNA from various S. aureus strains was prepared using a Trizol isolation kit (Invitrogen) and a reciprocating shaker, as described previously (Manna & Cheung, 2001, 2003; Manna et al., 2004; Schmidt et al., 2001). The cells were allowed to grow to different phases of growth based on OD600 (measured in a Spectronic 20D+ spectrophotometer) prior to RNA extraction. Samples containing the same amount of total cellular RNA (10 μg), as determined by measuring the A260, were analysed by Northern blotting, as described previously (Manna & Cheung, 2001, 2003; Manna et al., 2004; Schmidt et al., 2001). Ten sarA-family genes were either amplified by PCR or excised with restriction endonucleases from plasmids containing the desired genes. For detection of specific transcripts, gel-purified DNA probes were radiolabelled with [α-32P]dCTP using the random-primed DNA labelling kit (Roche Diagnostics), and hybridized under aqueous-phase conditions at 65 °C. Blots were subsequently washed and radioactive band(s) detected using a PhosphoImager. The intensity of the bands was quantified by ImageQuant software (Amersham Life Sciences). Each experiment was repeated at least three times with samples isolated on different dates.
Purification of Sar-family proteins and production of anti-Sar-family polyclonal antibodies.
Procedures for the cloning and purification of 6×His-tag fusion SarA, SarR, SarS, SarT, MgrA, SarZ and SarX proteins have been previously described (Ballal et al., 2009; Cheung et al., 2001; Chien et al., 1999; Manna & Cheung, 2001, 2006a; Manna et al., 2004; Schmidt et al., 2003). To clone and purify SarV (116-residue), Rot (133-residue) and SarU (247-residue) proteins, the 360, 420 and 750 bp DNA fragments containing the full-length sarV, rot and sarU coding regions, respectively, were amplified by PCR using chromosomal DNA from S. aureus RN6390 as the template and primers containing flanking restriction sites, NdeI and BamHI, to facilitate in-frame cloning into the expression vector pET14b (Novagen). The recombinant plasmids containing the respective genes were confirmed by restriction digestion and DNA sequencing and transformed into Escherichia coli BL21 (DE3) pLysS. The His6–SarV, Rot and SarU protein expression and purification were done in a manner similar to that described for other Sar-family proteins (Ballal et al., 2009; Cheung et al., 2001; Chien et al., 1999; Manna & Cheung, 2001, 2006a; Manna et al., 2004; Schmidt et al., 2003). Purity of the purified SarV, SarU and Rot proteins was checked in a 12 % polyacrylamide–SDS gel and they were found to be more than 98 % pure. The concentration of the purified protein was determined using the Bio-Rad protein estimation kit (Bio-Rad) and BSA as the standard.
Antisera against Sar-family proteins were prepared by immunizing four (BALB/c×SJL/J) F1 mice or two New Zealand rabbits with 100 μg of each protein, as described previously (Manna & Cheung, 2001). Western blotting was performed to monitor the titres of the immune sera.
Cell extract preparation and Western blot analysis.
Cell extracts from the early exponential (OD600 0.7), exponential (OD600 1.1) and post-exponential (OD600 1.7) phases of growth were prepared from six tested staphylococcal strains. Cells were grown as described above in 10 ml TSB medium. After harvesting, the cells were resuspended in 0.5 ml cell lysis buffer (25 mM Tris/HCl, pH 8.0, 5 mM EDTA, pH 8.0, 100 mM NaCl), and treated with lysostaphin. Lysostaphin-treated cells were briefly sonicated and centrifuged at 40 000 r.p.m. in a rotor type 80 Ti (Beckman) for 30 min at 4 °C to remove cell debris. The concentration of total proteins from clear lysates was determined using the Bio-Rad protein estimation kit and BSA as the standard.
Equal amounts of total cellular proteins were separated on 12 % polyacrylamide-SDS gels and transferred onto nitrocellulose membranes as previously described (Manna & Cheung, 2001). Blots were incubated at room temperature with an appropriate dilution (1 : 500 or 1 : 1000) of anti-Sar-family protein-specific monoclonal or polyclonal antibody for 2 h, followed by another 1 h incubation with a 1 : 10 000 dilution of goat anti-mouse or anti-rabbit horseradish peroxidase (HRP) conjugate (Pierce). Immunoreactive bands were detected with an ECL detection kit (Pierce). SeeBlue prestained protein standards (Invitrogen) were used for molecular mass estimations. The intensities of the bands were quantified using ImageQuant software (Amersham Life Sciences). Each experiment was repeated at least three times with different samples isolated on different dates.
RESULTS AND DISCUSSION
Expression of SarA, MgrA and Rot in different strains of S. aureus
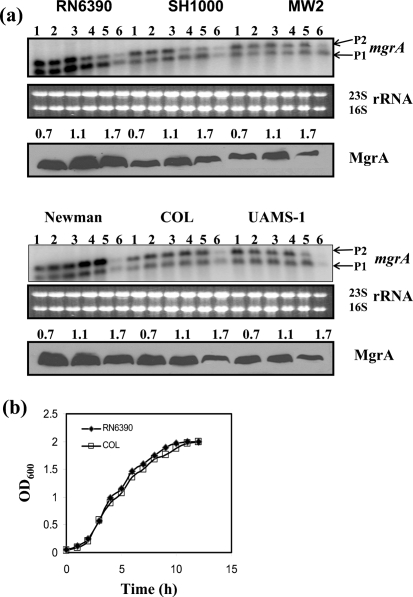
Prior to this study, preliminary transcriptional characterization of the sarA-family genes had been done mostly in the strains derived from strain 8325-4. In this study, a comparative analysis in expression of all 10 sarA-family genes was performed in both commonly used laboratory and clinical strains. Transcription of the mgrA gene originates from two different promoters (P1 and P2), but the translational product is MgrA (17 kDa). Expression of the mgrA transcripts was similar in all tested strains of S. aureus, except RN6390 and Newman (Fig. 1). During post-exponential phase, the P1 transcript (560 nt) was the major transcript in RN6390, while the P2 transcript (750 nt) was the major transcript in Newman. Transcription of mgrA decreased at the stationary (or overnight) phase in all strains. Western blot analysis of cell extracts from three different phases of growth with anti-MgrA monoclonal antibody clearly showed that MgrA was produced in all strains, but the level of expression varied between strains and at different phases of growth (Fig. 1). A substantial reduction (approximately twofold) in expression of MgrA was observed in the post-exponential phase of growth (OD ∼1.7) as compared with the exponential phase of growth (OD ∼1.1), where maximal expression of MgrA was observed. The level of MgrA protein observed in strain RN6390 was two- to threefold higher than those of UAMS-1 and MW2, and 1.5- to twofold more than those of the COL and SH1000 strains, whereas the Newman strain showed slightly lower production than strain RN6390. This correlates well with the higher transcription of mgrA observed in RN6390 and Newman strains compared with other strains. Taken together, Northern and Western analyses suggest that mgrA expression depends on both the growth phase and the strain of S. aureus being analysed.
Fig. 1.
Expression of the mgrA gene in the different wild-type strains at the various phases of growth. (a) Northern blots were hybridized with 565 bp DNA fragments containing the coding region of the mgrA gene. A total of 10 μg RNA was loaded in each lane. Lanes 1–6, total cellular RNA from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Manna & Ray, 2007). P1 (0.56 knt) and P2 (0.75 knt) indicate the two transcripts of the mgrA locus (Ingavale et al., 2003). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent the Western blot analyses for MgrA with anti-MgrA antibody in the different wild-type strains. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect MgrA expression. (b) Growth curves for the wild-type RN6390 and COL strains. There was no noticeable difference in growth of other strains. The OD600 of various cultures was measured in a Spectronic 20D spectrophotometer.
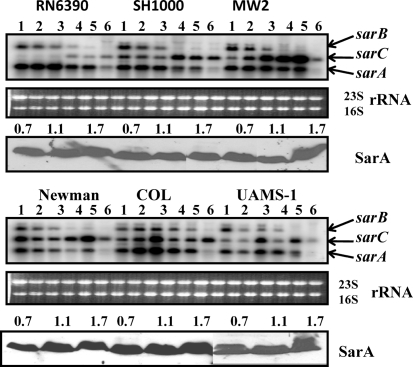
In various strains of S. aureus and Staphylococcus epidermidis, the sarA gene is the most well-studied gene of the sarA family. Typically, expression of SarA is derived from three overlapping sarA transcripts, originating from two σA (P2, sarB and P1, sarA) and one σB (P3, sarC) promoters with common 3′ ends in a growth-dependent manner (Bayer et al., 1996; Manna et al., 1998). Northern and Western analyses of expression of the sarA gene in six strains of S. aureus are shown in Fig. 2. Expression of the sarB and sarC transcripts was growth phase-dependent, sarB transcription decreased, and sarC transcription increased as cells progressed from the lag to the stationary phase of growth. Levels of sarC transcripts were consistent with the strain background: lower levels of the sarC transcript were observed in the low SigB-producing strain RN6390 (Cheung & Manna, 2005) compared with other higher SigB-producing strains. Analyses of the sarA transcript suggest that maximal transcription occurs in RN6390, SH1000 and COL strains in early to exponential phase, after which a slight decline is observed. In MW2, the levels of the sarA transcript remained almost unchanged until post-exponential phase. In all strains, the sarA transcript was substantially decreased at the overnight or the stationary phase of growth. To determine the expression of SarA in different phases of growth in these strains, Western blot analyses were performed with the cell extracts using anti-SarA polyclonal antibodies (Fig. 2). In spite of differential expression of three transcripts, there was no major growth phase-dependent difference (<1.5-fold among strains) in production of SarA in a particular strain, indicating that expression of SarA is constitutive. Furthermore, while the sigB-dependent sarC transcript is drastically reduced in RN6390, similar levels of SarA protein are observed in this strain as in others that make more sarC transcript. This implies that overall production of SarA is dependent on translation of combined sar transcripts. These findings are consistent with reported results demonstrating similar overall production of SarA in RN6390 and UAMS-1 strains (Blevins et al., 2002).
Fig. 2.
Expression of the sarA gene in the different wild-type strains at various phases of growth. Northern blots were hybridized with 450 bp DNA fragments containing the coding region of the sarA gene. A total of 10 μg cellular RNA was loaded in each lane. Lanes 1–6, total cellular RNA from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). The sarB (1.15 knt), sarC (0.8 knt) and sarA (0.56 knt) transcripts originated from the P2, P3 and P1 promoters of the sarA locus (Bayer et al., 1996). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent Western blots of intracellular extracts from the different wild-type strains probed with anti-SarA polyclonal antibodies. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect SarA expression.
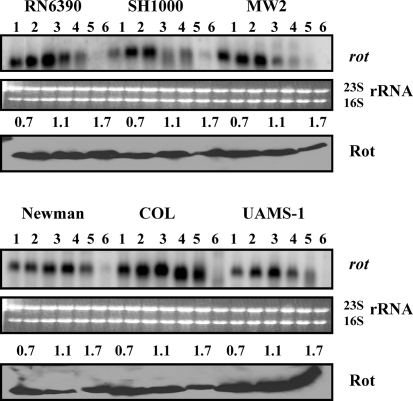
To investigate expression of rot in different strains of S. aureus, transcription and translational analyses were performed. As shown in Fig. 3, in most strains, a distinct increase in the level of rot transcription was observed from the lag phase to the exponential or early post-exponential phase, after which it declined. However, in COL, rot transcription levels did not decrease during the post-exponential phase. A diffuse pattern of rot transcription was observed during the late phases of growth in all strains, and appeared to be due to expression of multiple transcripts, which is mediated by four closely spaced σA-dependent promoters that give rise to four overlapping transcripts of 550, 620, 660 and 700 nt (Manna & Ray, 2007). Despite the four different transcripts, Western results show the production of a single protein product of 16.5 kDa, Rot protein (Fig. 3). Western analysis with anti-Rot polyclonal antibodies demonstrated no difference in Rot expression during the three phases of growth in UAMS-1 and RN6390 strains, whereas a significant decrease (two- to 10-fold) in Rot production was observed in other strains. It should be noted that Rot expression was observed in all phases of growth in all strains when blots were exposed to X-ray film for a longer period of time (data not shown). The transcription of rot is growth phase-dependent, with maximal transcription occurring during the exponential phase of growth, which correlates well with synthesis of Rot protein in most of the strains, except RN6390 and UAMS-1. The probable reason for higher expression of Rot (1.5- to twofold) in UAMS-1 compared with RN6390 could be the decreased production of agr RNAIII molecules. RNAIII modulates the production of Rot at the translational level by binding to two complementary UUGGGA sequences, one of which is located at the ribosome-binding site (SD) and the other 44 nt upstream of the SD site on the rot transcript. Base pairing between RNAIII and these two regions inhibits the translation of rot mRNA (Geisinger et al., 2006). Transcription of agr, in particular RNAIII, production is maximal in RN6390 and decreased in MW2, SH1000, Newman, UAMS-1 and COL (data not shown). Surprisingly, RN6390 produces a significant amount of Rot compared with COL and other strains. Therefore, regulation of translation of the rot transcripts may not depend fully on RNAIII, as proposed elsewhere (Geisinger et al., 2006). Among other known regulators, SarA has been shown to repress transcription of the rot gene in strain RN6390 (Manna & Ray, 2007), but the effect of SarA on the expression of rot is minimal, as no substantial difference was observed in SarA expression in these strains. Overall the results suggest that expression of SarA, MgrA and Rot is sustained throughout all phases of growth in all of these strains, and is consistent in several repeated independent sets of experiments.
Fig. 3.
Analysis of rot gene expression in the different wild-type strains at various phases of growth. Northern blots were hybridized with 450 bp DNA fragments containing the coding region of the rot gene to detect the ∼0.6 knt rot transcript. Lanes 1–6, 10 μg cellular RNA per lane from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent Western blots of intracellular extracts from the different wild-type strains probed with anti-Rot polyclonal antibodies. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect Rot expression.
Expression of the sarZ and sarX genes in various S. aureus strains
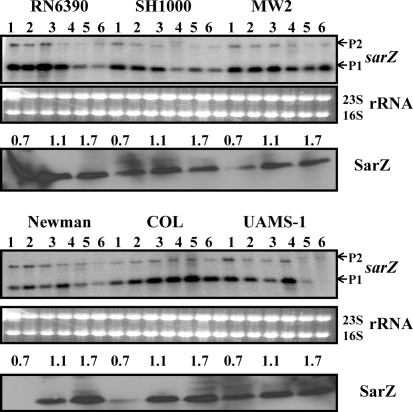
Characterization of sarX and sarZ has been reported recently in several S. aureus strains, RN6390, SH1000 and Newman, and also in S. epidermidis strain 1457 (Ballal et al., 2009; Chen et al., 2009; Kaito et al., 2006; Tamber & Cheung, 2009; Wang et al., 2008). To determine expression profiles of both genes in other backgrounds, Northern and Western blot analyses were performed. Results from Northern analysis suggested that the expression of the sarZ transcripts (0.55 knt and 1.5 knt) depends on growth phase (Fig. 4). Maximal transcription was observed in the exponential to early post-exponential phases of growth, and thereafter a decrease in sarZ transcription was found in most of the strains, except MW2 and COL, where substantial levels of transcript were retained in the post-exponential and stationary phases of growth. Western blot analysis of cell extracts from different phases of growth probed with anti-SarZ polyclonal antibodies did not correlate very well with SarZ expression in many strains, especially during the early or post-exponential phase of growth. Newman, SH1000 and UAMS-1 showed reduced sarZ transcription during post-exponential phase, but SarZ protein production was not decreased in this phase compared with exponential phase (Fig. 4). It is plausible that additional factors are present in these strains that are responsible for a higher stability of the SarZ protein, in spite of decreased sarZ transcription. Recently we have shown that sarZ transcription is partially activated by MgrA (Ballal et al., 2009). Interestingly, RN6390 shows increased sarZ transcription and SarZ production (more than twofold) in the early phase of growth compared with both later phases of growth and with SH1000. This correlates well with the higher level of MgrA expression in this strain compared with SH1000.
Fig. 4.
Expression of the sarZ gene in the different wild-type strains at various phases of growth. Northern blots were hybridized with 550 bp DNA fragments containing the coding region of the sarZ gene. A total of 10 μg cellular RNA was loaded in each lane. Lanes 1–6, total cellular RNA from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). P1 and P2 indicate the major (0.55 knt) and minor (1.5 knt) transcripts of the sarZ locus (Ballal et al., 2009). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent Western blots of intracellular extracts from the different wild-type strains probed with anti-SarZ polyclonal antibodies. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect SarZ expression.
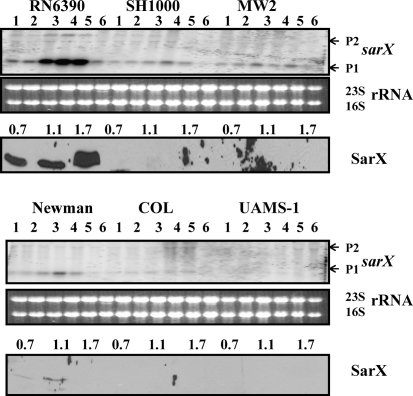
Transcription of the sarX gene was found to be strongly growth phase-dependent in RN6390, where maximal transcription was observed at the post-exponential phase of growth (Fig. 5). Surprisingly, transcriptional analysis of SH1000, which belongs to the same lineage as RN6390 with an intact rsbU gene (Horsburgh et al., 2002), showed that only a low level of the sarX transcript was detectable. Similarly, low or undetectable levels of sarX transcripts were observed in clinical strains, MW2, Newman, COL and UAMS-1 (Fig. 5). Western analysis with anti-SarX polyclonal antibodies demonstrated SarX expression only in RN6390, with maximal expression in the post-exponential phase of growth. The expression of sarX is significantly increased (>12-fold) in strain RN6390 compared with other strains. A low level of SarX expression was also observed in strain Newman in the early phases of growth. Overall, the results demonstrated drastically reduced sarX expression in the relatively high SigB-producing strains, which suggests that SigB responsive factor(s) are involved in modulation of sarX transcription in S. aureus, although the promoter structure of the major sarX transcript is σA-dependent (Manna & Cheung, 2006a). MgrA is a positive regulator of sarX transcription, and MgrA expression is clearly observed in all strains tested, indicating that lack of SarX expression is not due to absence of MgrA.
Fig. 5.
Expression of the sarX gene in the different wild-type strains at various phases of growth. Northern blots were hybridized with 550 bp DNA fragments containing the coding region of the sarX gene. A total of 10 μg cellular RNA was loaded in each lane. Lanes 1–6, total cellular RNA from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). P1 and P2 indicate the major (0.5 knt) and minor (1.5 knt) transcripts of the sarX locus (Manna & Cheung, 2006a). The region containing 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent Western blots of intracellular extracts from the different wild-type strains probed with anti-SarX polyclonal antibodies. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect SarX expression.
Expression of the sarS and sarR genes
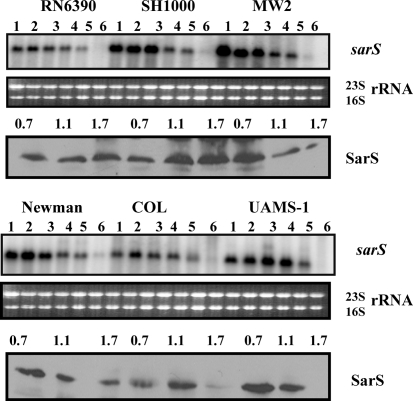
The sarS locus has been previously shown to produce either three [two minor (P2, 1500 nt and P3, 3000 nt) and one major P1 (∼952 nt) sarS transcripts (Tegmark et al., 2000)] or a major sarS transcript (Cheung et al., 2001), depending on the Northern hybridization conditions used. Among these three transcripts, the highly transcribed P1 and the weakly transcribed P2 are σA- and σB-dependent, respectively, whereas the nature of the weakly transcribed largest transcript (P3) is unknown (Tegmark et al., 2000). Northern analysis showed increased sarS transcription in the early phases of growth in all strains, after which it declined (Fig. 6). In spite of detection of several transcripts of the sarS locus, the major gene product of the sarS locus was a 29.9 kDa SarS protein. Western analysis of the cell extracts showed that SarS production decreased in the later phases of growth, particularly in the clinical isolates MW2, Newman, COL and UAMS-1. Both of the laboratory strains (RN6390 and SH1000) retained more SarS expression in the later phases of growth, which was consistently observed in several repeated experiments. SarS is an activator for expression of its adjacent spa (protein A) gene and is repressed by mgrA, sarA and sarZ inactivation (Ballal et al., 2009; Cheung et al., 2001; Tegmark et al., 2000), while activated by sarT and rot inactivation (Said-Salim et al., 2003; Schmidt et al., 2003). SarS expression is marginally lower in some of the strains, which may correlate with the higher expression of some of the sarS-repressive SarA paralogues. For example, transcription of sarS is lower (1.5- to twofold) in RN6390 compared with SH1000, which correlates well with higher expression of the SarS-repressive paralogues MgrA (1.5- to twofold) and SarZ (1.5- to threefold) in RN6390 as compared with SH1000.
Fig. 6.
Analysis of expression of the sarS gene in the different wild-type strains at various phases of growth. Northern blots were hybridized with 800 bp DNA fragments containing the coding region of the sarS gene to detect the 0.95 knt sarS major transcript. Lanes 1–6, 10 μg cellular RNA per lane from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control. The third and sixth panels from the top represent Western blots of intracellular extracts from the different wild-type strains probed with anti-SarS polyclonal antibodies. Equivalent amounts of extracts (20 μg) from the different phases of growth (OD600 ∼0.7, early exponential; OD600 ∼1.1, exponential; OD600 ∼1.7, post-exponential) were used to detect SarS expression.
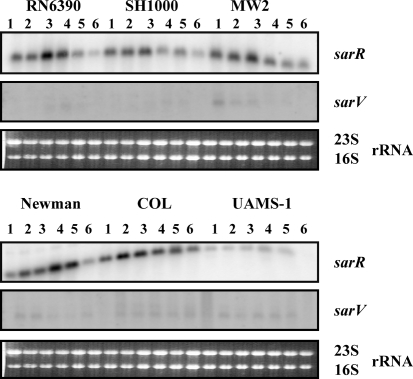
Results from Northern analysis of the sarR gene (Fig. 7) showed that maximal transcription occurred in the early to mid exponential phases of growth in most of the strains. In UAMS-1, relative to other strains, lower levels of the sarR transcript (∼500 nt) were observed in all phases of growth. Western analysis with anti-SarR polyclonal antibodies also showed SarR production to be preferentially increased during the exponential phase of growth as compared with other phases of growth in all strains analysed (data not shown).
Fig. 7.
Northern blot analysis of the sarR (0.5 knt) and sarV (0.5 knt) transcripts in the different wild-type S. aureus strains. Blots were probed with 400 bp sarR and 360 bp sarV fragments containing the entire coding region of the sarR and sarV genes. A total of 10 μg cellular RNA was loaded into each lane. Lanes 1–6, total cellular RNA from the growing cultures at OD600 0.3, 0.7, 1.1, 1.4, 1.7 and overnight (stationary) (Fig. 1b). The region containing the 23S and 16S rRNA of the ethidium bromide-stained gel used for blotting is shown as a loading control.
Undetectable or low levels of transcription of sarU, sarT and sarV genes in different S. aureus strains
Prior to this analysis, it had been shown that transcription of sarU and sarT genes is undetectable in several other strains of S. aureus, including RN6390, whereas transcription of sarV is very low in RN6390 (Manna & Cheung, 2003; Manna et al., 2004; Schmidt et al., 2001). Northern analyses for the sarT and sarU genes with the total RNA from all six strains from various phases of growth failed to detect any 0.8 knt sarT or 1.2 knt sarU transcripts for these two genes (data not shown). Similarly, Northern analysis of the sarV gene showed a very low level of expression of the sarV transcript (∼500 nt) in all these strains (Fig. 7). Western analysis with cell extracts from different strains and anti-SarV polyclonal antibodies revealed no detectable expression of SarV protein (data not shown). Therefore, these results suggest that sarT and sarU are not expressed in any strains, while sarV is weakly transcribed in both laboratory and clinical strains. In RN6390, it has been demonstrated that transcription of the sarV and sarT genes is repressed by SarA and MgrA, whereas SarT represses sarU transcription (Manna & Cheung, 2003; Manna et al., 2004; Schmidt et al., 2001). It is possible that in clinical isolates these genes are not expressed due to the repressive effects of other SarA paralogues and the growth conditions used. It is likely that under different sets of environmental conditions, some of these would be expressed. Indeed, a detectable level of sarT transcription is observed when SH1000 and MW2 are grown in media with a low pH (our unpublished observation). It should be noted that UAMS-1 does not carry the sarT or sarU gene (Cassat et al., 2006). Overall, the experimental results presented in this report are consistent across several repeated sets of experiments.
Overall, the comparative results presented in this report clearly suggest that most of the sarA paralogue genes show a similar pattern of expression in both laboratory and clinical strains, with the notable exception of the sarX gene. Results from numerous studies have clearly established the role of SarA paralogues in the regulation of a large number of target genes involved in virulence and other important processes. However, not much is known about the roles that these genes play in modulating staphylococcal virulence in vivo, the precise mechanisms by which the sarA family of genes are regulated (i.e. internal and external signals), or how they regulate their target genes at the molecular level. This study characterized the expression of these paralogues in different strains of S. aureus in an attempt to define a role for these regulatory pathways, in particular, those mediated by the SarA protein family.
Acknowledgments
The authors thank Dr Ambrose Cheung, Dartmouth Medical School, for S. aureus strains and some of the Sar paralogue antibodies. We would also like to thank Dr Victor Huber for critical reading of the manuscript. This work was supported by the 2010-initiative start-up fund and South Dakota Biomedical Research Infrastructure Network (SD BRIN) (2P20 RR016479) subawarded to A. C. M.
References
- Alekshun, M. N., Levy, S. B., Mealy, T. R., Seaton, B. A. & Head, J. F. (2001). The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat Struct Biol 8, 710–714. [DOI] [PubMed] [Google Scholar]
- Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., Nagal, Y., Iwama, N., Asano, K. & other authors (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. [DOI] [PubMed] [Google Scholar]
- Bae, T., Banger, A. K., Wallace, A., Glass, E. M., Aslund, F., Schneewind, O. & Missiakas, D. M. (2004). Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101, 12312–12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballal, A., Ray, B. & Manna, A. C. (2009). sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J Bacteriol 191, 1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, M. G., Heinrichs, J. H. & Cheung, A. L. (1996). The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol 178, 4563–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, J. S., Beenken, K. E., Elasri, M. O., Hurburt, B. K. & Smeltzer, M. S. (2002). Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun 70, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner, S., Monteil, H. & Prevost, G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28, 183–200. [DOI] [PubMed] [Google Scholar]
- Cassat, J. E., Dunman, P. M., McAleese, F., Murphy, E., Projan, S. J. & Smeltzer, M. S. (2005). Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J Bacteriol 187, 576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat, J., Dunman, P. M., Murphy, E., Projan, S. J., Beenken, K. E., Palm, K. J., Yang, S.-J., Rice, K. C., Bayles, K. W. & Smeltzer, M. S. (2006). Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152, 3075–3090. [DOI] [PubMed] [Google Scholar]
- Chen, P. R., Bae, T., Williams, W. A., Duguid, E. M., Rice, P. A. & Schneewind, O. (2006). An oxidative-sensing mechanism is used by the global regulator MgrA. Nat Chem Biol 2, 591–595. [DOI] [PubMed] [Google Scholar]
- Chen, P. R., Nishida, S., Poor, C. B., Cheng, A., Bae, T., Kuechenmeister, L., Dunman, P. M., Missiakas, D. & He, C. (2009). A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol 71, 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. L. & Manna, A. C. (2005). The role of the distal sarA promoters in SarA expression. Infect Immun 73, 4391–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. L., Koomey, J. M., Butler, C. A., Projan, S. J. & Fischetti, V. A. (1992). Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci U S A 89, 6462–6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. L., Schmidt, K. A., Bateman, B. & Manna, A. C. (2001). SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect Immun 69, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H. & Xiong, Y.-Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40, 1–9. [DOI] [PubMed] [Google Scholar]
- Cheung, A. L., Nishina, K. A., Trotonda, M.-P. & Tamber, S. (2008). The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40, 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, Y.-T., Manna, A. C., Projan, S. J. & Cheung, A. L. (1999). SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 274, 37169–37176. [DOI] [PubMed] [Google Scholar]
- Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., Brown, E. L., Zagursky, R. J., Shlaes, D. & Projan, S. J. (2001). Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by agr and/or sarA loci. J Bacteriol 183, 7341–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger, E., Adhikari, R. P., Jin, R., Ross, H. F. & Novick, R. P. (2006). Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61, 1038–1048. [DOI] [PubMed] [Google Scholar]
- Giachino, P. S., Engelmann, S. & Bischoff, M. (2001). σB activity depends on RsbU in Staphylococcus aureus. J Bacteriol 183, 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, R. J. & Lowy, F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infections. Clin Infect Dis 46, S350–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh, M. J., Aish, J. L., White, I. J., Shaw, L., Lithgow, J. K. & Foster, S. J. (2002). σB modulates virulence determinants expression and stress resistance: characterization of a functional rsbU strain of Staphylococcus aureus 8325-4. J Bacteriol 184, 5457–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale, S. S., Van Wamel, W. & Cheung, A. L. (2003). Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol 48, 1451–1466. [DOI] [PubMed] [Google Scholar]
- Kaito, C., Morishita, D., Matsumota, Y., Kurokawa, K. & Sekimizu, K. (2006). Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol 62, 1601–1617. [DOI] [PubMed] [Google Scholar]
- Klevens, R. M., Morrison, M. A., Nadle, J., Petit, S., Gershman, K., Ray, S., Harrison, L. H., Lynfield, R. & other authors (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771. [DOI] [PubMed] [Google Scholar]
- Kornblum, J., Kreiswirth, B., Projan, S. J., Ross, H. & Novick, R. P. (1990). Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In Molecular Biology of the Staphylococci, pp. 373–402. Edited by R. P. Novick. New York: VCH Publishers.
- Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L., Oguchi, A., Aoki, K.-I. & other authors (2001). Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- Li, R., Manna, A. C., Cheung, A. L. & Zhang, G. (2003). Crystal structure of the SarS protein from Staphylococcus aureus. J Bacteriol 185, 4219–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, J. A., Moore, C. E., Day, N. P., Peacock, S. J., Witney, A. A., Stabler, R. A., Husain, S. E., Butcher, P. D. & Hinds, J. (2006). Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol 188, 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Manna, A., Li, R., Martin, W. E., Murphy, R. C., Cheung, A. L. & Zhang, G. (2001). Crystal structure of the SarR protein from Staphylococcus aureus. Proc Natl Acad Sci U S A 98, 6877–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Manna, A. C., Pan, C.-H., Kriksunov, I. A., Thiel, D. J., Cheung, A. L. & Zhang, G. (2006). Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc Natl Acad Sci U S A 103, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy, F. D. (1998). Staphylococcus aureus infections. N Engl J Med 339, 520–532. [DOI] [PubMed] [Google Scholar]
- Luong, T. T., Newell, S. W. & Lee, C. Y. (2003). MgrA, a novel global regulator in Staphylococcus aureus. J Bacteriol 185, 3703–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong, T. T., Dunman, P. M., Murphy, E., Projan, S. J. & Lee, C. Y. (2006). Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol 188, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, A. & Cheung, A. L. (2001). Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect Immun 69, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, A. C. & Cheung, A. L. (2003). sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect Immun 71, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, A. C. & Cheung, A. L. (2006a). Expression of SarX, a negative regulator of agr and exoproteins synthesis, is activated by MgrA in Staphylococcus aureus. J Bacteriol 188, 4288–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, A. C. & Cheung, A. L. (2006b). Transcriptional regulation of the agr locus and the identification of DNA-binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol Microbiol 60, 1289–1301. [DOI] [PubMed] [Google Scholar]
- Manna, A. C. & Ray, B. (2007). Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology 153, 1538–1545. [DOI] [PubMed] [Google Scholar]
- Manna, A. C., Bayer, M. G. & Cheung, A. L. (1998). Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol 180, 3828–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, A. C., Ingavale, S. S., Maloney, M., Van Wamel, W. & Cheung, A. L. (2004). Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 186, 5267–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt, D., Francois, P., Vandaux, P. & Foster, T. J. (1994). Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol 11, 237–248. [DOI] [PubMed] [Google Scholar]
- McNamara, P. J. & Bayer, A. S. (2005). A rot mutation restores parental virulence to an agr-null Staphylococcus aureus strain in a rabbit model of endocarditis. Infect Immun 73, 3806–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, P. J., Milligan-Monroe, K. C., Khalili, S. & Proctor, R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 182, 3197–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48, 1429–1449. [DOI] [PubMed] [Google Scholar]
- Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B. & Moghazeh, S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12, 3967–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtin, T. M., Gillaspy, A. F., Schumacher, M. A., Brennan, R. G., Smeltzer, M. S. & Hurlburt, B. K. (1999). Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol Microbiol 33, 307–316. [DOI] [PubMed] [Google Scholar]
- Said-Salim, B., Dunman, P. M., McAleese, F. M., Macapagal, D., Murphy, E., McNamara, P. J., Arvidson, S., Foster, T. J., Projan, S. J. & Kreiswirth, B. N. (2003). Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185, 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, K. A., Manna, A. C., Gill, S. & Cheung, A. L. (2001). SarT, a repressor of α-hemolysin in Staphylococcus aureus. Infect Immun 69, 4749–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, K. A., Manna, A. C. & Cheung, A. L. (2003). SarT influences the expression of sarS in S. aureus. Infect Immun 71, 5139–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamber, S. & Cheung, A. L. (2009). SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun 77, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegmark, K., Karlsson, A. & Arvidson, S. (2000). Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 37, 398–409. [DOI] [PubMed] [Google Scholar]
- Truong-Bolduc, Q. C., Zhang, X. & Hooper, D. C. (2003). Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J Bacteriol 185, 3127–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Li, M., Dong, D., Bach, T. H., Sturdevant, D. E., Vuong, C., Otto, M. & Gao, Q. (2008). SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J Infect Dis 197, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]