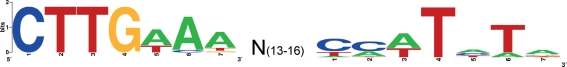Abstract
Francisella tularensis is a highly infectious pathogen that infects animals and humans to cause the disease tularemia. The primary targets of this bacterium are macrophages, in which it replicates in the cytoplasm after escaping the initial phagosomal compartment. The ability to replicate within macrophages relies on the tightly regulated expression of a series of genes. One of the most commonly used means of coordinating the regulation of multiple genes in bacteria consists of the association of dedicated alternative sigma factors with the core of the RNA polymerase (RNAP). In silico analysis of the F. tularensis LVS genome led us to identify, in addition to the genes encoding the RNAP core (comprising the α1, α2, β, β′ and ω subunits), one gene (designated rpoD) encoding the major sigma factor σ70, and a unique gene (FTL_0851) encoding a putative alternative sigma factor homologue of the σ32 heat-shock family (designated rpoH). Hence, F. tularensis represents one of the minority of bacterial species that possess only one or no alternative sigma factor in addition to the main factor σ70. In the present work, we show that FTL_0851 encodes a genuine σ32 factor. Transcriptomic analyses of the F. tularensis LVS heat-stress response allowed the identification of a series of orthologues of known heat-shock genes (including those for Hsp40, GroEL, GroES, DnaK, DnaJ, GrpE, ClpB and ClpP) and a number of genes implicated in Francisella virulence. A bioinformatic analysis was used to identify genes preceded by a putative σ32-binding site, revealing both similarities to and differences from RpoH-mediated gene expression in Escherichia coli. Our results suggest that RpoH is an essential protein of F. tularensis, and positively regulates a subset of genes involved in the heat-shock response.
INTRODUCTION
Francisella tularensis is a Gram-negative, facultative intracellular pathogen that causes tularemia in humans and a large number of animal species. Human infections can occur after ingestion of contaminated water or food, through arthropod bites, from contaminated animal carcasses, or by inhalation of bacteria. Due to its high infectivity and the severity of disease, F. tularensis has been classified by the US Centers for Disease Control as a potential biological weapon (for reviews, see McLendon et al., 2006; Sjostedt, 2006). The pathogenicity of F. tularensis is largely dependent on its ability to replicate inside host cells, and in particular in macrophages in vivo. Several factors important for virulence have been identified, and in particular genes located in the Francisella pathogenicity island (FPI) (de Bruin et al., 2007; Golovliov et al., 2003; Lai et al., 2004; Ludu et al., 2008; Nano et al., 2004; Nano & Schmerk, 2007; Santic et al., 2007; Tempel et al., 2006; Weiss et al., 2007), but the molecular mechanisms underlying disease are not yet fully understood.
Regulation of gene expression in bacteria occurs primarily at the transcriptional level. The association of dedicated alternative sigma factors to the core of the RNA polymerase (RNAP) provides a simple and efficient way for bacteria to rapidly adapt to various environmental changes. The RNAP holoenzyme contains the subunits of the core molecule (comprising two α subunits, the β, β′ and ω subunits) and a sigma factor enabling the holoenzyme to specifically recognize promoter elements and initiate transcription at these sites. The regulon of a single sigma factor can comprise hundreds of genes. Many sigma factors are induced in response to environmental changes and are often controlled by anti-sigma factors (for reviews see Gourse et al., 2006; Gruber & Gross, 2003). The number of sigma factors encoded by different bacterial species varies considerably. For example, seven sigma factors have been identified in Escherichia coli to date, whereas Streptomyces coelicolor is thought to encode at least 61. Some bacteria contain only a single sigma factor, but the majority express several, and the number generally relates to the size of the genome and the environmental variation that allows growth of the organism (Kill et al., 2005; Rodrigue et al., 2006).
Another ubiquitous mode of bacterial response to environmental changes involves the function of two-component signal transduction systems (TCS). Each TCS consists of a membrane-bound sensor that monitors environmental variations and a dedicated cytoplasmic response regulator that mediates an adaptive response, i.e. a transcriptional regulation of gene expression. Strikingly, Francisella subspecies do not possess a single complete TCS pair (Mohapatra et al., 2007 and references therein), and analyses of the F. tularensis genomes have revealed the presence of only a single gene encoding a protein homologous to an alternative sigma factor, thereby placing F. tularensis within the relatively small group of bacterial species that express only a single or no alternative sigma factor. The absence of any complete TCS and the presence of only one gene encoding an alternative sigma factor may reflect the fact that the Francisella genome is relatively small and/or is thought to be deteriorating, based on the high number of pseudogenes and incomplete biosynthetic pathways (Larsson et al., 2005). The absence of multiple alternative sigma factors or TCS suggests that F. tularensis has a limited ability to respond to and proliferate in diverse environmental conditions.
Given the small number of putative regulatory proteins encoded by F. tularensis, we were interested in examining the function of the only alternative sigma factor that may help this organism in survival and in the stress response. In the present study, we show that the only alternative sigma factor encoded by the F. tularensis genome, RpoH, is a genuine σ32 unit, and our data suggest that rpoH is an essential gene of F. tularensis. To gain further insight into the RpoH-dependent mechanisms of gene expression in F. tularensis we performed a transcriptomic analysis of the heat-stress response of F. tularensis LVS and evaluated the role of RpoH in the regulation of the bacterial genome in an RpoH-overproducing strain. In addition, a bioinformatic analysis determined a consensus binding sequence for RpoH and identified genes preceded by a putative RpoH-binding site. Our results demonstrate similarities between the heat-stress response in F. tularensis and the well-studied response in E. coli, but also illuminate differences between these two organisms. A number of heat-induced genes have previously been demonstrated to be of importance for the ability of F. tularensis to survive and/or multiply inside host cells, suggesting that stress responses are of significance for the virulence of Francisella.
METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1 and oligonucleotide sequences in Supplementary Table S1. F. tularensis LVS was grown on pre-made chocolate agar enriched with PolyVitex (bioMérieux), chocolate agar plates prepared from GC medium base, IsoVitalex vitamins and haemoglobin (BD Biosciences), or in Schaedler + vitamin K3 broth (Shaedler-K3, bioMérieux) at 37 °C or room temperature. E. coli strains were grown on Luria–Bertani (LB) plates or in LB broth (Difco) at 37 °C or room temperature aerobically.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description or genotype | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F−φ80lacZ ΔM15 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 (lacZYA-argF) U169 | Laboratory strain collection |
| CAG48315 | λ− phtpg–lacZ ΔrpoH : : Kan zhf50 : : Tn10, pRB11 | Zhou et al. (1988) |
| MG1655 | K-12 | Arnaud Gutierrez |
| MG1655/pRB11 | MG1655 containing plasmid pRB11 expressing E. coli rpoH | This study |
| MG1655/pBADrpoH | MG1655 containing plasmid pBAD expressing F. tularensis rpoH | This study |
| MG1655ΔrpoH/pRB11 | MG1655/pRB11 transduced with P1 lysate from CAG48315 | This study |
| MG1655ΔrpoH/pBADrpoH | MG1655/pBADrpoH transduced with P1 lysate from CAG48315 | This study |
| F. tularensis strains | ||
| LVS | F. tularensis subspecies holarctica live vaccine strain | Laboratory strain collection |
| LVS/pFNLTP6gro-0851 | LVS with plasmid pFNLTP6gro_0851 expressing rpoH from the groES promoter | This study |
| LVS/pFNLTP6gro | LVS with plasmid pFNLTP6gro with no insert | This study |
| Plasmids | ||
| pRB11 | pBR322 derivative with E. coli rpoH gene downstream of IPTG-inducible tac promoter | Yura et al. (2007) |
| pBAD | araBAD promoter, araC, pBR322 origin, AmpR | Invitrogen |
| pBADrpoH | F. tularensis rpoH gene cloned downstream of arabinose-inducible promoter of pBAD | This study |
| pFNLTP6gro | E. coli–F. tularensis shuttle vector with groE operon promoter, KmR AmpR, constructed from pFNLTP6gro-gfp by deleting gfp gene | Meibom et al. (2009); Maier et al. (2006) |
| pFNLTP6gro-0851 | E. coli–F. tularensis shuttle vector with F. tularensis rpoH downstream of groE operon promoter, KmR AmpR | This study |
For F. tularensis, antibiotics were used at 5 μg ml−1 (kanamycin) and 1.75 μg ml−1 (chloramphenicol). For E. coli, antibiotics were used at the following concentrations: 25 μg ml−1 (kanamycin), 2.5 μg ml−1 (chloramphenicol) and 50 μg ml−1 (ampicillin).
The plasmid used for overexpression of rpoH, pFNLTP6gro-0851, was constructed by amplifying a 957 bp fragment (corresponding to the sequence 78 bp upstream of the rpoH gene and the entire gene) using primers 0851_F and 0851_R (Supplementary Table S1), followed by digestion with NdeI and NotI and cloning into plasmid pFNLTP6gro (Maier et al., 2004; Meibom et al., 2009) downstream of the groE operon promoter of F. tularensis. pFNLTP6gro-0851 was introduced into the LVS strain by electroporation.
The plasmid pBADrpoH was constructed by amplifying the LVS rpoH gene using primers 0851_F2 and 0851_R2, followed by digestion with EcoRI and cloning into plasmid pBAD downstream of the arabinose-inducible pBAD promoter. The rpoH gene was verified to be in the right orientation by PCR using primers 5′pBAD and 0851_R2.
Functional complementation.
E. coli K-12 strain MG1655 was transformed with either pRB11 (Yura et al., 2007) or pBADrpoH. Phage P1 was used to transduce the ΔrpoH : : Kan zhf50 : : Tn10 mutation from strain CAG48315 (Zhou et al., 1988) into MG1655/pRB11 and MG1655/pBADrpoH. For the transduction lysate preparation, an overnight culture of CAG48315 was diluted 100-fold in fresh LB supplemented with 5 mM CaCl2 and 0.2 % glucose, and grown at 37 °C with shaking for 1 h before adding 200 μl P1 phage lysate into 5 ml culture. When the culture had lysed completely, several drops of chloroform were added and the transduction lysate was kept at 4 °C. For the transduction, the recipient strains (MG1655/pRB11 and MG1655/pBADrpoH) were grown overnight in LB supplemented with 50 μg ampicillin ml−1 and 0.2 % arabinose. The cells were then harvested and resuspended in LB supplemented with 50 μg ampicillin ml−1, 0.2 % arabinose, 100 mM MgSO4 and 5 mM CaCl2. A 100 μl volume of bacterial cells and 100 μl transduction lysate were mixed and incubated at 37 °C with shaking for 30 min. After addition of 200 μl 1 M sodium citrate (pH 5.5), cells were incubated at 37 °C with shaking in 1 ml LB containing 50 μg ampicillin ml−1 and 0.2 % arabinose for 1 h, and plated on LB agar supplemented with 50 μg ampicillin ml−1, 25 μg kanamycin ml−1 and 0.2 % arabinose. Colonies appearing after overnight incubation were verified to have the ΔrpoH : : Kan zhf50 : : Tn10 mutation by PCR using primers KanaRtn903_F and KanaRtn903_R. The first strain obtained after transduction, MG1655ΔrpoH/pRB11, was then grown on LB agar supplemented with 50 μg ampicillin ml−1 and 25 μg kanamycin ml−1 at room temperature, 30 and 37 °C. The second strain, MG1655ΔrpoH/pBADrpoH, was grown on LB agar supplemented with 50 μg ampicillin ml−1 and 25 μg kanamycin ml−1 in the presence and absence of 0.2 % arabinose at room temperature, 30 and 37 °C.
RNA extraction.
RNA was extracted from exponential-phase bacteria with TRIzol reagent (Invitrogen). After extraction with chloroform, the aqueous phase was purified using the RNeasy Clean-up protocol (Qiagen). Contaminating DNA was removed from RNA preparations with RNA-free DNase I (Ambion).
Microarray experiments and data analyses.
To prepare RNA for microarray analyses, F. tularensis LVS, F. tularensis LVS+pFNLTP6gro, and F. tularensis LVS+pFNLTP6gro-0851 were grown at 37 °C with shaking to OD600 ∼0.3–0.4. For rpoH overexpression experiments, bacterial cells were then harvested and RNA was extracted. For heat-shock-response experiments, the culture of F. tularensis LVS was split into two: one part of the culture was placed at 37 °C with shaking, whereas the other was incubated at 44 °C with shaking. After 30 min incubation, bacterial cells were harvested and RNA was extracted.
Reverse transcription and cDNA fluorescent labelling were performed for each sample on 3 μg of RNA according to the protocol provided by The Institute for Genomic Research (TIGR; http://pfgrc.jcvi.org). Briefly, the RNA was reverse-transcribed using Superscript II reverse transcriptase in the presence of amino-allyl-dUTP, dNTP and random primers, followed by coupling of cyanine dye (Cy3 or Cy5). Microarray hybridization and post-hybridization washings were performed according to the TIGR protocol. Briefly, after pre-hybridization of microarray slides, hybridization was performed in hybridization buffer (40 % formamide, 5× saline sodium citrate (SSC), 0.1 % SDS, 0.6 μg salmon sperm DNA μl−1) at 42 °C for 18 h in a hybridization chamber (Corning).
The F. tularensis microarrays, provided by the Pathogen Functional Genomics Resource Center (PFGRC; Rockville, MD, USA), contain 2331 oligonucleotides (70-mer) with four replicates per oligonucleotide, representing all the ORFs of F. tularensis Schu S4 and F. tularensis LVS.
Microarrays were scanned with a Genepix 4000B scanner (Molecular Devices). To determine signal fluorescence intensities for each spot, TIFF images were analysed using the Spotfinder and Midas software (TIGR). Statistical analyses were performed using the Significance Analysis of Microarrays (SAM) software (Tusher et al., 2001). The number of genes with significantly modified expression was defined by performing a one-class analysis using a false discovery rate (FDR) of 0 %.
Two RNA extractions from independent experiments were done for each condition and each sample set was hybridized to two microarrays.
Real-time quantitative RT-PCR (qRT-PCR).
To validate the microarray results, four ORFs were selected for qRT-PCR analysis with the same RNA samples used in the microarray hybridizations. For the heat-shock time-course experiment, RNA samples were isolated before temperature shift from 37 to 44 °C and after 10, 20, 30, 45 and 60 min incubation at 44 °C. For the analyses, 1 μg RNA was reverse-transcribed using random hexamers and Superscript II reverse transcriptase (Invitrogen) according to the protocol provided by the manufacturer. Real-time PCR was performed with gene-specific primers using an ABI PRISM 7700 and SYBR green PCR master mix (Applied Biosystems). To calculate the amount of gene-specific transcript, a standard curve was plotted for each primer set using a series of diluted genomic DNA from LVS. To compare the transcript amounts for the different conditions, the amounts of each gene transcript were normalized to DNA helicase (FTL_1656), as this gene has been shown to change little in expression during growth (Brotcke et al., 2006). The expression of each gene was determined from three replicates in a single qRT-PCR experiment.
Bioinformatics analyses.
The RpoH-binding site consensus sequence was identified as described previously (Slamti et al., 2007) using BioProspector (Liu et al., 2001) to search for conserved motifs with a structure similar to those of the RpoH-binding sites in E. coli and Vibrio cholerae. The search parameters and input sequences were adjusted until the motif identified corresponded to the previously predicted RpoH-binding sites upstream of F. tularensis grpE and groES (Meibom et al., 2008) (Supplementary Tables S3 and S4). The sequences 200 bp upstream of the first 400 annotated F. tularensis ORFs were used as the background model. The final search parameters were as follows: motif, 2 blocks; width of the first motif block, 7; width of the second motif block, 7; minimum gap between the blocks, 13; maximum gap between the blocks, 16. Identification of putative RpoH-binding sites was accomplished in two steps. First, four position frequency matrices (PFMs) were constructed. These were composed of the PFMs of blocks 1 and 2 identified by BioProspector separated by 13–16 columns, respectively, in which the frequencies of all four nucleotides were set to be identical. Second, the program patser (van Helden, 2003) was used to search the 200 bp upstream of all F. tularensis genes for sequences corresponding to each of these four matrices. The minimum patser score threshold was set to 9. In cases where two putative binding sites had the same block 1 or block 2 sequences but different gap lengths, the lower-scoring site was discarded.
RESULTS
Identification of a functional rpoH gene of F. tularensis LVS
The in silico analysis of the LVS genome allowed us to identify all the genes involved in the synthesis of core RNAP. As reported by Charity et al. (2007), the F. tularensis genome contains two genes, designated rpoA1 (FTL_0261) and rpoA2 (FTL_0616), which encode two different α subunits of RNAP that differ from one another in regions that are predicted to be critical for dimer formation, promoter recognition, and activator interaction. FTL_1744 and FTL_1743 (rpoB and rpoC) encode the β and β′ subunits, respectively, and FTL_1533 (rpoZ) the ω subunit.
The major sigma factor (σ70) is encoded by FTL_1050 (rpoD). Strikingly, we identified only one gene, FTL_0851, encoding a putative alternative sigma factor sharing significant sequence similarity with other members of the family of σ32 heat-shock factors (hence designated rpoH). The rpoH sequence of LVS encodes a protein of 292 aa, with 53.4 % amino acid identity with σ32 of E. coli K-12, and is highly conserved among the F. tularensis subspecies. Multiple sequence alignments of σ32 family members have led to the identification of a highly conserved stretch of nine amino acids Q(R/K)(K/R)LFFNLR that is designated the RpoH box (Nakahigashi et al., 1995). It is of note that this box is perfectly conserved in the F. tularensis σ32 sequences, strongly suggesting that it encodes a functional σ32 factor.
Functional complementation of E. coli rpoH
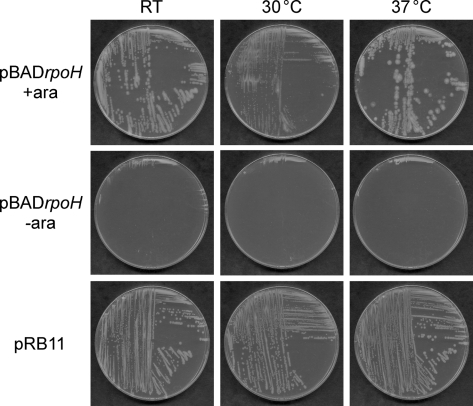
We first tested the ability of the LVS rpoH gene to functionally complement the temperature-sensitive growth of an E. coli ΔrpoH mutant lacking σ32, which can grow only at or below 20 °C (Zhou et al., 1988). For this, we introduced the ΔrpoH mutation from strain CAG48315 (Zhou et al., 1988; Table 1) into E. coli strain MG1655 by transduction. The recipient strain contains a plasmid encoding the F. tularensis rpoH gene (pBADrpoH) under the control of an arabinose-inducible promoter. The resulting strain did not grow on agar plates without supplement at any temperature (room temperature, 30 or 37 °C; Fig. 1, middle row). However, the F. tularensis rpoH gene promoted growth of the mutant strain when arabinose was added to the medium (Fig. 1, top row), showing that F. tularensis rpoH can functionally complement the lack of σ32 in E. coli. As a control, we transduced the ΔrpoH mutation into a recipient strain containing a plasmid encoding the E. coli rpoH gene (pRB11; Yura et al., 2007) (Fig. 1, bottom row), and this strain exhibited the same growth pattern as the MG1655ΔrpoH/pBADrpoH strain grown in the presence of arabinose. These results show that F. tularensis rpoH encodes a genuine σ32 unit of RNAP.
Fig. 1.
The rpoH gene from F. tularensis functionally complements an E. coli ΔrpoH mutant. E. coli stain MG1655ΔrpoH harbouring plasmid pBADrpoH (encoding rpoH from F. tularensis) or pRB11 (encoding rpoH from E. coli) was streaked on LB agar containing ampicillin and 0.2 % arabinose, where indicated (+ara), and incubated for 24 h at 30 and 37 °C, or for 48 h at room temperature (RT).
F. tularensis transcriptomic response to heat stress
We next studied gene expression changes in F. tularensis LVS in response to heat shock. For this, we extracted RNA from bacterial cultures grown in Schaedler medium (supplemented with vitamin K3) after a temperature upshift for 30 min at 44 °C. After cDNA synthesis and labelling, control (the same culture kept at 37 °C) and heat-shocked samples were paired and hybridized on the microarrays. Statistical analysis using SAM software (Tusher et al., 2001) indicated that 181 genes exhibited significant (FDR 0 %) differential expression at a twofold or greater level in response to a temperature upshift from 37 to 44 °C (Table 2 and Supplementary Table S2). The wide distribution of putative functional roles attributed to the differentially expressed genes indicates the extent of the molecular response that enables F. tularensis LVS to survive and eventually adapt to thermal stress. The transcripts of 112 genes were more abundant after the temperature upshift (Table 2). Among these, 15 genes had annotated functions in protein fate. Well-known heat-shock proteins, such as Hsp40, GroEL, GroES, DnaK, DnaJ, GrpE, ClpB, ClpX, ClpP and HtpG, form the major part of this group. The major sigma factor RpoD (σ70) was found to be upregulated, with a fold-induction of 4.6. In contrast, rpoH did not appear to be upregulated after a temperature upshift. About 40 % of the upregulated genes were genes encoding proteins of unknown function or pseudogenes. Transposases composed 21 % of the upregulated genes, but this could reflect cross-hybridizations on the microarrays as these transposases are highly homologous. Interestingly, five genes of the F. tularensis FPI were found to be downregulated: FTL_0115/1161, FTL_0116/_1162 and FTL_0124/_1170, encoding proteins of unknown function, and pdpB1 and iglB1 (intracellular growth locus B)(Supplementary Table S2). One third of the downregulated genes had annotated functions in metabolism, biosynthesis of cofactors or ribonucleotide biosynthesis. As for the upregulated genes, genes encoding proteins of unknown function or pseudogenes represented a significant portion of the downregulated genes. Furthermore, several genes encoding proteins with transport functions were downregulated after heat shock.
Table 2.
Genes significantly increased in expression (at least twofold) after temperature upshift
| Locus* | Gene product | Gene | Cellular role category† | Fold change‡ |
|---|---|---|---|---|
| FTL_0034 | Hypothetical protein | Hypothetical proteins: conserved | 2.02 | |
| FTL_0088 | Acetyltransferase protein | Unknown function | 2.38 | |
| FTL_0093 | Chitinase | Metabolism | 10.24 | |
| FTL_0094 | ClpB protein | clpB | Degradation of proteins, peptides and glycopeptides | 5.51 |
| FTL_0144 | Hypothetical protein | Unknown function: general | 2.77 | |
| FTL_0163 | MFS transporter | Transport and binding proteins | 4.31 | |
| FTL_0180 | Acyltransferase | htrB | Metabolism | 2.25 |
| FTL_0186 | MFS transport protein, fragment | Transport and binding proteins | 4.01 | |
| FTL_0201 | Hypothetical protein | Unknown function | 2.15 | |
| FTL_0202 | Conserved hypothetical membrane protein | Cell envelope: other | 3.29 | |
| FTL_0203 | Hypothetical membrane protein | Unknown function: general | 2.00 | |
| FTL_0219 | Hypothetical protein | Unknown function: general | 2.86 | |
| FTL_0267 | Heat-shock protein 90 | htpG | Protein folding and stabilization | 3.01 |
| FTL_0281 | Heat-shock protein, hsp40 | Protein folding and stabilization | 7.13 | |
| FTL_0359 | Type IV pili fibre building block protein | Unknown function | 2.55 | |
| FTL_0365 | − | Pseudogene | 2.92 | |
| FTL_0390 | Type IV pili fibre building block protein | Unknown function: general | 1.99 | |
| FTL_0645 | Hypothetical lipoprotein | Cell envelope | 3.20 | |
| FTL_0662 | Prophage repressor protein | Mobile and extrachromosomal element functions | 3.31 | |
| FTL_0663 | Hypothetical protein | Hypothetical proteins: conserved | 4.02 | |
| FTL_0758 | − | Pseudogene | 2.24 | |
| FTL_0826 | − | Pseudogene | 2.09 | |
| FTL_0844 | LysR transcriptional regulator family protein | Regulatory functions | 2.35 | |
| FTL_0867 | Hypothetical protein | Hypothetical proteins: conserved | 2.78 | |
| FTL_0891 | Trigger factor | tig | Protein folding and stabilization | 4.38 |
| FTL_0892 | ATP-dependent Clp protease subunit P | clpP | Degradation of proteins, peptides and glycopeptides | 3.70 |
| FTL_0893 | ATP-dependent protease ATP-binding subunit | clpX | Degradation of proteins, peptides and glycopeptides | 2.87 |
| FTL_0894 | DNA-binding, ATP-dependent protease La | lon | Degradation of proteins, peptides and glycopeptides | 2.53 |
| FTL_0908 | − | Pseudogene | 2.07 | |
| FTL_0909 | Hypothetical protein | Hypothetical proteins: conserved | 2.29 | |
| FTL_0911 | − | Pseudogene | 2.34 | |
| FTL_0912 | − | Pseudogene | 1.97 | |
| FTL_0964 | ATP-dependent protease ATP-binding subunit | hslU | Protein folding and stabilization | 4.33 |
| FTL_0965 | ATP-dependent protease peptidase subunit | hslV | Degradation of proteins, peptides and glycopeptides | 5.13 |
| FTL_0996 | AhpC/Tsa family protein | Detoxification | 2.01 | |
| FTL_0997 | Hypothetical protein | Hypothetical proteins: conserved | 3.13 | |
| FTL_1008 | − | Pseudogene | 2.01 | |
| FTL_1048 | Hypothetical protein | Unknown function: general | 2.23 | |
| FTL_1049 | DNA primase | dnaG | DNA replication, recombination and repair | 2.21 |
| FTL_1050 | RNAP sigma-70 factor | rpoD | Transcription | 4.58 |
| FTL_1125 | Transcription regulator | hipA | Regulatory functions | 2.75 |
| FTL_1126 | Transcription regulator | Regulatory functions | 2.26 | |
| FTL_1190 | Chaperone protein GrpE (heat-shock protein family 70 cofactor) | grpE | Protein folding and stabilization | 6.32 |
| FTL_1191 | Chaperone protein DnaK (heat-shock protein family 70 protein) | dnaK | Protein folding and stabilization | 2.23 |
| FTL_1192 | Chaperone protein DnaJ (heat-shock protein 70 family cofactor) | dnaJ | Protein folding and stabilization | 6.19 |
| FTL_1194 | Pirin family protein | Unknown function: general | 4.17 | |
| FTL_1221 | Hypothetical membrane protein | Cell envelope | 5.11 | |
| FTL_1222 | Conserved hypothetical protein | Hypothetical proteins: conserved | 4.58 | |
| FTL_1226 | − | Pseudogene | 2.65 | |
| FTL_1257 | − | Pseudogene | 2.31 | |
| FTL_1259 | − | Pseudogene | 3.41 | |
| FTL_1260 | − | Pseudogene | 2.27 | |
| FTL_1265 | 2-Amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase | folK | Biosynthesis of cofactors, prosthetic groups and carriers: folic acid | 5.01 |
| FTL_1266 | Lipase/esterase | Metabolism | 5.82 | |
| FTL_1267 | Hypothetical protein | Hypothetical proteins: conserved | 4.01 | |
| FTL_1269 | − | Pseudogene | 2.07 | |
| FTL_1270 | − | Pseudogene | 2.68 | |
| FTL_1361 | Cold-shock protein | cspA | Hypothetical proteins | 2.30 |
| FTL_1365 | Hypothetical protein | Hypothetical proteins: conserved | 2.49 | |
| FTL_1366 | Membrane fusion protein, putative | Transport and binding proteins | 2.13 | |
| FTL_1403 | Conserved hypothetical membrane protein | Cell envelope | 2.90 | |
| FTL_1411 | Hypothetical protein | ybaB | Hypothetical proteins: conserved | 2.31 |
| FTL_1412 | Recombination protein RecR | recR | DNA replication, recombination and repair | 2.07 |
| FTL_1517 | Hypothetical protein | Hypothetical proteins: conserved | 2.09 | |
| FTL_1535 | 2-Dehydro-3-deoxyphosphooctonate aldolase | Cell envelope | 1.99 | |
| FTL_1555 | Hypothetical membrane protein | Cell envelope | 4.32 | |
| FTL_1575 | Hypothetical membrane protein | Cell envelope | 4.09 | |
| FTL_1600 | Periplasmic l-asparaginase II precursor | ansB | Metabolism | 2.04 |
| FTL_1691 | Hypothetical protein | Hypothetical proteins: conserved | 2.02 | |
| FTL_1692 | Hypothetical protein | Cell envelope: surface structures | 2.13 | |
| FTL_1714 | Chaperone protein, GroEL | groL | Protein folding and stabilization | 3.05 |
| FTL_1715 | Chaperonin protein, GroES | groS | Protein folding and stabilization | 3.12 |
| FTL_1770 | Type IV pili nucleotide-binding protein | Transport and binding proteins | 3.01 | |
| FTL_1771 | Type IV pili nucleotide-binding protein | pilT | Transport and binding proteins | 3.03 |
| FTL_1776 | Hypothetical membrane protein | Cell envelope | 3.01 | |
| FTL_1777 | − | Pseudogene | 2.13 | |
| FTL_1778 | − | Pseudogene | 3.19 | |
| FTL_1793 | Hypothetical protein | Hypothetical proteins: conserved | 2.06 | |
| FTL_1805 | ATPase | Unknown function | 2.92 | |
| FTL_1806 | MFS transport protein | Transport and binding proteins | 2.91 | |
| FTL_1807 | Histidyl-tRNA synthetase | hisS | tRNA aminoacylation | 3.12 |
| FTL_1808 | Ribosome-binding factor A | Transcription | 2.96 | |
| FTL_1810 | Transcription elongation factor NusA | nusA | Transcription | 4.04 |
| FTL_1811 | Hypothetical protein | Unknown function: general | 2.74 | |
| FTL_1814 | − | Pseudogene | 2.76 | |
| FTL_1878 | Two-component sensor protein KdpD | kdpD | Regulatory functions: other | 2.14 |
| FTL_1894 | Transposase, fragment | Mobile and extrachromosomal element functions | 4.16 | |
| FTL_1913 | Sua5/YciO/YrdC family protein | Unknown function | 2.06 | |
| FTL_1945 | Hypothetical protein | Unknown function: general | 1.95 | |
| FTL_1957 | Heat-shock protein | Protein folding and stabilization | 8.57 | |
| FTL_XXXX§ | Transposase | isftu2 | Mobile and extrachromosomal element functions | 2.0–6.9 |
| FTL_YYYY|| | Transposase | isftu1 | Mobile and extrachromosomal element functions | 2.3–3.2 |
*Genes in bold type have a putative RpoH-binding sequence located before the start codon (see Supplementary Table S3 for detailed information). Gene numbers marked in italic type have been identified in screens or selections for determinants required for intracellular survival and/or virulence (Kraemer et al., 2009; Maier et al., 2007; Su et al., 2007; Tempel et al., 2006; Weiss et al., 2007).
†Cellular role category according to TIGR.
‡Fold change calculated as the average expression at 44 °C relative to 37 °C calculated from two independent experiments with duplicate hybridizations.
§A number of isftu2-type transposases: FTL_0079, 0220, 0270, 0367, 0441, 0468, 0568, 0575, 0591, 0622, 0861, 0868, 1301, 1368, 1471, 1516, 1655, 1680 and 1885.
||Three isftu1-type transposases: FTL_0128, 0417 and 0607.
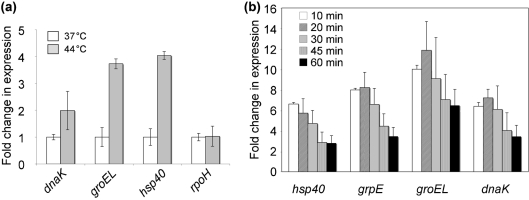
To verify the accuracy of the microarray data, we performed qRT-PCR analyses on transcripts of four chosen genes. For these experiments, we used one of the two RNA sample sets that were used for the microarray experiments. As seen in Fig. 2(a), the relative levels of rpoH, dnaK, groEL and hsp40 transcripts fully supported the microarray data, with dnaK, groEL and hsp40 expressed at higher levels at 44 °C than at 37 °C and the expression of rpoH not changed by the temperature upshift. To investigate the heat-shock response in a temporal manner, we performed qRT-PCR analyses on four heat-shock-induced genes (groEL, dnaK, grpE and hsp40) over a period of 60 min incubation at 44 °C (Fig. 2b). For all four genes we observed increased expression at all times (10–60 min) compared with before heat shock. However, the transcript levels appeared to be less elevated at the later time points (45 and 60 min).
Fig. 2.
Quantification by RT-PCR of transcript levels of selected genes after heat shock. (a) Validation of microarray experiment. Transcript levels were normalized to that of DNA helicase (FTL_1656) and the fold difference (relative to 37 °C) and sds are shown for the dnaK, groEL, hsp40 and rpoH genes. (b) Change in expression of four heat-shock genes during 60 min incubation at 44 °C. Transcript levels were normalized to that of DNA helicase (FTL_1656) and the fold difference (relative to sample taken before temperature shift) and sds are shown for the dnaK, groEL, hsp40 and grpE genes. Data are from triplicate samples.
Role of rpoH in F. tularensis transcriptional regulation
In spite of our repeated efforts, we were unable to inactivate the rpoH gene of LVS. We tried to inactivate rpoH either by insertional mutagenesis or by allelic exchange at temperatures ranging from 20 to 37 °C, but were unsuccessful. It is thus likely (although not formally proven) that rpoH is essential in F. tularensis, as it is thought to be in other bacteria, such as V. cholerae and Neisseria gonorrhoeae (Laskos et al., 2004; Slamti et al., 2007).
To overcome the lack of an rpoH mutant, we evaluated the effect of overproduction of RpoH on the LVS transcriptome. For this, we compared the transcriptomes of LVS (transformed with the plasmid pFNLTP6gro without inserted gene) to that of an LVS overexpressing RpoH (i.e. transformed with the plasmid pFNLTP6gro_0851, expressing FTL_0851 from the groES promoter). We extracted RNA from exponentially grown bacterial cultures, and after cDNA synthesis and labelling, control and RpoH-overexpressing samples were paired and hybridized to the microarrays. Statistical analysis indicated that only seven genes exhibited significant increased expression (FDR 0 %) in response to overexpression of RpoH (Table 3). As expected, rpoH itself was upregulated 21-fold. The six other upregulated genes all encoded well-known heat-shock proteins (Hsp40, Hsp90, GroES, GroEL, DnaK and DnaJ). The transcriptional control by RpoH therefore seems to be very specific to heat-shock proteins.
Table 3.
Genes showing a change in expression in the presence of a plasmid overexpressing RpoH
| Locus | Gene product* | Gene | Cellular role category† | Fold change‡ |
|---|---|---|---|---|
| FTL_0851 | RNAP sigma factor | rpoH | Transcription: transcription factors | 20.6 |
| FTL_0281 | Heat-shock protein, Hsp40 | Protein fate: protein folding and stabilization | 2.9 | |
| FTL_0267 | Heat-shock protein 90 | htpG | Protein fate: protein folding and stabilization | 2.9 |
| FTL_1191 | Chaperone protein DnaK (heat-shock protein family 70 protein) | dnaK | Protein fate: protein folding and stabilization | 2.6 |
| FTL_1192 | Chaperone protein DnaJ (heat-shock protein 70 family cofactor) | dnaJ | Protein fate: protein folding and stabilization | 2.0 |
| FTL_1714 | Chaperone protein, GroEL | groEL | Protein fate: protein folding and stabilization | 2.2 |
| FTL_1715 | Chaperone protein, GroES | groES | Protein fate: protein folding and stabilization | 2.3 |
*Genes with significantly changed expression, upregulated at least twofold, are included in the table and displayed according to their functional category.
†Cellular role category according to TIGR.
‡Values are average fold change between LVS/pFNLTP6gro-0851 and LVS/pFNLTP6gro from two independent experiments with duplicate hybridization. Each microarray contains four spots for each gene.
Consensus sequence for Francisella RpoH-dependent promoter
To identify an RpoH-binding site consensus sequence, we analysed the 200 bp sequence upstream of all 112 heat-upregulated genes listed in Table 2 using Bioprospector (Liu et al., 2001). These analyses failed to identify an RpoH consensus binding sequence, which we reasoned was likely due to the fact that a relatively small proportion of heat-regulated genes identified in our microarray analyses are directly regulated by RpoH, making it difficult for Bioprospector to identify conserved binding sites. To address this problem, we repeated our Bioprospector analyses using only those upregulated genes annotated by NCBI as chaperones, chaperonins, proteases or heat-shock proteins, classes of genes known to be directly regulated by RpoH in other bacteria (Supplementary Tables S3 and S4). This yielded several putative binding sites, several of which corresponded to previously predicted F. tularensis RpoH-binding sites (Meibom et al., 2008). A PFM representing the sites identified by Bioprospector was constructed and the program patser (van Helden, 2003) was used to interrogate the upstream regions of all 1754 ORFs in the NCBI annotation of LVS for putative RpoH-binding sites. A total of 72 non-overlapping putative RpoH-binding sites with a score of 9 or above were identified upstream of 71 genes (Supplementary Table S5), including 13 of the genes upregulated by heat shock (Table 4 and marked in bold type in Table 2). From these 13 putative sites a consensus binding sequence was determined (Fig. 3) that shares significant homology with the RpoH-binding site consensus sequences in E. coli and V. cholerae (Slamti et al., 2007).
Table 4.
Heat-induced genes with a potential σ32-binding sequence in the promoter region
| Distance* | Length† | Binding site sequence | Locus | Gene product‡ |
|---|---|---|---|---|
| 34 | 27 | CTTGAAAAACAGCAAAGTATCCTTAAG | FTL_0094 | ClpB protein |
| 81 | 30 | CTTGAAAAAACTAACTTCCACTCTCATATA | FTL_0267 | Heat-shock protein 90, HtpG |
| 84 | 27 | CTTGTATTTATATAAAAAGACCTTATA | FTL_0281 | Heat-shock protein, Hsp40 |
| 7 | 28 | CTTGAAAATGATTAGAAAAAGTATTTTA | FTL_0663 | Hypothetical protein |
| 101 | 29 | CTTGTCATATATTTAATAATAAACATATA | FTL_0891 | Trigger factor |
| 162 | 28 | CTTGAAGTGATGTTTCATATACCTTCAT | FTL_0894 | DNA-binding, ATP-dependent protease La |
| 48 | 29 | CTTGTAAAATTTAATATCAATTCCATTTA | FTL_0965 | ATP-dependent protease, peptidase subunit, HslV |
| 82 | 29 | CTTGAAAATACAGAATTAATCACTATCTA | FTL_1190 | Chaperone protein GrpE (heat-shock protein family 70 cofactor) |
| 92 | 29 | CTTGAAAAGATTATAAATATGCCCATCTA | FTL_1191 | Chaperone protein DnaK (heat-shock protein family 70 protein) |
| 68 | 30 | CTTGAAAATTAAAAAAAAGCCCCCAATTTC | FTL_1265 | 2-Amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase |
| 98 | 29 | CTTGAAAATTTTTTTTTTGACTCAATATC | FTL_1715 | Chaperonin protein, GroES |
| 40 | 29 | CTTGTATTTAGCATGATCGTATCATTCTT | FTL_1805 | ATPase |
| 75 | 27 | CTTGAAATTTTAAAAACCGATCATATA | FTL_1957 | Heat-shock protein |
*Distance from ATG start codon.
†Length (nucleotides) of the putative σ32-binding site.
‡Gene product according to TIGR.
Fig. 3.
Consensus sequence of the F. tularensis σ32-binding site displayed with WebLogo (http://weblogo.berkeley.edu). The height of each column indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each nucleic acid at that position. N(13–16) indicates the gap between the two blocks.
DISCUSSION
The bacterial heat-shock response results in a transient induction of a set of highly conserved heat-shock proteins, including chaperones and proteases. The alternative σ-factor σ32 (RpoH) is the primary regulator that controls the transcription of genes during heat shock and some other general stress conditions. The activity of the heat-shock proteins ensures the correct folding of proteins during normal growth, and is essential for resistance to stress by minimizing the damage caused by the accumulation of misfolded and aggregated proteins. Our examination of the heat-shock response in F. tularensis resulted in the identification of many bacterial heat-shock proteins, some of which have been previously reported to increase in amount after heat or oxidative stress (Ericsson et al., 1994; Lenco et al., 2005) and in response to mammalian body temperature (Horzempa et al., 2008), in addition to various other genes.
Role of σ32 in heat-stress response
Control of σ32 amount and activity in E. coli is complex and is performed at transcriptional, post-transcriptional and post-translational levels. The heat-shock response in E. coli is induced as a result of increased σ32 levels, primarily due to an increase in rpoH mRNA translation and increased stability and activity of σ32 (Arsène et al., 2000), whereas control of rpoH transcription seems to be of only minor importance. Similarly, we did not observe an increase in transcription of rpoH in F. tularensis after temperature upshift. In contrast, transcription of σ70 was increased after heat shock, suggesting that σ70-containing RNAPs are required for the heat-shock response in Francisella, perhaps in combination with gene-specific regulators.
To more directly define the genes dependent on RpoH, we performed transcriptional profiling of F. tularensis LVS harbouring a plasmid expressing RpoH from the groE promoter and compared it with bacteria carrying an empty plasmid. This allowed us to identify six genes (in addition to rpoH itself) that were expressed at increased levels in the strains overexpressing RpoH. These genes encode the heat-shock proteins Hsp40, HptG, DnaK, DnaJ, GroES and GroEL, which were all induced after heat stress as well. The previous studies of the σ32 regulon in E. coli, V. cholerae and N. gonorrhoeae identified these genes also (Gunesekere et al., 2006; Nonaka et al., 2006; Slamti et al., 2007; Zhao et al., 2005), demonstrating common features of RpoH regulation in different species. However, the number of genes identified in F. tularensis was unexpectedly low when compared with other bacterial species. To date, there is no promoter that allows the specific induction of transcription in Francisella, and our experiments were therefore performed under continuous overexpression of RpoH. Based on the knowledge of the regulation of σ32 in E. coli, we believe this could lead to underestimation of the genes directly controlled by RpoH. In the current model for regulation of σ32 in E. coli, called the ‘unfolded protein titration model’, the stability and activity of σ32 rely on the level of unfolded protein relative to the chaperone level (for review see Guisbert et al., 2008). When unfolded protein levels are low compared with chaperones, σ32 is predominantly bound to the chaperones and is inactive, whereas a high level of unfolded proteins will titrate the chaperones away from σ32, thereby rendering σ32 active. In our experimental set-up, unfolded protein levels are expected to be low (since no stress is imposed on the bacteria), whereas the chaperones will be expressed as a result of RpoH overexpression. If the same mechanism of σ32 regulation exists in F. tularensis as in E. coli, our set-up will lead to chaperone-mediated inactivation of σ32, and this could be responsible for the low number of genes we observe to be induced in the RpoH-overexpressing strain.
Defining an RpoH consensus box
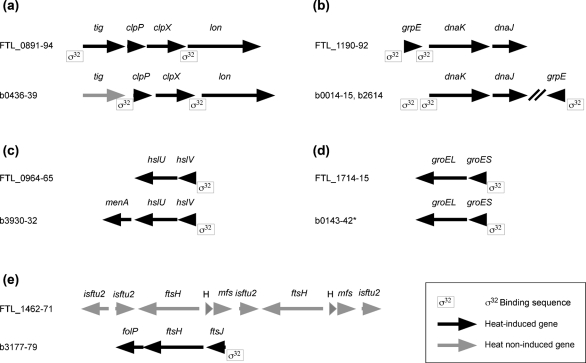
Using the data we obtained from the transcriptional profiling, we performed a bioinformatics analysis to determine a consensus sequence for binding of σ32 (Fig. 3). We identified a putative σ32-binding site in the promoter region of 13 genes that were induced after heat shock. Among these are all the genes induced by RpoH overexpression, or the first gene in the presumed two-gene transcripts dnaK-dnaJ and groES-groEL, further suggesting that these genes are directly under σ32 control. In the remainder of the heat-induced genes, we did not observe any putative σ32-binding sites, indicating that some of these are not controlled directly by σ32. The consensus binding sequence for σ32 resembles the consensus sequence from V. cholerae and E. coli (Nonaka et al., 2006; Slamti et al., 2007), and several of the genes and putative operons that are preceded by a σ32-binding site in F. tularensis are also found in E. coli, although small variations are often observed (Nonaka et al., 2006) (Fig. 4). We observed heat-mediated upregulation of the four consecutive genes tig-clpP-clpX-lon, and bioinformatics analysis identified a σ32-binding site upstream of the tig and lon genes (Fig. 4a). However, in E. coli, only clpP, clpX and lon are induced by heat shock, and a σ32-binding site is found upstream of clpP and lon. In both F. tularensis and E. coli, the transcription unit dnaK-dnaJ is induced by heat shock and preceded by a σ32-binding site. In F. tularensis, these genes are adjacent to grpE, which is also heat-induced and preceded by a σ32-binding site, although in E. coli grpE is located elsewhere on the genome (Fig. 4b). The organization of the two-gene transcriptional unit groES-groEL seems to be identical in the two species (Fig. 4d), as is the case with the hslV-hslU genes, with the exception that the E. coli genes are followed by a third heat-induced gene, menA (Fig. 4c). The protease FtsH carries out degradation of σ32 in E. coli and is itself induced by heat shock, and the transcription unit contains a σ32-binding site (Fig. 4e). Interestingly, in F. tularensis we did not observe increased expression of the ftsH gene after heat shock, and the genetic region encoding FtsH is not predicted to contain a σ32-binding site (Fig. 4e). The region containing ftsH seems to have been duplicated in the LVS strain, whereas it is only found in one copy in the other strains of F. tularensis for which sequence information is available. Currently, we do not know whether the activity of σ32 in F. tularensis is controlled by a mechanism similar to the ‘unfolded protein titration model’ and whether degradation of σ32 is performed by the FtsH protease. In any case, the regulatory mechanism must differ from the one in E. coli, since FtsH is not part of the heat-shock response in F. tularensis.
Fig. 4.
Schematic representations of the organization of selected genes containing putative σ32-binding sequences in F. tularensis and E. coli. Black arrows represent genes that are induced by heat stress. Grey arrows represent genes that are not induced by heat stress. Gene symbols are indicated above arrows and locus numbers to the left in each panel (numbers preceded by FTL_ designate the loci in F. tularensis LVS and numbers preceded by b the loci in E. coli MG1655). isftu, transposase; H, hypothetical protein; mfs, major facilitator transporter. The locations of σ32-binding sites in E. coli are from Nonaka et al. (2006).
We also observed σ32-binding sites next to a number of genes that we did not find to be upregulated by RpoH overexpression or after heat shock. It is possible that some of these were missed in the transcriptomic analysis of heat stress or were not induced under constant overexpression of RpoH. We performed transcriptomic analysis after 30 min of heat exposure, and it is probable that some genes are induced only early in the heat-shock response. Our qRT-PCR analysis of the temporal expression of four heat-shock genes showed an increased expression at all times from 10 to 60 min of heat exposure, although with less elevated transcript levels at the later time points. These results indicate that the heat-shock response is initiated promptly and continues for up to 1 h, but do not rule out the possibility that some genes could have increased expression only early in the heat-shock response. Interestingly, a number of the genes containing putative σ32-binding sites (that are not induced by heat stress) are induced by oxidative stress (N. Grall, unpublished results), indicating that not only the heat-shock response is mediated by σ32. Finally, we anticipate that some of the putative sites do not represent true σ32-binding sites, although they were identified in our bioinformatics analysis.
A relationship between heat-stress response and virulence
Remarkably, a considerable number of the genes that were upregulated by heat stress (20 genes, see Table 2) have been identified in screens or selections for genes required for intracellular multiplication and/or survival. These genes include clpB (Gray et al., 2002; Maier et al., 2007; Meibom et al., 2008; Su et al., 2007; Tempel et al., 2006; Weiss et al., 2007), hspG (Weiss et al., 2007), FTL_0663 (Kraemer et al., 2009; Su et al., 2007), dnaK (Tempel et al., 2006) and FTL_1806 [encoding major facilitator superfamily (MFS) transport protein] (Maier et al., 2007; Weiss et al., 2007). The identification of these genes, which are important for intracellular survival, may indicate that stress responses play a central role in the pathogenesis of F. tularensis and its ability to survive inside host cells. The intracellular environment is hostile to the bacterium, and induction of stress responses may be imperative to its survival within host cells.
Given the significant number of genes (18 %) that are induced under heat stress and are implicated in the virulence of Francisella, it will be interesting to further study the molecular mechanisms underlying the stress response. In particular, an understanding of how the activity of σ32 is controlled, including determining which proteins and signals mediate this control, will lead to a much better understanding of how σ32-mediated stress responses contribute to survival within the mammalian host.
Acknowledgments
We thank Dr Virgil Rhodius, University of California, San Francisco, for kindly providing strain CAG48315 and plasmid pRB11, and Arnaud Gutierrez, CNRS UMR 7625, Université Pierre et Marie Curie, Paris, France, for providing strain MG1655 and P1 phage. DNA microarrays were obtained through the National Institute of Allergy and Infectious Diseases (NIAID) Pathogen Functional Resource Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, National institutes of Health (NIH), Department of Health and Human Services (DHHS) and operated by TIGR. These studies were supported by Institut National de la Santé et de la Recherche Médicale (INSERM) and Centre National de la Recherche Scientifique (CNRS). N. G. was supported by a fellowship from Assistance Publique-Hôpitaux de Paris (AP-HP). J. L. was supported by award number K99AI076608 from the NIAID.
Abbreviations
FDR, false discovery rate
FPI, Francisella pathogenicity island, MFS, major facilitator superfamily
PFM, position frequency matrix
qRT-PCR, quantitative RT-PCR
RNAP, RNA polymerase
TIGR, The Institute for Genomic Research
Footnotes
The microarray data discussed in this paper have been deposited in ArrayExpress under the accession numbers E-MEXP-2048 and E-MEXP-2037.
Five supplementary tables, listing primers used in this study, genes significantly decreased in expression after temperature upshift, genes that were used in BioProspector analysis, putative binding sites found by BioProspector and used for building of matrices, and genes with a potential σ32-binding sequence in their promoter region, are available with the online version of this paper.
References
- Arsène, F., Tomoyasu, T. & Bukau, B. (2000). The heat shock response of Escherichia coli. Int J Food Microbiol 55, 3–9. [DOI] [PubMed] [Google Scholar]
- Brotcke, A., Weiss, D. S., Kim, C. C., Chain, P., Malfatti, S., Garcia, E. & Monack, D. M. (2006). Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun 74, 6642–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity, J. C., Costante-Hamm, M. M., Balon, E. L., Boyd, D. H., Rubin, E. J. & Dove, S. L. (2007). Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog 3, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin, O. M., Ludu, J. S. & Nano, F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson, M., Tarnvik, A., Kuoppa, K., Sandstrom, G. & Sjostedt, A. (1994). Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect Immun 62, 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov, I., Sjostedt, A., Mokrievich, A. & Pavlov, V. (2003). A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett 222, 273–280. [DOI] [PubMed] [Google Scholar]
- Gourse, R. L., Ross, W. & Rutherford, S. T. (2006). General pathway for turning on promoters transcribed by RNA polymerases containing alternative σ factors. J Bacteriol 188, 4589–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, C. G., Cowley, S. C., Cheung, K. K. & Nano, F. E. (2002). The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol Lett 215, 53–56. [DOI] [PubMed] [Google Scholar]
- Gruber, T. M. & Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57, 441–466. [DOI] [PubMed] [Google Scholar]
- Guisbert, E., Yura, T., Rhodius, V. A. & Gross, C. A. (2008). Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesekere, I. C., Kahler, C. M., Powell, D. R., Snyder, L. A. S., Saunders, N. J., Rood, J. I. & Davies, J. K. (2006). Comparison of the RpoH-dependent regulon and the general stress response in Neisseria gonorrhoeae. J Bacteriol 188, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa, J., Carlson, P. E., Jr, O'Dee, D. M., Shanks, R. M. & Nau, G. J. (2008). Global transcriptional response to mammalian temperature provides new insight into Francisella tularensis pathogenesis. BMC Microbiol 8, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kill, K., Binnewies, T. T., Sicheritz-Ponten, T., Willenbrock, H., Hallin, P. F., Wassenaar, T. M. & Ussery, D. W. (2005). Genome update: sigma factors in 240 bacterial genomes. Microbiology 151, 3147–3150. [DOI] [PubMed] [Google Scholar]
- Kraemer, P. S., Mitchell, A., Pelletier, M. R., Gallagher, L. A., Wasnick, M., Rohmer, L., Brittnacher, M. J., Manoil, C., Skerett, S. J. & Salama, N. R. (2009). Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun 77, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, X. H., Golovliov, I. & Sjostedt, A. (2004). Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb Pathog 37, 225–230. [DOI] [PubMed] [Google Scholar]
- Larsson, P., Oyston, P. C., Chain, P., Chu, M. C., Duffield, M., Fuxelius, H. H., Garcia, E., Hälltorp, G., Johansson, D. & other authors (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet 37, 153–159. [DOI] [PubMed] [Google Scholar]
- Laskos, L., Ryan, C. S., Fyfe, J. A. M. & Davies, J. K. (2004). The RpoH-mediated stress response in Nesseria gonorrhoeae is regulated at the level of activity. J Bacteriol 186, 8443–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenco, J., Pavkova, I., Hubalek, M. & Stulik, J. (2005). Insights into the oxidative stress response in Francisella tularensis LVS and its mutant ΔiglC1+2 by proteomics analysis. FEMS Microbiol Lett 246, 47–54. [DOI] [PubMed] [Google Scholar]
- Liu, X., Brutlag, D. L. & Liu, J. S. (2001). BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138. [PubMed]
- Ludu, J. S., de Bruin, O. M., Duplantis, B. N., Schmerk, C. L., Chou, A. Y., Elkins, K. L. & Nano, F. E. (2008). The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J Bacteriol 190, 4584–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T. M., Havig, A., Casey, M., Nano, F. E., Frank, D. W. & Zahrt, T. C. (2004). Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol 70, 7511–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T. M., Pechous, R., Casey, M., Zahrt, T. C. & Frank, D. W. (2006). In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl Environ Microbiol 72, 1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T. M., Casey, M. S., Becker, R. H., Dorsey, C. W., Glass, E. M., Maltsev, N., Zahrt, T. C. & Frank, D. W. (2007). Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect Immun 75, 5376–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon, M. K., Apicella, M. A. & Allen, L. A. (2006). Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol 60, 167–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom, K. L., Dubail, I., Dupuis, M., Barel, M., Lenco, J., Stulik, J., Golovliov, I., Sjostedt, A. & Charbit, A. (2008). The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol 67, 1384–1401. [DOI] [PubMed] [Google Scholar]
- Meibom, K. L., Forslund, A. L., Kuoppa, K., Alkhuder, K., Dubail, I., Dupuis, M., Forsberg, A. & Charbit, A. (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun 77, 1866–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra, N. P., Soni, S., Bell, B. L., Warren, R., Ernst, R. K., Muszynski, A., Carlson, R. W. & Gunn, J. S. (2007). Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun 75, 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi, K., Yanagi, H. & Yura, T. (1995). Isolation and sequence analysis of rpoH genes encoding σ32 homologs from Gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res 23, 4383–4390. [PMC free article] [PubMed] [Google Scholar]
- Nano, F. E. & Schmerk, C. (2007). The Francisella pathogenicity island. Ann N Y Acad Sci 1105, 122–137. [DOI] [PubMed] [Google Scholar]
- Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., Ludu, J. S., Letendre, G. W., Meierovics, A. I. & other authors (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 186, 6430–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, G., Blankschien, M., Herman, C., Gross, C. A. & Rhodius, V. A. (2006). Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev 20, 1776–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L. & Manganelli, R. (2006). The σ factors of Mycobacterium tuberculosis. FEMS Microbiol Rev 30, 926–941. [DOI] [PubMed] [Google Scholar]
- Santic, M., Molmeret, M., Barker, J. R., Klose, K. E., Dekanic, A., Doric, M. & Kwaik, Y. A. (2007). A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol 9, 2391–2403. [DOI] [PubMed] [Google Scholar]
- Sjostedt, A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect 8, 561–567. [DOI] [PubMed] [Google Scholar]
- Slamti, L., Livny, J. & Waldor, M. K. (2007). Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J Bacteriol 189, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, J., Yang, J., Zhao, D., Kawula, T. H., Banas, J. A. & Zhang, J.-R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun 75, 3089–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel, R., Lai, X. H., Crosa, L., Kozlowicz, B. & Heffron, F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect Immun 74, 5095–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., Tibshirani, R. & Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden, J. (2003). Regulatory sequence analysis tools. Nucleic Acids Res 31, 3593–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D. S., Brotcke, A., Henry, T., Margolis, J. J., Chan, K. & Monack, D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104, 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura, T., Guisbert, E., Poritz, M., Lu, C. Z., Campbell, E. & Gross, C. A. (2007). Analysis of σ32 mutants defective in chaperone-mediated feedback control reveals unexpected complexity of the heat shock response. Proc Natl Acad Sci U S A 104, 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., Liu, M. & Burgess, R. R. (2005). The global transcriptional response of Escherichia coli to induced σ32 protein involves σ32 regulon activation followed by inactivation and degradation of σ32 in vivo. J Biol Chem 280, 17758–17768. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. N., Kusukawa, N., Erickson, J. W., Gross, C. A. & Yura, T. (1988). Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor σ32. J Bacteriol 170, 3640–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]