Abstract
Surface adhesion plays an essential part in the survival of the commensal organism Streptococcus gordonii in the oral cavity as well as during opportunistic infections such as endocarditis. At least two types of cell surface protein involved in adhesion are found on the surface of Gram-positive bacteria: those anchored via an LPXTG motif by the enzyme sortase A (SrtA) and those associated with the cell surface by, as yet, unknown mechanisms. In srtA− mutants, LPXTG-containing proteins have been shown to be released rather than cross-linked to the cell wall. We have therefore used 2D gel electrophoresis of released proteins from an srtA− mutant as well as the wild-type strain, followed by peptide identification by MS, to identify a set of novel proteins predicted to be present on the surface of S. gordonii DL1. This includes two large LPXTG-linked proteins (SGO_0707 and SGO_1487), which both contain tandemly repeated sequences similar to those present in known fibrillar adhesins. A 5′-nucleotidase and a protein with a putative collagen-binding domain, both containing LPXTG motifs, were also identified. Anchorless proteins with known chaperone, stress response and elongation factor functions, apparently responsible for bacterial binding to keratinocytes and saliva-coated surfaces in the absence of the LPXTG-linked adhesins, were also associated with the cell surface. These data reveal a range of proteins to be present on the S. gordonii DL1 cell surface, the expression of which plays an important role in adhesion to epithelia and which represent likely candidates for novel virulence factors in S. gordonii.
INTRODUCTION
Streptococcus gordonii and other members of the Mitis group of viridans streptococci are important members of the commensal oral microflora (Schachtele et al., 2007). They are found at multiple sites within the oral cavity (Aas et al., 2005) and play a major role as pioneer species in the development of dental plaque. Central to the survival of S. gordonii in the oral cavity is the ability of the organisms to adhere to surfaces. Adherence to teeth as well as mucosae occurs through interactions between macromolecules on the bacterial cell wall and proteins or glycoproteins on the oral surfaces. Although the surface adhesins appear to show redundant function and can substitute for one another (Nobbs et al., 2007), the specific adhesion functions of the great majority of streptococcal surface proteins are, as yet, unknown.
In the oral cavity, both the teeth and keratinocytes are covered with a pellicle composed of salivary proteins. Based on Western blotting, the buccal mucosal pellicle has been shown to be composed primarily of the high-molecular-mass glycoproteins MG1 (MUC5B) and MG2 (MUC7), as well as amylase, cystatin and acidic proline-rich proteins (Bradway et al., 1992). A recent study using shotgun proteomics showed the acquired enamel pellicle to contain 130 protein species (Siqueira et al., 2007), suggesting that many more proteins than previously identified are present in the pellicles on mucosal surfaces. Salivary proteins can serve as receptors for oral streptococci (Murray et al., 1992) and S. gordonii expresses several cell surface proteins capable of interacting with salivary components. For example, S. gordonii strain DL1 surface adhesins SspA/B and CshA/B both mediate interactions with salivary agglutinin (gp340), while a 20 kDa protein encoded by the abpA gene binds to salivary amylase (Rogers et al., 1998). GspB and Hsa, two homologous serine-rich cell surface glycoproteins also isolated from S. gordonii DL1, mediate bacterial interactions with sialylated molecules such as salivary MUC7, secretory IgA and gp340 (Takahashi et al., 2002; Takamatsu et al., 2006). Several S. gordonii adhesins can also mediate binding to connective tissue proteins or cell surface receptors. For instance, CshA can bind to fibronectin, and SspA/B recognize both type 1 collagen and β-1 integrins, while GspB and Hsa can interact with sialylated receptors on human platelets. Thus, expression of these adhesins may be an important virulence factor in the pathogenesis of invasive opportunistic infections such as infective endocarditis (Kerrigan et al., 2007; Takahashi et al., 2004; Takamatsu et al., 2005; Yajima et al., 2008).
Surface proteins of Gram-positive bacteria can be grouped into at least two different types: those containing the peptidoglycan anchor motifs such as LPXTG and others that appear to be anchorless and associated with the cell surface by currently unknown mechanisms (Chhatwal, 2002). Cell surface proteins containing the conserved peptide motif (LPXTG) are cleaved between the threonine and glycine residues, and are covalently attached to cell wall peptidoglycan by the cysteine protease–transpeptidase sortase A (SrtA) (see Paterson & Mitchell, 2004 for a review). In streptococci, inactivation of the sortase A gene (srtA) results in a failure to cross-link LPXTG cell wall-associated proteins, including β-galactosidase and neuraminidase A from Streptococcus pneumoniae (Kharat & Tomasz, 2003) and P1 antigen from Streptococcus mutans (Lee & Boran, 2003), to the cell wall. In a recent study (Nobbs et al., 2007), SspA and SspB proteins were recovered in elevated amounts from the medium in srtA− mutants of S. gordonii DL1, reflecting reduced cross-linking to the cell surface. Insufficient levels of anchored LPXTG proteins were accompanied by a reduction in the capacity of the mutant to bind to salivary agglutinin. Since the adhesins of S. gordonii are crucial to the ability of these organisms to adhere and colonize intraorally and intravascularly, the aim of this study was to use sortase A-negative mutants, in which the LPXTG proteins are released rather than cross-linked to the cell wall, to investigate the repertoire of cell surface adhesion proteins in S. gordonii DL1. Knowledge of the complexity and diversity of these proteins could suggest new targets to block the development of opportunistic infections.
METHODS
Bacterial strains and culture conditions.
The srtA gene in S. gordonii DL1 (Challis) was deleted by allelic exchange with the erythromycin-resistance determinant ermAM, as we have described previously (Nobbs et al., 2007). The srtA− mutant was complemented to give srtA+ by transformation using a plasmid containing the full srtA sequence (Nobbs et al., 2007). The wild-type and mutant strains were routinely grown in modified Actinomyces defined medium (M-ADM) for 16–20 h at 37 °C in 5 % CO2 in air. Erythromycin was added to the medium at a concentration of 50 μg ml−1 for the srtA− and srtA+ mutants, and kanamycin at a concentration of 250 μg ml−1 for the srtA+ mutant.
Binding of S. gordonii to oral keratinocytes.
Immortalized normal human oral keratinocytes (OKF6/TERT-2) (Dickson et al., 2000), obtained from Dr James Rheinwald (Brigham and Women's Hospital, Boston, USA), were seeded into collagen type IV-coated μ-slide flow-cell chambers (ibidi) in keratinocyte serum-free medium (Gibco) supplemented with 0.2 ng ml−1 human recombinant epidermal growth factor (EGF), 25 μg bovine pituitary extract ml−1, and 0.3 mM CaCl2 containing 1 IU penicillin ml−1 and 1 μg streptomycin ml−1 (DF-K medium). Cells were then grown to near confluence in high-density medium (HDM) consisting of equal volumes of DF-K medium and a mixture of Dulbecco's modified Eagle's medium and Hams F-12 at a 1 : 1 ratio (v/v) supplemented with 25 μg bovine pituitary extract ml−1, 3 mM l-glutamine, 0.2 ng human recombinant EGF ml−1, 1 IU penicillin ml−1 and 1 μg streptomycin ml−1.
Overnight cultures of S. gordonii DL1 (wild-type, srtA− or srtA+) were centrifuged (12 000 g, 3 min, 4 °C) and resuspended in DF-K medium at OD600 ∼ 0.07–0.08. Keratinocyte monolayers were washed three times with DF-K medium (120 μl per channel) to remove the antibiotics and 100 μl bacterial suspensions were added to each channel. Slides were then maintained at 37 °C, 5 % CO2 in air for 0–4 h. The channels were then flushed twice with 120 μl DF-K medium per channel to remove unbound bacteria. Finally, 40 μl BacLight Gram stain (Molecular Probes) was introduced into each channel and the slides were viewed with a confocal microscope (Nikon TE2000E) equipped with an Argon 488 nm laser. Images for analysis were made of bacteria attached to both the cell and the collagen substratum.
Adhesion to saliva-coated flow-cell surfaces.
Sterile saliva for coating was prepared by treatment of whole saliva (pooled from four healthy male volunteers) with 2.5 mM DTT, followed by centrifugation (30 000 g, 20 min), dilution 1 : 4 with sterile H2O and filtration through a 0.22 μm pore-size filter. Glass flow-cells were coated with the saliva preparation, air-dried for 15 min and then rinsed by circulating M-ADM (Bowden et al., 1976) over the surface at a rate of 200 ml per hour for 10 min. Bacteria grown to exponential growth phase in M-ADM were washed and adjusted to OD600 0.8 and then circulated in the flow-cell system at a rate of 42 ml h−1 for 120 min. After rinsing for 30 min with M-ADM, adhered cells were visualized using the LIVE/DEAD BacLight stain and the number of cells was counted. Three independent experiments were carried out and the mean and se calculated for each strain.
Preparation of cell surface and released proteins.
Cell surface proteins were prepared from the bacteria by shaking (250 r.p.m.) in lysis buffer [40 mM Tris, pH 9.5, containing 5 M urea, 2 M thiourea, 2 % CHAPS, 2 % caprylyl sulphobetaine, 2 mM tributylphosphine and 0.5 % IPG buffer (GE Healthcare Life Sciences)], and mixing vigorously every 10 min for 1 h. The samples were then centrifuged at 6000 g for 10 min at 4 °C, and the supernatants collected and stored at −20 °C until use. For isolation of extracellular proteins, the culture medium (M-ADM) was harvested from batch cultures of cells in exponential growth phase by centrifugation, and proteins were precipitated overnight at 4 °C by the addition of 10 % TCA. After centrifugation at 16 000 g for 30 min at 4 °C, the resulting protein pellet was resuspended in acetone and sonicated for 3× 10s. The samples were then centrifuged at 16 000 g for 30 min at 4 °C and the supernatant was discarded. The resulting pellet was dissolved in 500 μl 2D gel electrophoresis rehydration buffer (8 M urea, 2 % CHAPS, 10 mM DTT, 2 % IPG buffer; GE Healthcare Life Sciences) and stored at −20 °C until use.
2D PAGE.
The protein concentration in the extracts was determined using a 2D Quant kit (GE Healthcare Life Sciences). A volume corresponding to 20 μg protein was diluted with rehydration buffer and placed in a reswelling cassette with 18 cm pH 4–7 linear IPG strips (GE Healthcare Life Sciences) on top. Rehydration was carried out at room temperature for 30 h under silicone oil. IEF was carried out using a Multiphor II (GE Healthcare Life Sciences) with cooling water at 15 °C supplied by Pharmacia Multitemp II. The focusing was initiated at 150 V for 1 h and continued at 300 V for 3 h, 600 V for 3 h, 1200 V for 12 h and finally 3500 V for 20 h. After focusing, the IPG strips were stored at −80 °C. Before the second dimension was run, the IPG strips were equilibrated first in 50 mM Tris buffer, pH 6.8, containing 2 % SDS, 26 % glycerol and 16 mM DTT for 15 min, and then in 50 mM Tris buffer, pH 6.8, containing 2 % SDS, 26 % glycerol, 250 mM iodoacetamide and 0.005 % bromophenol blue for another 15 min. The equilibrated IPG strips were embedded on top of 7 or 14 % polyacrylamide gels (20×20×0.1 cm) using 0.5 % (w/v) molten agarose. SDS-PAGE was performed at a constant current of 15 mA per gel, at 10 °C, overnight in a PROTEAN II xi cell (Bio-Rad) with High-Range Rainbow molecular mass standards (GE Healthcare Life Sciences) run on the acidic side of the IPG strips. Gels were stained with silver according to the protocol from GE Healthcare Life Sciences. All gels were run in triplicate and only protein spots occurring in all gels were considered further. Proteins were considered to show increased or decreased production only when the trend was reproduced in all three gels.
Spot detection and identification of streptococcal proteins by liquid chromatography (LC)-MS/MS.
Gels were analysed using Delta2D software (Decodon). After identification of the spot boundaries, the integrated optical density (IOD) values were determined and expressed as a percentage of the total intensity per gel. For protein identification, gels were stained with colloidal Coomassie Brilliant Blue G (CCBB). The most abundant proteins were excised manually from 2D PAGE gels and the gel pieces were digested with trypsin (modified sequencing grade, Promega) using an Investigator ProGest robotic workstation (Genomic Solutions). Briefly, proteins were reduced with DTT (60 °C, 20 min), S-alkylated with iodoacetamide (25 °C, 10 min) then digested with trypsin (37 °C, 8 h). The peptide extract was dried by rotary evaporation (SC110 Speedvac, Savant Instruments) and dissolved in 0.1 % formic acid. Peptide solutions were analysed using an HCTultraPTM Discovery system (Bruker Daltonics) coupled to an UltiMate 3000 LC system (Dionex). Tryptic peptides were separated on a monolithic capillary column (200 μm internal diameter×5 cm, Dionex) with a gradient of 5 % acetonitrile in water containing 0.5 % formic acid to 37 % acetonitrile in water containing 0.45 % formic acid over 12 min at a flow rate of 2.5 μl ml−1. Peptide fragment mass spectra were acquired in data-dependent AutoMS(2) mode with a scan range of 300–1500 m/z, three averages, and up to three precursor ions selected from the MS scan (100–2200 m/z). Precursors were actively excluded within a 1.0 min window, and all singly charged ions were excluded. Peptide peaks were detected and deconvoluted automatically using Data Analysis software (Bruker). Mass lists in the form of Mascot Generic files were created automatically and used as the input for Mascot MS/MS ion searches of the NCBInr database using the Matrix Science web server (www.matrixscience.com). Parameters were set to 0.5 Da peptide mass tolerance, methionine oxidation and carboxyamidomethyl cysteine modification.
In silico and data analysis.
Primary amino acid sequences of identified proteins were investigated for the presence of known functional domains or tandem repeat domains using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) and Smart databases. The full repertoire of LPXTG-containing proteins was determined by searching the protein database for S. gordonii (PubMed).
To study the adhesion of S. gordonii to keratinocytes, three channels were used for each bacterial strain at each time point. Images of 10 fields selected at random were captured using the NIS Elements software, and numbers of bacteria on the cell surface (stained red) and numbers of epithelial cells (stained green) in each field were counted. Bacteria internalized into cells were not stained and therefore not accounted for in this study. The average number of bacteria per cell in each field was then determined and the mean and se for the 30 fields at each time point were then calculated. Experiments were carried out three times, and finally the mean and se for the three independent experiments were calculated. For the studies of adhesion to saliva-coated glass surfaces or the collagen type IV substrate, images of 10 fields selected at random were captured using the NIS Elements software. Image analysis was performed using the bioImage_L software (Chávez de Paz, 2009), at www.bioimagel.com, in which the area covered by stained cells in each field was determined by a colour segmentation algorithm which also enumerated the number of bacteria.
RESULTS
Adhesion of wild-type and srtA− mutant S. gordonii to oral keratinocytes and saliva-coated surfaces
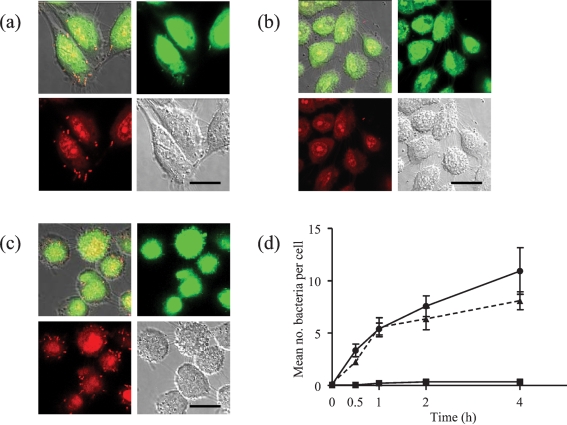
To investigate how the repertoire of bacterial cell surface proteins influences the binding of S. gordonii to surfaces, wild-type DL1 and sortase A-negative mutant strains were compared for adhesion to surfaces modelling those found in the oral cavity. First, the number of bacteria adhering to oral keratinocytes over time was assessed after staining with LIVE BacLight bacterial Gram stain and imaging by confocal scanning laser microscopy. The wild-type DL1 bound well to the keratinocytes, particularly around the filapodial projections of the cells (Fig. 1a); bacteria adhered to most cells in each field, suggesting that the keratinocytes were relatively homogeneous in their binding capacities. Adhesion of S. gordonii to the type IV collagen substratum was very low (mean value <1 bacterial cell 100 μm2 after 4 h of incubation). For the srtA− strain, only occasional bacteria were seen adhering to the keratinocytes (Fig. 1b), while the srtA+ strain, in which a functional copy of the sortase gene had been reinserted, showed a similar level of binding to that of the wild-type (Fig. 1c). At time zero, no binding to the keratinocytes was seen for any of the three strains (Fig. 1d). However, the numbers of bound wild-type bacteria increased from 0 to an average of 11 bacteria per keratinocyte over 4 h of incubation (Fig. 1d). In contrast, adhesion of the srtA− strain at all times of observation was significantly lower (P<0.0001), and the mean number of bound bacteria remained at <1 per keratinocyte after 4 h (Fig. 1d). The complemented mutant (srtA+) and wild-type strains bound similarly until 1 h of incubation, when the levels diverged, but over time the difference in binding was not significant.
Fig. 1.
Interaction of S. gordonii with cultured oral keratinocytes after 2 h incubation. (a) Wild-type DL1, (b) srtA− mutant of DL1 and (c) complemented mutant (srtA+) were incubated with OFK6 TERT-2 oral keratinocytes in ibidi mini-flow cells for 2 h. After washing, adherent bacteria were stained using the BacLight Gram stain as described in Methods. Panels show the colour channel images superimposed upon differential interference contrast (DIC) images of the same cells, as well as each of the images (green, red and DIC) separately. Bars, 10 μm. (d) The mean number and se of bacteria per keratinocyte in each field for the wild-type (•), srtA− mutant (▴) and complemented mutant (▪) were calculated and plotted after 0, 0.5, 1, 2 and 4 h.
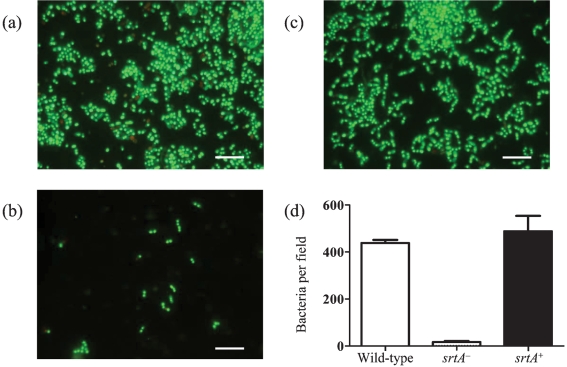
To investigate how well the different strains could bind to an experimental salivary pellicle, the numbers of bacteria adhering to glass surfaces conditioned with saliva were studied in a flow-cell system. Bacteria were recirculated for 120 min and the numbers of attached cells were estimated after staining with a LIVE/DEAD BacLight bacterial viability kit (Fig. 2). The mean binding from three independent experiments for wild-type DL1 was 440±38 bacteria per 4.8 mm2, which was significantly higher (P<0.001) than that for the srtA− mutant (20±2 bacteria per 4.8 mm2). There was no significant difference between the binding of the wild-type and that of the srtA+ complemented mutant (488±65 bacteria per 4.8 mm2). These data suggest that LPXTG-containing proteins, linked to the cell surface by sortase A, play an important role in the binding of S. gordonii to oral surfaces.
Fig. 2.
Adhesion of S. gordonii to salivary pellicles on glass surfaces after 2 h incubation. (a) Wild-type DL1, (b) srtA− mutant of DL1 and (c) complemented mutant (srtA+) were recirculated over saliva-coated glass surfaces for 2 h as described in Methods. (d) The numbers of adherent bacteria were enumerated after staining with the LIVE/DEAD BacLight kit. Bars, 10 μm.
Identification of LPXTG-containing cell surface proteins from S. gordonii
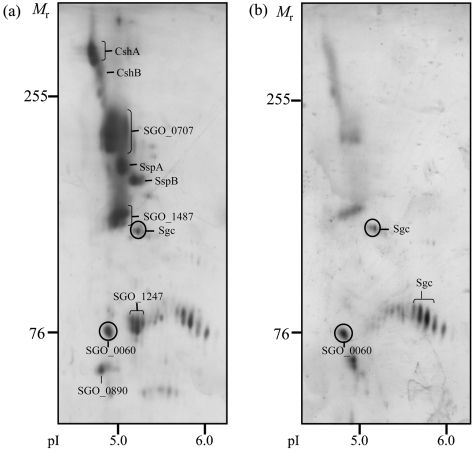
To identify cell surface proteins from S. gordonii DL1 which may be involved in bacterial binding, proteins released into the culture supernatants from the srtA− mutant of DL1 were isolated and subjected to 2D-PAGE in 7 % gels (Fig. 3a). A total of 10 prominent spots, with molecular sizes ranging from >255 to ∼60 kDa, were identified in three replicate gels. Spots from corresponding Coomassie-stained gels were excised, and the proteins were identified by LC-MS/MS and peptide matching against the genomic sequence database for S. gordonii DL1 (Challis) using the Mascot search engine. Of the proteins identified, eight out of 10 contained the LPXTG consensus motif for linkage to cell wall peptidoglycan by sortase A (Table 1). At least 17 peptides were matched to each protein and the coverage was 18–52 %. The largest species, with Mr greater than 255 kDa, were shown to correspond to the previously described fibrillar proteins from S. gordonii, CshA and CshB. The members of the antigen I/II family from S. gordonii, SspA and SspB, were also present at Mr 164–172 kDa.
Fig. 3.
Released proteins from S. gordonii resolved on 2D-PAGE gels and stained with silver. Proteins released into the medium from (a) the srtA− mutant or (b) wild-type strain DL1 were separated by IEF (pI range 4–7) in the first dimension and by SDS-PAGE in 7 % gels in the second dimension. The migration positions of molecular mass markers and pI values are shown. The identities of the major spots are presented in Table 1.
Table 1.
Identity of LPXTG-containing proteins from S. gordonii separated by 2D-PAGE
| Accession number* | Gene symbol† | Observed migration‡ | Theoretical migration§ | Number of peptides matched|| | Coverage¶ | Ratio mean spot volume, srtA−/wild-type# | Protein characteristics** | ||
|---|---|---|---|---|---|---|---|---|---|
| pI | Mr | pI | Mr | ||||||
| SGO_0854 | cshA | 4.7 | 343 | 4.7 | 264 | 43 | 24 | 3.6 | Adhesin |
| SGO_1148 | cshB | 4.7 | 299 | 4.7 | 244 | 32 | 32 | ||
| SGO_0707 | 4.9 | 223 | 5.0 | 180 | 43 | 32 | 3.8 | Large tandem repeat-containing protein | |
| SGO_0210 | sspA | 5.0 | 190 | 4.9 | 172 | 45 | 30 | 4.3 | Adhesin |
| SGO_0211 | sspB | 5.1 | 177 | 5.0 | 164 | 30 | 23 | 2.3 | Adhesin |
| SGO_1487 | 4.9 | 144 | 4.9 | 191 | 27 | 18 | 3.7 | Large tandem repeat-containing protein | |
| SGO_1247 | 5.1 | 82 | 5.1 | 78 | 43 | 52 | 3.8 | Enzyme – 5′-nucleotidase | |
| SGO_0890 | 4.8 | 71 | 5.5 | 69 | 17 | 23 | 5.8 | Unknown | |
*Accession number in the TIGR database for S. gordonii.
†Gene symbol in the TIGR database for S. gordonii Challis CH1.
‡Calculated from the data presented in Fig. 3. Mr values are in kDa.
§As given in the TIGR database for S. gordonii.
||Number of tryptic peptides obtained.
¶The percentage amino acid coverage (peptides observed/theoretical value from sequence data).
#Ratio of IOD for each protein derived from srtA− cells to that for the same protein derived from wild-type DL1 cells (mean, n=3, for each strain).
**Putative functions assigned from the TIGR database or characteristics determined through manual comparison of sequences.
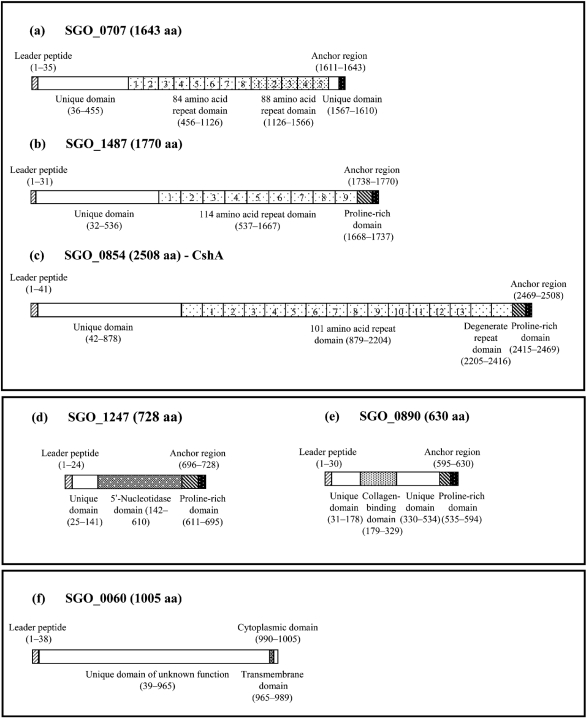
In addition to these known proteins, four novel proteins corresponding to the ORFs, SGO_0707, SGO_1487, SGO_1247 and SGO_0890, were identified (Table 1). None of these has previously been characterized, but all contain an N-terminal signal peptide and are predicted to be cell wall-anchored due to the presence of an LPXTG motif at the C terminal. As seen from the 2D-PAGE gels, SGO_0707 migrated between CshA/B and SspA/B, while SGO_1487 appeared at a position corresponding to a lower molecular mass (Fig. 3a). The average IOD of the silver-stained spots for SGO_0707 (22 % of the total for the srtA− mutant) and SGO_1487 (15 % of the total for the srtA− mutant) suggests that the proteins were released in at least similar abundance to CshA/B (12 %) and SspA/B (11 %). Examination of the amino acid sequence of both SGO_0707 (Fig. 4a) and SGO_1487 (Fig. 4b) revealed striking structural similarities to CshA (Fig. 4c). All the proteins are predicted to contain a C-terminal anchor region consisting of a cytoplasmic tail of positively charged amino acids and the LPXTG motif, separated by a hydrophobic membrane-spanning sequence. Immediately adjacent to the anchor domain, both CshA and SGO_1487 contain short proline-rich sequences. The N-terminal extracellular domain of each of the proteins is dominated by a series of tandemly repeated sequences containing 84–114 amino acids. In both CshA and SGO_0707, the N-terminal and tandem repeat domains are separated by a non-repeat domain which shows no homology to any known functional domain or to any recognized binding domain for eukaryotic cell receptors or matrix proteins. In SGO_1487, however, amino acids 185–318 within the non-repeat domain show homology to collagen-binding protein A (SSA_1663) from Streptococcus sanguinis.
Fig. 4.
Schematic diagrams of the putative domain structures of novel S. gordonii cell surface proteins. The two largest proteins, SGO_0707 (a) and SGO_1487 (b), shared structural features such as the LPXTG-containing anchor domain and tandem repeats with the large fibrillar protein CshA (c). Two smaller proteins, SGO_1247 (d) and SGO_0890 (e), contained the LPXTG anchor domains but lacked tandem repeats. The final novel protein SGO_0600 (f) contained a transmembrane sequence as well as a large extracellular domain of unknown function. Leader peptide (N-terminal consensus sequence for membrane insertion) (cross-hatched), unique domains (non-repetitive sequences) (white), repeat domains (tandemly repeated amino acid sequences of different lengths or degenerate tandem repeat domains) (lightly stippled), proline-rich domains (cross-hatched), domain with putative 5′-nucleotidase activity (white stippling on grey), domain with putative collagen-binding function (densely stippled), transmembrane domain (hydrophobic amino acid sequence) (cross-checked), and anchor domain (LPXTG-containing sequence) (white stippling on black).
Two smaller LPXTG-containing proteins with molecular masses of around 78 and 70 kDa were identified as the ORFs SGO_1247 and SGO_0890, respectively. The deduced amino acid sequences of both these proteins contain N-terminal signal peptides as well as the characteristic Gram-positive anchor and proline-rich domains (Fig. 4d, e). No tandemly repeated amino acid sequences were identified within the extracellular domains.
The most prominent feature of SGO_1247 identified from the KEGG protein database was the presence of a 5′-nucleotidase motif within the extracellular domain. An LPXTG-linked protein containing this motif has also been identified in Streptococcus pyogenes (Janulczyk & Rasmussen, 2001). The N-terminal domain of SGO_0890 is predicted to contain collagen-binding domains and Cna protein B-type domains, which are thought to act as molecular ‘spacers’ to lift proteins away from the cell wall. No peptides were obtained from the absolute N-terminal regions of any of the LPXTG-containing proteins, confirming that translocation over the cell membrane and cleavage of the N-terminal signal peptide occur normally, despite the inactivation of sortase A. For comparison, the presence of the LPXTG proteins was also investigated in the culture supernatant from the wild-type DL1 strain (Fig. 3b). 2D-PAGE on 7 % gels showed that all of the proteins mentioned above were also released but in significantly lower amounts (at least a twofold difference) from the wild-type strain than from the srtA− mutant (Table 1).
An in silico analysis revealed the presence of 21 predicted LPXTG-linked cell wall proteins in the S. gordonii genome, of which only five have previously been characterized (Table 2). They range in theoretical molecular mass from 69 to 397 kDa and represent different functional classes of proteins. In addition to the adhesins and large tandem repeat proteins identified in this study, one other very high molecular mass protein (SGO_2005) was found. In addition, a range of putative cell-wall-linked enzymes, mainly proteases, as well as proteins of unknown function were identified. While it cannot be completely ruled out that the proteins could not be detected in our study for technical reasons, the data suggest that only a subpopulation of the possible LPXTG-linked proteins are expressed under the experimental conditions used here.
Table 2.
LPXTG-containing proteins from S. gordonii identified by in silico analysis
| Accession number* | Gene symbol† | Theoretical molecular weight‡ | Protein characteristics/function§ |
|---|---|---|---|
| SGO_2005 | 397 | Unknown | |
| SGO_0854 | cshA | 264 | Adhesin |
| SGO_1148 | cshB | 244 | |
| SGO_0966 | hsa | 203 | Adhesin |
| SGO_1487 | 191 | Large tandem repeat-containing protein | |
| SGO_0707 | 180 | Large tandem repeat-containing protein | |
| SGO_0208 | 174 | Enzyme – glycosyltransferase | |
| SGO_0210 | sspA | 172 | Adhesin |
| SGO_0317 | 165 | Enzyme – serine protease | |
| SGO_0316 | 165 | Enzyme – serine protease | |
| SGO_0211 | sspB | 164 | Adhesin |
| SGO_0388 | 120 | Enzyme – zinc carboxypeptidase | |
| SGO_1415 | 117 | Enzyme – X-prolyl dipeptidylaminopeptidase | |
| SGO_0107 | 115 | Unknown | |
| SGO_0430 | 96 | Unknown | |
| SGO_2004 | 87 | Unknown | |
| SGO_1651 | 84 | Unknown | |
| SGO_1247 | 78 | Enzyme – 5′-nucleotidase | |
| SGO_1650 | 76 | Unknown | |
| SGO_1182 | 74 | Unknown | |
| SGO_0890 | 69 | Unknown |
*Accession number in the TIGR database for S. gordonii.
†Gene symbol in the TIGR database for S. gordonii Challis CH1.
‡As given in the TIGR database for S. gordonii.
§Protein characteristics or function as determined from the TIGR database or through manual comparison of sequences.
Release of non-LPXTG-containing proteins
In addition to the LPXTG-linked proteins, two other prominent protein spots were consistently present in the culture supernatants from the sortase A-negative mutant and wild-type DL1. The higher-molecular-mass species was identified as the product of ORF SGO_0566, which corresponds to the subtilisin-like serine protease challisin, a secreted enzyme of S. gordonii (Table 3). Interestingly, the amino acid sequence data show 35 % homology to two predicted LPXTG-linked serine proteases (SGO_0316 and SGO_0317; see Table 2) from S. gordonii, which were not identified in this study. The second non-LPXTG protein found was the hypothetical protein SGO_0060 (Fig. 4f). The presence of a putative transmembrane domain indicates that this protein is membrane-anchored, and although its function is as yet unknown, the central unique domain shows a high degree of homology with putative phage infection proteins from a number of other streptococcal species.
Table 3.
Identities of released surface proteins and secreted species from S. gordonii separated by 2D-PAGE
| Gene symbol* | Accession number† | Observed migration‡ | Theoretical migration§ | Number of peptides matched|| | Coverage¶ | Ratio mean spot volume wild-type/srtA−# | Protein identity | Putative cellular location** | Functional category†† | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pI | Mr | pI | Mr | ||||||||
| sgc | SGO_0566 | 5.3 | 83.5 | 5.3 | 164.5 | 33 | 21 | 2.0 | Challisin | Secreted | Serine protease enzyme |
| SGO_0060 | 4.5 | 76.0 | 5.1 | 109.1 | 51 | 46 | 1.1 | Hypothetical protein | Membrane-anchored | Unknown | |
| dnaK | SGO_0402 | 4.7 | 68.0 | 4.7 | 64.7 | 32 | 50 | 1.4 | DnaK | Surface-associated | Protein chaperone |
| pyk | SGO_1339 | 5.1 | 55.7 | 4.9 | 54.8 | 30 | 67 | 2.0 | Pyruvate kinase | Surface-associated | Metabolic enzyme |
| tuf | SGO_0761 | 5.1 | 52.7 | 4.9 | 44.0 | 32 | 61 | 1.8 | EF-Tu | Surface-associated | Transcription and translation |
| fusA | SGO_0206 | 5.1 | 80.4 | 4.9 | 76.8 | 36 | 61 | 2.0 | EF-G | Surface-associated | Transcription and translation |
| gsp-781 | SGO_2107 | 4.7 | 45.4 | 5.1 | 41.4 | 33 | 53 | 1.2 | Glucan-binding protein B | Surface-associated | General stress protein |
*Gene symbol in the TIGR database for S. gordonii Challis CH1.
†Accession number in the TIGR database for S. gordonii.
‡Calculated from the data presented in Fig. 5. Mr values are in kDa.
§As given in the TIGR database for S. gordonii.
||Number of tryptic peptides obtained.
¶The percentage amino acid coverage (number of peptides observed/theoretical value from sequence data).
#Ratio of IOD for each protein derived from wild-type DL1 cells to that for the same protein derived from DL1 srtA− cells (mean, n=3, for each strain).
**Putative protein type as ascertained from this study.
††Putative functions assigned from the TIGR database or through manual comparison of sequences.
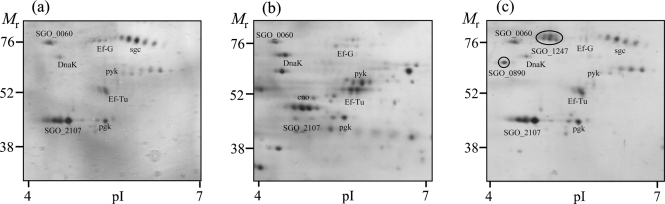
To obtain better resolution of the released proteins with a lower molecular mass, culture supernatants from the wild-type strain were subjected to 2D-PAGE on 14 % gels (Fig. 5a). Eight major proteins were resolved with molecular masses of less than 76 kDa. Two of these were the elongation factors G and Tu (EF-G and EF-Tu, respectively), and a third corresponded to the molecular chaperone heat-shock protein 70 or DnaK. In addition, the glycolytic enzymes pyruvate kinase (pyk) and phosphoglycerate kinase (pgk) were present. A protein showing isoelectric heterogeneity at around Mr=76 kDa was identified as a lower-molecular-mass isoform of the serine protease challisin. Another major protein detected in the extracellular supernatant was the general stress protein gsp-781, which appeared as a cluster of at least three isoforms at 47 kDa. To investigate whether these proteins, as well as being released, were also associated with the bacterial surface, cell surface proteins were removed from wild-type DL1 by shaking in a detergent buffer. 2D-PAGE revealed that all the proteins, with the exception of challisin, could be isolated from the cell surface (Fig. 5b). In the culture supernatant from the sortase A-negative mutant (Fig. 5c), levels of all the non-LPXTG surface-associated proteins were similar to those in the wild-type, and the ratio of the mean IOD values between the wild-type and srtA− mutant was ≤2. These data thus suggest that DnaK, EF-Tu and EF-G, pyruvate kinase and phosphoglycerate kinase, general stress protein-781 and the hypothetical protein SGO_0060 were all present in the supernatant as the result of release from the cell surface, and that their release was independent of sortase A.
Fig. 5.
Released proteins and cell surface proteins from wild-type, and released proteins from the srtA− mutant of S. gordonii, resolved using 2D-PAGE gels and stained with silver. Proteins (a) released into the medium by and (b) recovered from the cell surface of wild-type, and (c) proteins released into the medium by the srtA− mutant of S. gordonii were resolved by IEF (pI range 4–7) in the first dimension and by SDS-PAGE in 14 % gels in the second dimension. The migration positions of molecular mass markers and pI values are shown. The identities of the major spots are presented in Table 3.
DISCUSSION
Retention of commensal species on surfaces through interactions with cell surfaces and salivary pellicles is essential for their success in the oral cavity. We have shown that binding of S. gordonii to both oral keratinocytes and saliva-coated surfaces is markedly reduced, although not completely abolished, by inactivation of the sortase A gene. This is in agreement with other studies, in which sortase A-negative mutants exhibit reduced binding to proteins such as fibronectin and purified salivary agglutinin as well as a decreased ability to colonize the oral cavity of mice (Bolken et al., 2001; Nobbs et al., 2007). Thus, proteins linked through the LPXTG motif to the peptidoglycan cell wall appear to be critical for interaction of S. gordonii with oral surfaces.
In sortase A-negative mutants of S. gordonii, proteins are translocated normally across the cell membrane, but released into the culture supernatant in the absence of covalent cross-linkage to peptidoglycan. Using the sortase A-negative strain as a model, we identified a total of eight LPXTG-containing proteins from S. gordonii, four of which have not been described previously. Recently, the S. gordonii (Challis, CH1 strain) genome has been completely sequenced, providing a valuable resource for investigation of domain structures and binding motifs within newly identified proteins (Vickerman et al., 2007). Comparison of structural elements within the largest novel species, SGO_0707 and SGO_1487, revealed that both contain a membrane-spanning sequence and a sequence of positively charged amino acids at the C-terminal end. The N-terminal extracellular domain contains a series of tandemly repeated amino acid sequences as well as a non-repeat domain. This structural pattern is very similar to that of the fibril-forming protein CshA (McNab et al., 1999), suggesting that SGO_0707 and SGO_1487 also represent fibrillar adhesins. In Staphylococcus aureus, the presence of multiple tandem repeats in cell surface adhesins has been shown to be essential for adherence to and invasion of host tissues (Hussain et al., 2008). The non-repeat domains differed between the two proteins, with a collagen-binding domain in SGO_1487, while no homology to any matrix or host cell protein-binding domains was found in either CshA or SGO_0707.
Both SGO_0707 and SGO_1487 were abundantly present in the culture supernatants at levels seemingly higher than those of CshA/B and SspA/B (Nobbs et al., 2007). However, the relative levels of the proteins in the sortase-A negative mutant probably cannot be compared with abundance on the cell wall of the wild-type, since we have reported that inactivation of the sortase gene causes compensatory upregulation of some LPXTG-containing protein genes, including cshA/B and sspA/B (Nobbs et al., 2007). Thus, the bacteria may recognize the absence of some adhesins and respond by adjusting the levels of others. Indeed, certain adhesins in S. gordonii can transcriptionally regulate levels of others (J. Kreth and others, unpublished data). Another LPXTG-linked protein, SGO_0890, which contains a predicted collagen-binding motif, was also present. This protein had anchor and extracellular proline-rich domains in common with SGO_0707 and SGO_1487, but lacked the extracellular tandem repeats. The final protein identified belonged to the group of putative LPXTG-linked surface-associated enzymes, in this case a 5′-nucleotidase (SGO_1247). Other examples of cell wall-associated 5′-nucleotidases have been reported in streptococci (Janulczyk & Rasmussen, 2001), and enzymes including a β-galactosidase, an N-acetylhexosaminidase and several proteases are also amongst the predicted LPXTG-containing proteins in S. gordonii, suggesting that a range of enzymes can be expressed on the bacterial cell surface. It is not currently known whether these enzymes are active, but if so, they may play important roles in bacterial survival through the degradation of external metabolic substrates.
From the in silico analysis it is clear that we have only detected a subpopulation of the predicted LPXTG-linked proteins from S. gordonii in this study, and known proteins such as Hsa were not found. It appears likely that S. gordonii has a repertoire of cell wall-linked proteins which are not expressed continuously but rather are upregulated individually or in clusters to allow the bacteria to adapt to the prevailing extracellular conditions.
Challisin, a secreted enzyme, was also detected in the culture supernatants from both the wild-type and the mutant DL1. Challisin has been described as a secreted serine protease from S. gordonii which can inactivate competence-stimulating peptide (CSP) released from S. mutans (Wang & Kuramitsu, 2005). Serine proteases are released from a variety of streptococcal species, including S. pyogenes, in which they can degrade complement proteins and facilitate entry into blood vessels, but the role of challisin in S. gordonii remains to be determined. SGO_0060 appeared to be present in equal amounts in the culture supernatant from the wild-type and mutant DL1, and release of this protein was, therefore, independent of sortase A. As expected, the putative protein sequence did not contain an LPXTG motif, although a transmembrane domain was present. This structure is consistent with a non-wall-anchored protein released from the cell membrane. The extracellular domain contained no homology to any known protein and its function is unknown.
Analysis on 14 % gels showed that the low-molecular-mass proteins released into the culture supernatant from both the wild-type and mutant DL1 were few in number and most were also present on the cell surface. These included known stress proteins such as DnaK and general stress protein-781, EF-Tu and EF-G, as well as the glycolytic enzymes pyruvate kinase and phosphoglycerate kinase. In contrast to SGO_0060, none of these proteins contains a transmembrane domain. Data from earlier studies suggest that these proteins are released and can reassociate with the cell surface (Wilkins et al., 2003). Reassociated surface proteins could be cross-linked to the cell wall or other proteins by the action of transglutaminases. While their functions are currently unknown, these reassociated surface proteins could contribute to adhesion and may account for the low level of interaction seen between the sortase A-negative mutant and the keratinocyte cell surface or the salivary pellicle.
In conclusion, we have detected a set of novel proteins that are predicted to be on the surface of S. gordonii. Our data suggest that a range of proteins with different functions are anchored to the cell wall of S. gordonii DL1 via the LPXTG motif, and that expression of these proteins plays an important role in the adhesion of S. gordonii to oral surfaces. Other surface proteins with diverse putative functions have no anchorage motif. Further studies are required to functionally characterize these proteins in colonization of the oral cavity as well as in pathogenic processes such as bacteraemia, sepsis and infective endocarditis.
Acknowledgments
We thank Angela H. Nobbs and Yongshu Zhang, who constructed the sortase A mutants, as well as Agnethe Henriksson and Madeleine Blomqvist for excellent technical assistance. Support for this project from NIH/NIDCR R01DE08590 (M. C. H.), The Swedish Research Council K2005-06X-12266-07A (G. S.), The Knowledge Foundation, Sweden (G. S., J. R. D.), and The Swedish Dental Association (J. R. D.) is gratefully acknowledged. The Aberdeen Proteome Facility is funded jointly by the Scottish Higher Education Funding Council (SHEFC), the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the University of Aberdeen, UK.
Abbreviations
EF-G, elongation factor G
EF-Tu, elongation factor Tu
IOD, integrated optical density
LC, liquid chromatography
References
- Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I. & Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolken, T. C., Franke, C. A., Jones, K. F., Zeller, G. O., Jones, C. H., Dutton, E. K. & Hruby, D. E. (2001). Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect Immun 69, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden, G. H., Hardie, J. M. & Fillery, E. D. (1976). Antigens from Actinomyces species and their value in identification. J Dent Res 55, A192–A204. [DOI] [PubMed] [Google Scholar]
- Bradway, S. D., Berget, E. J., Scannapieco, F. A., Ramasubbu, N., Zawack, S. & Levine, M. J. (1992). Formation of salivary-mucosal pellicle: the role of transglutaminase. Biochem J 284, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez de Paz, L. E. (2009). Image analysis software based on color segmentation for characterization of viability and physiological activity in biofilms. Appl Environ Microbiol 75, 1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatwal, G. S. (2002). Anchorless adhesins and invasions of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol 10, 205–208. [DOI] [PubMed] [Google Scholar]
- Dickson, M. A., Hahn, W. C., Ino, Y., Ronfard, V., Wu, J. Y., Weinberg, R. A., Louis, D. N., Li, F. P. & Rheinwald, J. G. (2000). Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits normal life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 20, 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M., Haggar, A., Peters, G., Chhatwal, G. S., Herrmann, M., Flock, J. I. & Sinha, B. (2008). More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host invasion but not for leukocyte activation. Infect Immun 76, 5615–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulczyk, R. & Rasmussen, M. (2001). Improved pattern for genome-based screening identifies novel cell wall-attached proteins in Gram-positive bacteria. Infect Immun 69, 4019–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrigan, S. W., Jakubovics, N. S., Keane, C., Maguire, P., Wynne, K., Jenkinson, H. F. & Cox, D. (2007). Role of Streptococcus gordonii surface proteins SspA/SspB and Hsa in platelet function. Infect Immun 75, 5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharat, A. S. & Tomasz, A. (2003). Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect Immun 71, 2758–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. F. & Boran, T. L. (2003). Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect Immun 71, 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab, R., Forbes, H., Handley, P. S., Loach, D. M., Tannock, G. W. & Jenkinson, H. F. (1999). Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J Bacteriol 181, 3087–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, P. A., Prakobphol, A., Lee, T., Hoover, C. I. & Fisher, S. J. (1992). Adherence of oral streptococci to salivary glycoproteins. Infect Immun 60, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs, A. H., Vajna, R. M., Johnson, J. R., Zhang, Y., Erlandsen, S. L., Oli, M. W., Kreth, J., Brady, L. J. & Herzberg, M. C. (2007). Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 153, 4088–4097. [DOI] [PubMed] [Google Scholar]
- Paterson, G. K. & Mitchell, T. J. (2004). The biology of Gram-positive sortase enzymes. Trends Microbiol 12, 89–95. [DOI] [PubMed] [Google Scholar]
- Rogers, J. D., Haase, E. M., Brown, A. E., Douglas, C. W. I., Gwynn, J. P. & Scannapieco, F. A. (1998). Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology 144, 1223–1233. [DOI] [PubMed] [Google Scholar]
- Schachtele, C. F., Nobbs, A., Zhang, Y., Costalonga, M. & Herzberg, M. C. (2007). Oral streptococci: commensals and opportunistic pathogens. In The Molecular Biology of Streptococci, pp. 411–462. Edited by R. Hakenbeck & S. Chhatwal. Norfolk, UK: Horizon Scientific Press.
- Siqueira, W. L., Zhang, W., Helmerhorst, E. J., Gygi, S. P. & Oppenheim, F. G. (2007). Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res 6, 2152–2160. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Konishi, K., Cisar, J. O. & Yoshikawa, M. (2002). Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun 70, 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Yajima, A., Cisar, J. O. & Konishi, K. (2004). Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect Immun 72, 3876–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu, D., Bensing, B. A., Cheng, H., Jarvis, G. A., Siboo, I. R., Lopéz, J. A., Grifiss, J. M. & Sullam, P. M. (2005). Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Iα. Mol Microbiol 58, 380–392. [DOI] [PubMed] [Google Scholar]
- Takamatsu, D., Bensing, B. A., Prakobphol, A., Fisher, S. J. & Sullam, P. M. (2006). Binding of the streptococcal glycoproteins GspB and Hsa to human salivary proteins. Infect Immun 74, 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman, M. M., Iobst, S., Jesionowski, A. M. & Gill, S. R. (2007). Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol 189, 7799–7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. Y. & Kuramitsu, H. K. (2005). Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, J. C., Beighton, D. & Homer, K. A. (2003). Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl Environ Microbiol 69, 5290–5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima, A., Urano-Tashiro, Y., Shimazu, K., Takashima, E., Takahashi, Y. & Konishi, K. (2008). Hsa, an adhesin of Streptococcus gordonii DL1, binds to α2–3-linked sialic acid on glycophorin A of the erythrocyte membrane. Microbiol Immunol 52, 69–77. [DOI] [PubMed] [Google Scholar]