Abstract
Programmed −1 ribosomal frameshifting (PRF) is a distinctive mode of gene expression utilized by some viruses, including human immunodeficiency virus type 1 (HIV-1), to produce multiple proteins from a single mRNA. −1 PRF induces a subset of elongating ribosomes to shift their translational reading frame by 1 base in the 5′ direction. The appropriate ratio of Gag to Gag-Pol synthesis is tightly regulated by the PRF signal which promotes ribosomes to shift frame, and even small changes in PRF efficiency, either up or down, have significant inhibitory effects upon virus production, making PRF essential for HIV-1 replication. Although little has been reported about the cellular factors that modulate HIV-1 PRF, the cis-acting elements regulating PRF have been extensively investigated, and the PRF signal of HIV-1 was shown to include a slippery site and frameshift stimulatory signal. Recently, a genome-wide screen performed to identify cellular factors that affect HIV-1 replication demonstrated that down-regulation of eukaryotic release factor 1 (eRF1) inhibited HIV-1 replication. Because of the eRF1 role in translation, we hypothesized that eRF1 is important for HIV-1 PRF. Using a dual luciferase reporter system harboring a HIV-1 PRF signal, results showed that depletion or inhibition of eRF1 enhanced PRF in yeast, rabbit reticulocyte lysates, and mammalian cells. Consistent with the eRF1 role in modulating HIV PRF, depleting eRF1 increased the Gag-Pol to Gag ratio in cells infected with replication-competent virus. The increase in PRF was independent of a proximal termination codon and did not result from increased ribosomal pausing at the slippery site. This is the first time that a cellular factor has been identified which can promote HIV-1 PRF and highlights HIV-1 PRF as essential for replication and an important but under exploited antiviral drug target.
Keywords: Drug Design, Ribosome Function, Translation Regulation, Translation Release Factors, Viral Replication, HIV-1, eRF1, Programmed Ribosomal Frameshift
Introduction
The ability of ribosomes to maintain the correct translational open reading frame (ORF)2 is fundamental to the integrity and fidelity of protein synthesis. However, there are a number of examples in which elongating ribosomes are programmed to shift their translational ORF 1 base in either the 5′ or 3′ direction (−1 or +1 ribosomal programmed ribosomal frameshift (PRF), respectively) to translate multiple proteins from a single mRNA (1–5). HIV-1 is one example of a virus that uses this process as an integral part of its replication cycle.
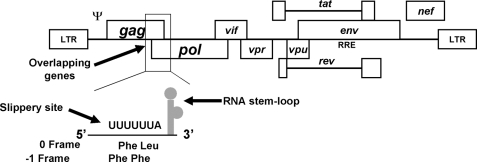
The HIV-1 Gag and Pol proteins are synthesized as p55Gag or p160Gag-Pol polyprotein precursors using the same translation start codon but different ORFs in the full-length viral mRNA (6, 7). The gag ORF is located at the 5′ end of the viral mRNA, whereas the pol ORF is 3′ of the HIV-1 PRF element and out-of-frame with the gag ORF (Fig. 1). Therefore, the enzymatic protein products of the pol gene are only translated through a −1 PRF event (8–10). Importantly, the frequency of the PRF is controlled precisely via the interaction between viral RNA cis-acting elements and host translation factors. The frequency of the PRF in HIV-1 is ∼5% of translational events (9, 11–13) and is essential for the morphogenesis and maturation of retrovirus particles because fluctuations in the Gag/Gag-Pol ratio are deleterious to virus replication (14–16). −1 PRF occurs in all viruses via a similar mechanism (17, 18). Two basic RNA elements have been identified which are absolutely required to generate and regulate PRF: a slippery site and a segment of RNA secondary structure called the frameshift stimulatory signal (11, 17, 19, 20) (Fig. 1). The slippery site consists of a stretch of seven nucleotides that do not have a uniform sequence but span three amino acid codons and conform to the heptameric sequence X XXY YYZ (the gag ORF is indicated by spaces) where X is any nucleotide, Y is an A or U, and Z is A, U, or C (21–24). The slippery sequence of HIV-1 is U UUU UUA. The PRF-stimulatory signal of HIV-1 is contained within a 69-base segment (11) which forms an extended stem-loop and helps regulate the probability of the PRF event (9, 11, 19, 20, 25–29). This contrasts with most other viral systems containing −1 frameshift signals as they have been reported to form pseudoknots 3′ to the slippery site.
FIGURE 1.
HIV-1 genome and the frameshifting region. The HIV-1 genome is shown with an expanded view of the gag-pol transframe region, which includes the elements important for ribosomal frameshifting. The frameshift signal is composed of the heptameric slippery site and an RNA stem-loop structure. LTR, long terminal repeat.
The mechanism of PRF is hypothesized to occur at least in part because a translocating ribosome pauses over a slippery site sequence as a consequence of a 3′ RNA structure (30). The RNA secondary structure is thought to promote a torsional stress that leads to the ribosome shifting one nucleotide in the −1 direction. Following the shift in reading frame, the ribosome continues translation in the new ORF to produce the p160Gag-Pol polyprotein of HIV-1. In the absence of PRF, the elongating ribosomes maintain their reading frame and encounter the gag gene stop codon resulting in the production of the p55Gag polyprotein. The rules governing PRF appear to be universal in eukaryotes because it has been shown that the PRF signals from mammalian viruses function in yeast cells (8, 10, 31).
The model described above suggests that alteration of PRF efficiency will result when certain aspects of the translation elongation process are modified. We hypothesize that a subset of translation factors involved in translational fidelity, mRNA-rRNA interactions, or rRNA-tRNA interactions can have a role in regulating PRF. Such factors may include elongation factors, nonsense-mediated decay proteins, translation termination factors, ribosomal proteins, as well as factors that modulate the activity of these processes such as kinases or phosphatases. In addition, RNA-binding proteins or factors that can remodel RNA structures (ATPases/helicases) might also affect PRF.
Eukaryotic release factor 1 (eRF1) was originally identified in a screen in the yeast Saccharomyces cerevisiae and was identified as a suppressor of nonsense mutations and designated SUP45 (32). Subsequent genetic and biochemical analyses demonstrated that SUP45 was a translational release factor which then also became known as eRF1 (32). eRF1 adopts a tertiary structure mimicking tRNAs (33) allowing it to recognize all three stop codons when they enter the ribosomal A site. eRF1 then mediates peptidyl-tRNA hydrolysis at the peptidyl transferase center of the ribosome to terminate translation. Mutation of eRF1 in yeast has been previously reported to promote stop codon readthrough and to increase the frequency of +1 frameshift suppression (32).
A recent genome-wide screen for cellular factors involved in the HIV-1 lifecycle found that “knockdown” of eRF1 inhibited HIV-1 replication (34). Because of eRF1 function in translation, we were interested in examining whether eRF1 played a role in modulating HIV-1 PRF. In this report we demonstrate that depletion of eRF1 increases PRF in HIV-1. This phenomenon was caused by eRF1 depletion directly and not by another factor affected by eRF1 depletion. Moreover, the finding was not dependent on ribosomal pausing at the slippery site, and it occurred independently of a stop codon proximal to the slippery site. This result identifies for the first time a cellular factor that is important for HIV-1 PRF, suggests an additional role for eRF1 in translation control and indicates that it is important to identify other factors that modulate HIV-1 PRF to exploit this essential step in HIV-1 replication for therapeutic intervention.
EXPERIMENTAL PROCEDURES
Plasmids
The backbone of the dual luciferase construct was taken from p2Luci (35), which harbors the Renilla luciferase gene followed by the firefly luciferase gene in the 0 translational reading frame. To make EK189 containing the yeast PGK1 promoter driving dual luciferase expression along with the corresponding 3′ untranslated region of the PGK1 gene, the dual luciferase markers were PCR-amplified from p2Luci and cloned into the BamHI-SalI-digested backbone of EK135, for which the map will be supplied upon request. The 80-bp HIV-1 PRF signal from the HXB2 strain (11) was PCR-amplified and inserted into the BglII–BamHI site of EK189 producing EK190 (slip). In similar fashion, modified versions of the PRF signal, including one with an extra T in the slippery site and another with a termination codon between the Renilla luciferase gene and the PRF slippery site, were inserted into EK189 resulting in EK191 (PC) and EK193 (NC), respectively.
Plasmids used for in vitro transcription/translation assays were made by inserting PCR-amplified dual luciferase region of EK190 (slip), EK191 (PC), or EK193 (NC) into the XhoI–SacII site of pRS415 to produce dual luciferase mRNA under control of the T7 promoter and poly(A) sequence. Plasmids used for human tissue culture experiments were made by inserting HIV-1 HXB2 frameshift region into SalI–SacI site of p2Luc (35). Plasmid harboring mutated human eRF1 G183R (36) was made by PCR, and the fragment was inserted into ApaI–PstI site of pRS415.
Backbone of plasmids used for the Western blot analysis was made by inserting partial glutathione S-transferase sequence, frameshift region, hemagglutinin (HA) tag, and 3×FLAG tag made from annealing of two oligoucleotides or PCR, into pYD1 (Invitrogen). The resulting sequence between partial glutathione S-transferase and 3×FLAG were amplified by PCR and inserted into SpeI–EcoRI site of p414-GPD (37).
Yeast Strains
Yeast strains harboring the plasmid encoding temperature-sensitive Sup45p/eRF1 (I222S) (38) or its wild type counterpart were kindly provided by Dr. Tobias von der Haar and Dr. Gloria Merritt.
RNA Interference
A siRNA pool composed of three siRNAs against eRF1 (Ambion), s4838, s4839, and s4840 was employed to transfect HeLa cells using Lipofectamine® 2000 (Invitrogen) according to the manufacturer's protocol. The final concentration of total siRNA was 50 nm. siRNA plus 0.2 μg of vector plasmid were cotransfected into HeLa cells. Seventy-two hours after transfection, the cells were collected, and RNA and protein were extracted and analyzed by real-time PCR and Western blotting for RNA and protein quantification. Dual luciferase activities of HeLa cells were assayed using the Promega Dual Luciferase® Reporter Assay system.
Real-time PCR
Total RNA was isolated from HeLa cells transfected with siRNA and/or vector and was purified using RNAqueous® phenol-free total RNA isolation kit (Ambion), supplemented with RQ1 RNase-free DNase (Promega). The absence of DNA contamination was confirmed by a control excluding reverse transcriptase (RT). cDNA was obtained using Applied Biosystems TaqMan® reverse transcription Reagents (Applied Biosystems), and aliquots were subjected to amplification using an IQTM SYBR Green kit (Bio-Rad). Each PCR was carried out as follows: preheat 15 min at 95 °C, then 40 cycles of 20 s at 95 °C, 30 s at 60 °C, followed by an extension at 72 °C for 2 min. Using the comparative Ct method, gene expression was calculated, and glyceraldehyde-3-phosphate dehydrogenase was used as a control gene. HeLa cells treated only with vector control were set as 100%, and the fold change in expression in siRNA and vector-treated HeLa cells were represented as bar graphs.
In Vitro Transcription/Translation Assay
In vitro transcription/translation assays were performed using TNT® Quick Coupled Transcription/Translation Systems (Promega), according to the manufacturer's protocol. The mutant human eRF1 expression vector was added to rabbit reticulocyte lysate with the amounts noted in the figures. Following incubation at 30 °C for 5 min, 1 μg of the dual luciferase construct harboring HIV-1 RFS elements or its variants were added. After additional incubation at 30 °C for one h, Renilla and firefly luciferase activities were determined with the Dual Luciferase® Reporter Assay system.
Determination of the PRF Ratio for Replication-competent Virus
Six micrograms of plasmid pNL4-3 was transfected into 293T cells in a 10-cm culture plate using FuGENE 6® HD transfection reagent (Roche Applied Science). After incubation at 37 °C for 48 h, virus supernatant was collected, filtered with a 0.45-μm filter, and used to inoculate HeLaT4 cell cultures in 6-well plates, which had been pretreated with siRNA for 12 h. Virus was harvested from the infected HeLaT4 cells after incubation at 37 °C for 48 h followed by analysis employing p24 ELISA and RT assays.
RT assays were performed following Willey et al. (39), except the incubation period was reduced to 45 min. p24 assays were carried out using RETRO-TEK HIV-1 p24 Antigen ELISA kit (Zeptametrics).
Toeprinting Assay
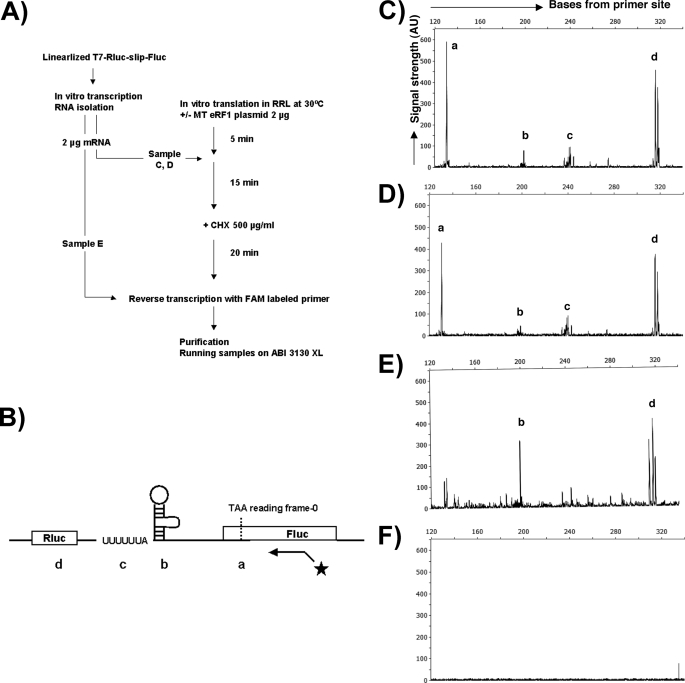
The general procedure followed was gleaned from Gould et al. (40). Two micrograms of mRNA produced from a linearized dual luciferase construct using the RiboMAX Large Scale RNA Production system-T7 (Promega) was added to rabbit reticulocyte lysates from TNT Quick-coupled Transcription/Translation System (Promega), with and without the plasmid encoding mutant human eRF1 (see Fig. 6A). After adding cycloheximide 15 min later and incubating at 30 °C for 20 min, mRNAs with attached ribosomes were reverse transcribed with a carboxyfluorescein (FAM)-labeled primer using Superscript III RT (Invitrogen). cDNA analysis was performed using an ABI 3130 XL DNA sequencer in conjunction with GeneMapper software (ABI).
FIGURE 6.
Toeprinting indicates that mutant eRF1 does not promote ribosomal stacking at the PRF signal. A, flowchart of toeprinting assay. mRNAs transcribed from the linearized dual luc HIV-1 construct in vitro were added to rabbit reticulocyte lysates with or without the mutant eRF1 plasmid. After adding cycloheximide to arrest the ribosomes, mRNAs were reverse transcribed with a FAM-labeled primer downstream of the PRF signal. B, schematic representation of where peaks a, b, c, and d from panels C–E map including location of the primer. The starred line with arrow represents the binding position of the FAM-labeled primer. Peaks a, b, and c correspond to pausing at the in-frame stop codon, the stem and loop, and the slippery site, respectively. Peak d maps to a likely secondary structure in rluc. C, fragment analysis data from the sample with mutant eRF1. D, sample without mutant eRF1 plasmid. E, sample without incubation in rabbit reticulocyte lysate. F, no-mRNA control. RRL, rabbit reticulocyte lysate; MT, mutant; CHX, cycloheximide; AU, arbitrary unit.
Dual Luciferase Assay of Yeast Strain Harboring Mutant eRF1
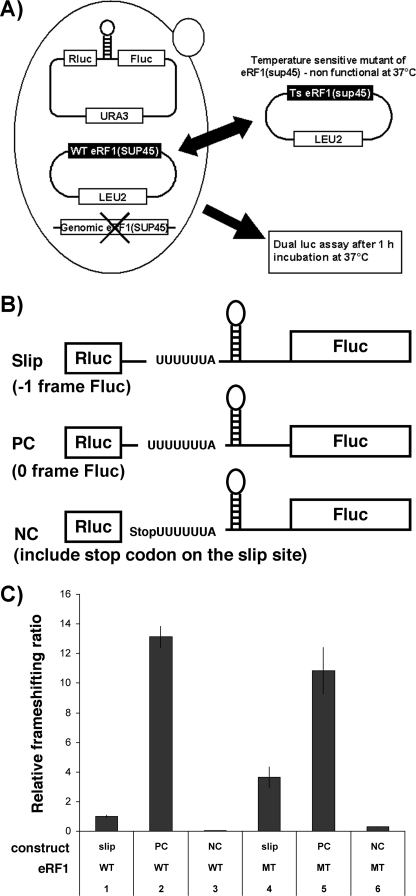
At least three independent, 2-ml liquid cultures of yeast strains harboring the Slip construct and its variant (see Fig. 4), and the wild type or mutant eRF1 genes were incubated at 27 °C until mid log phase. After switching the temperature to 37 °C for 1 h, 250 μl of the cultures were chilled with iced water, spun down, and resuspended into 250 μl of Passive Lysis buffer from the Promega Dual Luciferase® Reporter Assay system. The samples were freeze-thawed three times using liquid nitrogen and water baths to break cells and 10 μl of the cell lysates were used for dual luciferase assay using the Promega kit mentioned above.
FIGURE 4.
Effects of the temperature-sensitive mutant allele of eRF1 (Sup45 I222S) on PRF in yeast cells. A, yeast strain and strategy used for analyzing the effect of eRF1 mutation on the HIV-1 PRF. Yeast cells, from which endogenous eRF1 was deleted, were transformed with a plasmid harboring either wild type eRF1 or temperature-sensitive eRF1 alleles and various dual luciferase constructs. After a 1-h incubation at 37 °C to deactivate temperature-sensitive eRF1, fluc and rluc activities were determined and relative frameshifting ratios obtained. B, reporter constructs. Basic backbone of these constructs was made by inserting the HIV-1 PRF between the two luciferase genes with all of the constructs using the PGK1 promoter in the yeast pRS416 plasmid. Slip represents the reporter construct with fluc in the −1 reading frame relative to rluc. PC is a positive control vector in which rluc and fluc are in the same reading frame; NC represents a negative control in which a stop codon was inserted between rluc and fluc 5′ to the PRF signal. C, relative frameshifting ratio determined by dividing fluc activity by the rluc activity normalizing the samples to wild type eRF1 plus slip construct sample. Each value was average from three independent samples. WT, wild type; MT, mutant. Error bars, ±S.D.
Western Blot Analysis
Plasmids 0S-1, 00, and PC were introduced into yeast strains harboring plasmids encoding either the temperature-sensitive Sup45p/eRF1 (I222S) (38) or wild type Sup45p/eRF1. These yeast strains were switched to the nonpermissive temperature as described previously. Protein extracts were prepared followed by electrophoresis in a 15% SDS-polyacrylamide gel with Tris-glycine buffer according to Kushnirov (41).
RESULTS
Depletion of eRF1 Increases HIV-1 PRF
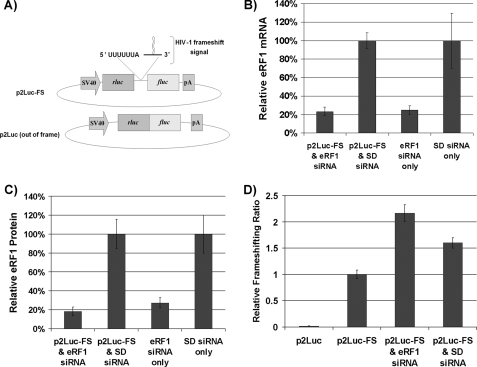
Recently, genome-wide screening utilizing RNA interference technology was employed to identify cellular factors involved in HIV-1 replication. We hypothesized that genes involved in translation identified in these screens might affect HIV-1 PRF. To test this hypothesis, seven genes (eRF1, SSB, EIF2C3, EIF3H, DMIT1L, PIGH, and PIGY) identified by Brass et al. (34) were examined to determine whether they played a role in HIV-1 PRF. The results demonstrated that knockdown of only one gene, eRF1, altered the PRF ratio. Initially, a loss-of-function analysis was performed using RNA interference (Fig. 2). HeLa cells were first treated with siRNAs for 72 h. Each gene was targeted with three different siRNAs simultaneously at a total concentration of 50 nm. The siRNAs targeting eRF1 knocked down the mRNA to approximately 20% of control levels with a corresponding depletion of eRF1 protein to ∼20% of the mock control (Fig. 2, B and C). The scrambled duplex negative control did not result in depletion of eRF1 mRNA or protein (Fig. 2, B and C). Next, HeLa cells were transfected simultaneously with siRNA and dual luciferase reporter constructs, p2Luc or p2Luc-FS, which contain the Renilla luciferase gene (rluc) in the 5′ position and the firefly luciferase gene (fluc) in the −1 frame in the 3′ position (Fig. 2). p2Luc-FS contains the HIV-1 PRF signal placed between the luciferase genes, whereas p2Luc serves as a negative control yielding a background frameshifting ratio. Renilla and firefly luciferase activities were measured 72 h after transfection, and the fluc/rluc ratio was calculated and normalized to the control ratio obtained with p2Luc-FS in mock siRNA-treated cells (Fig. 2). There was approximately a 2-fold increase in PRF in cells when eRF1 was depleted (Fig. 2D). A 2-fold increase in PRF strongly inhibits HIV-1 replication (42). These results suggest that eRF1 is involved in regulating HIV-1 PRF and that further investigation of eRF1 modulation of HIV-1 PRF was warranted.
FIGURE 2.
Depletion of eRF1 protein increases PRF in HeLa cells. HeLa cells harboring construct p2Luc-FS or p2Luc were treated with 50 nm siRNA, knocking down eRF1 expression. SD represents a scrambled duplex negative control. A, schematic representation of p2Luc-FS and p2Luc. p2Luc-FS contains the firefly luciferase gene (fluc) and the Renilla luciferase gene (rluc) separated by the HIV-1 PRF signal with fluc being in the −1 reading frame relative to rluc. p2Luc represents a negative control in which rluc and fluc are not separated by the PRF signal while fluc is still maintained in the −1 reading frame. B, eRF1 mRNA levels resulting from the indicated treatments measured via quantitative real-time PCR. C, eRF1 protein expression levels monitored via Western blotting with relative levels being quantified using a PhosphorImager (Molecular Dynamics). D, relative frameshift ratios were determined by measuring the fluc to rluc activities and normalizing to the values obtained from mock treated cells containing p2Luc-FS. This experiment was done in triplicate. Error bars, ±S.D.
eRF1 Knockdown Increases the Pol/Gag Ratio of Replication-competent Virus
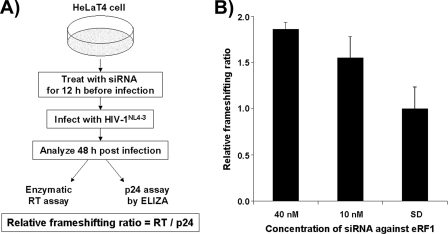
We next asked whether eRF1 depletion altered the Pol/Gag ratio in a live virus assay using the HIV-1NL4-3 strain. HeLaT4 cells were transfected with siRNA targeting eRF1 mRNA for 12 h followed by infection with HIV-1NL4-3 (Fig. 3). Forty-eight hours after infection, viral supernatant was harvested followed by measuring p24Gag levels via ELISA and by assaying for RT activity utilizing a standard enzymatic assay (39). It was found that the RT/p24Gag ratio increased by almost 2-fold in cells treated with eRF1 siRNAs at a concentration of 40 nm (Fig. 3). This result is in good agreement with those obtained using the dual luciferase model system (Fig. 2).
FIGURE 3.
Depletion of eRF1 protein promotes PRF of replication-competent virus. A, strategy for analyzing the effect of eRF1 knockdown on PRF of replication-competent HIV-1 in HeLaT4 cells. Replication-competent HIV-1NL4-3 was used to infect HeLaT4 cells in which eRF1 was knocked down with eRF1-specific siRNAs beforehand. Forty-eight hours after infection, supernatant was harvested from infected cells, and RT levels (for pol gene expression) were obtained by measuring enzymatic activity and normalizing to known amounts of RT protein while p24 levels (for gag gene expression) were assessed via ELISA. Relative frameshifting ratios were determined by dividing the RT concentrations by the p24 concentrations, normalizing to control samples treated with scrambled duplex (SD). B, relative frameshifting ratios obtained. Each value was averaged from three independent samples. Error bars, ±S.D.
Mutant eRF1 Increases HIV-1 PRF in Yeast
The results described above showed that the depleting eRF1 with low concentrations of siRNAs altered the PRF ratio whereas scrambled duplex controls had no effect upon PRF. It is possible, however, that these observations were due to “off-target” siRNA effects. To rule out this possibility, we examined whether inhibition of eRF1 activity enhances PRF in another system. We chose to utilize the yeast S. cerevisiae as a model system for further testing because it has been reported that the HIV-1 PRF signal could direct frameshifting in this system (10). A yeast strain was employed in which the endogenous eRF1 allele was inactivated and replaced with a plasmid expressing a temperature-sensitive mutant of the eRF1 gene (I222S) (38) (Fig. 4). The efficiency of PRF was determined using dual luciferase reporter constructs (Fig. 4). To deplete eRF1, the temperature-sensitive mutant was expressed in yeast cells, and 1 h before monitoring luciferase activities the temperature was changed from the permissive temperature, 27 °C, to the nonpermissive temperature, 37 °C. When the reporter harbored the HIV-1 PRF signal and fluc in the −1 frame, PRF increased more than 3-fold in the presence of mutant eRF1, whereas no significant change in the ratio was found using control plasmids PC (with rluc and fluc in the 0 reading frame) and NC (with a stop codon inserted between rluc and the slippery site) (Fig. 4C). These results clearly indicate that depletion of eRF1 observed above did not result from off-target siRNA effects and that the effect is observable in diverse biological systems. This result is consistent with our previous results indicating the eRF1 is important in modulating HIV −1 PRF and establishes a genetic system to investigate the universality of this phenomenon.
Transdominant Mutant of eRF1 Enhances PRF in Rabbit Reticulocyte Lysate
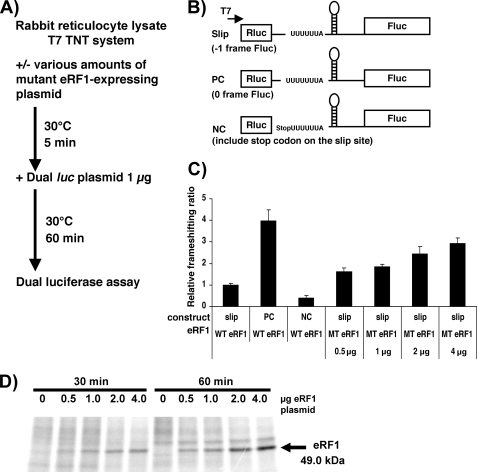
We also tested whether the effects upon PRF could be reproduced in rabbit reticulocyte lysates. The goal of this experiment was to determine whether the increase in frameshifting is directly due to eRF1 depletion rather than other genes whose expression is affected by eRF1 depletion. The advantage of using the TNT rabbit reticulocyte lysates (Promega) is that it is an in vitro translation assay that was treated with micrococcal nuclease, so expression of genetic information is tightly controlled by what plasmids are added to the reaction. A trans-dominant mutant of the human eRF1 gene, G183R, was used to repress endogenous eRF1 function (36). The G183R mutation is at the first glycine of the “GGQ motif ” in the M domain of eRF1 and is essential for hydrolysis of peptidyl-tRNA in the ribosome. Dual luciferase reporter constructs were again employed, including the HIV PRF (slip), PC, and NC variants as described in the previous section. Increasing amounts of the mutant eRF1 plasmid were introduced into the transcription/translation-coupled rabbit reticulocyte lysates as described in Fig. 5. The results demonstrated a dose-dependent increase in the level of PRF, reaching an approximately 3-fold increase of frameshifting efficiency at the highest concentration of the mutant eRF1 plasmid. The dose-dependent increase in frameshifting also correlated with increased expression of a de novo synthesized protein corresponding in molecular mass with mutant eRF1 (Fig. 5D). These results are consistent with the results above indicating that eRF1 is directly modulating HIV-1 PRF.
FIGURE 5.
eRF1 has a direct effect on frameshifting in a cell-free transcription/translation system. A, flowchart of strategy. B, dual luciferase constructs used. Similar to the constructs in Fig. 4, three constructs were made: slip for measuring PRF site directed frameshifting, PC as a positive in-frame control, and NC as a negative control with a stop codon between the reporter genes. The T7 promoter was used to express RNAs in this system. C, relative frameshifting ratios determined by dividing the fluc activity by the rluc activity normalizing to lysates with the slip construct in the presence of endogenous active eRF1. Each value was the average of three independent samples. WT and MT stand for endogenous wild type eRF1 and mutant eRF1 plasmid, respectively. Error bars, ±S.D. D, de novo synthesized proteins labeled with [35S]methionine followed by SDS-PAGE and autoradiography. Numbers shown on top of the image correspond to the amount of the plasmid encoding mutant-eRF1 and incubation time after adding the plasmid to the lysate.
Enhanced PRF Is Not Correlated with Increased Ribosomal Pausing
Previously, −1 ribosomal frameshifting was shown to be stimulated by ribosomal pausing at the slippery site in both the yeast L1 killer virus system as well as in a novel model system (43, 44). One possibility for the increase of PRF when eRF1 is depleted is that the rate of translational termination was decreased resulting in stacking of ribosomes 5′ to the stop codon increasing ribosomal pausing at the slippery site. To test this hypothesis, a toeprinting assay was carried out (45, 46). In this assay, a labeled DNA primer is annealed to an mRNA followed by primer extension with RT. If RT encounters a factor or complex bound to the mRNA at a specific location, the primer extension will be inhibited, and the position of the factor or complex can be ascertained from the size of the extended DNA product after resolution in a DNA sequencing gel. mRNA containing the HIV-1 PRF signal was produced using the RiboMAX large scale RNA production system-T7 and added to rabbit reticulocyte lysates (Promega) in the presence or absence of the trans-dominant eRF1 mutant human allele. Cycloheximide was subsequently added to the reaction to inhibit translation elongation and arrest ribosomes on their mRNA template (Fig. 6). A FAM-labeled DNA primer was then annealed to the mRNAs containing the PRF signal followed by DNA elongation with RT. The labeled DNA products were then analyzed using an ABI 3130 XL DNA sequencer.
The results demonstrated that in the absence of eRF1 mutant protein, four peaks were observed within 120–320 bases from the primer. Peak a corresponds to the TAA termination codon which was anticipated to reveal paused terminating ribosomes (Fig. 6). Peak b corresponds to the 3′ side of the PRF stem-loop also expected because the stem-loop should perturb RT elongation. Peak c corresponds to ribosomal pausing at the PRF slippery site due to the ribosome encountering the stem-loop. Peak d was an unexpected peak that mapped to sequence within the rluc gene. We do not know why this sequence caused a peak, although in the control sample without rabbit reticulocyte lysates it is still evident along with the peak at the 3′ side of the stem-loop, indicating that it involves RNA secondary structure. When mutant eRF1 was added to the system, it did not appreciably change the toeprint. As anticipated, none of the RT elongation arrest peaks is visible in the no-RNA control. Taken together, these results suggests that the stimulation of PRF by eRF1 depletion is not due to increased pausing of ribosomes at the termination signal stacking ribosomes behind it and increasing the pause rate over the slippery site.
−1 Frameshifting Does Not Occur at a Stop Codon Proximal to the Slippery Site upon eRF1 Depletion
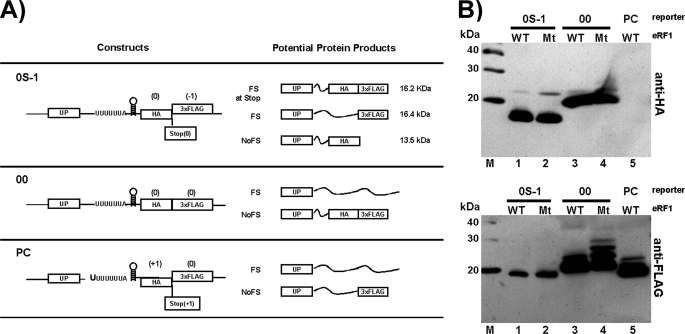
Another potential explanation for the effects observed above is that eRF1 depletion promotes −1 frameshifting at a stop codon proximal to the PRF signal. A previous report suggested that +1 frameshifting might occur at a stop codon in eRF1-depleted cells (32). To address this possibility, a frameshift construct harboring a reporter in which different epitopes were positioned to distinguish between frameshifting at the PRF versus at the stop codon. Construct 0S-1 contains a stop codon between the HA and 3×FLAG epitopes: the HA epitope is in the 0 reading frame and the 3×FLAG in the −1 reading frame (Fig. 7). 0S-1 was introduced into yeast strains harboring mutant or wild type eRF1 and the protein produced was monitored by Western blot analysis. If frameshifting occurs at the termination codon, then a 16.2-kDa fusion peptide harboring both the HA and 3×FLAG tags would be observed on blots probed with both anti-HA and anti-FLAG antibodies. In contrast, if frameshifting occurs at the slippery site of the PRF signal, a 16.4 kDa band would be observed in the anti-FLAG blot but not in the anti-HA blot (Fig. 7). Protein products produced without frameshifting at either the PRF signal or the stop codon will yield a 13.5-kDa product that can be visualized in the anti-HA blot but not the anti-FLAG blot.
FIGURE 7.
Depletion of eRF1 does not cause −1 frameshifting at a 0-frame stop codon proximal to the frameshift signal. A, constructs used in this assay encode HA and 3×FLAG epitopes 3′ to the HIV-1 PRF signal. The numbers −1, 0, and +1 above HA and 3×FLAG indicate their reading frame. Positions of a stop codon between HA and 3×FLAG tags are indicated for two constructs including their reading frame in parentheses. UP refers to the coding sequence upstream of the PRF signal and is a fusion between a portion of glutathione S-transferase and the Xpress epitope. The potential protein products obtained from the constructs in the event of frameshifting at a stop codon between HA and 3×FLAG (FS at Stop), frameshifting at the HIV-1 PRF (FS), and no frameshifting (NoFS) are indicated. Thin wavy lines represent peptides not encoding an epitope marker. It should be noted that construct PC was made by inserting an extra U at the slippery site. B, constructs were introduced into either the yeast wild type strain or the eRF1 mutant strain and cultured overnight at 27 °C followed by switching to 37 °C for 1 h and subsequent extraction of samples. The samples were then subjected to electrophoresis in a 15% polyacrylamide gel followed by transfer to polyvinylidene difluoride filters. The blots were then probed with either anti-HA or anti-FLAG antibodies as described under “Experimental Procedures.” WT and Mt stand for wild type and mutant, respectively. M stands for protein markers.
The results are consistent with frameshifting at the slippery site within the PRF signal and not at the stop codon for both wild type and mutant eRF1 (Fig. 7). Both the wild type and mutant eRF1 samples produced a 16.4 kDa band in the anti-FLAG blot, and these bands display the same mobility as the predominant band of the PC wild type control, which was designed as a control to produce the same 16.4-kDa protein as its major product (Fig. 7, anti-FLAG blot, lanes 1, 2, and 5). In addition, a 16.2-kDa protein, which would result from frameshifting at the stop codon and be detectable in the anti-HA blot, is not produced by 0S-1 in either the presence or absence of mutant eRF1 as evidenced by the absence of a 16.2 kDa band in the 0S-1 lanes which would have exactly the same mobility as the predominant band for the 00 controls (Fig. 7, anti-HA blot, lanes 1-4). The predominant proteins produced by 0S-1 and detected in the anti-HA blot are consistent with the 13.5-kDa protein product which should be produced when frameshifting does not occur. There are minor bands in lanes 1 and 2 of the anti-HA blot running at ∼22 kDa, suggesting that there is some translational readthrough downstream of the 3×FLAG epitope. It should be noted that the primary bands for control 00 (Fig. 7, lanes 3 and 4 of anti-FLAG blot) run a bit slower than the 0S-1 bands in the anti-FLAG blot (Fig. 7, lanes 1 and 2 of the anti-FLAG blot); this is likely due to the amino acid content being different at the decoded HA section of the gene products because in the case of 00, HA is in the 0 reading frame whereas in the case of 0S-1, HA is in the −1 reading frame so the amino acid content is different. Also, it should be pointed out that there are other bands running slower than the major bands in lanes 3 and 4 of the anti-FLAG blot (Fig. 7), again probably due to stop codon readthrough downstream of the 3×FLAG epitope which is exacerbated upon inhibition of eRF1. Taken together, the anti-HA and anti-FLAG blots are consistent with −1 frameshifting being enhanced at the HIV-1 slippery site upon eRF1 knockdown and not at a proximal termination codon (Fig. 7).
DISCUSSION
The results described here indicate that the translation termination factor eRF1 plays a direct role in HIV PRF based upon four lines of evidence. First, employing a dual luciferase reporter separated by the HIV-1 PRF signal, we showed that depletion of eRF1 in mammalian cells using RNA interference knockdown resulted in an ∼2-fold increase in PRF (Fig. 2). Second, RNA interference knockdown of eRF1 in cells infected with replication-competent HIV-1 increased the Gag-Pol/Gag ratio also about 2-fold as assessed by ELISA to monitor p24Gag expression and enzymatic activity to measure RT levels (Fig. 3). Third, when yeast cells harboring a conditional mutant of eRF1 were shifted to a nonpermissive temperature, −1 frameshifting was enhanced by a similar magnitude as above suggesting that the earlier findings were not due to off-target effects of siRNA and that the effect is global in nature (Fig. 4). Fourth, when a trans-dominant mutant of eRF1 was added to rabbit reticulocyte lysates, there was a dose-dependent increase in PRF, providing a fourth confirmation of the effect and implying that the effect is directly due to eRF1 inhibition and not due to expression of another gene product because of eRF1 depletion (Fig. 5). Interestingly, the increase in the frequency of PRF was not dependent upon a proximal termination codon suggesting that frameshifting was not being modulated at a stop codon proximal to the PRF signal (Fig. 7). Thus, eRF1 is the first protein factor identified that has a direct role in modulating HIV-1 PRF.
The change in PRF resulting from reduction of eRF1 may appear on the surface to be modest, However, the 2–3-fold change in frameshifting efficiency was shown to be well within the range that would lead to inhibition of viral replication. Previous results have demonstrated that a 2-fold change in HIV-1 PRF dramatically inhibits HIV-1 replication (14–16). Therefore, the inhibition of virus replication resulting from the knockdown eRF1 expression observed by Brass et al. (34) taken with the results presented here suggest that this inhibition is at least in part due to an alteration in the Gag/Pol ratio.
Several lines of evidence are consistent with the notion that eRF1 is a factor involved in PRF. For example, in Escherichia coli, Horsfield et al. (47) placed a slippery sequence UUU UUU A between marker genes and found that overexpression of defective RF2, an E. coli termination factor, increased the frameshifting ratio. Furthermore, Park et al. (48) demonstrated that a [PSI+] yeast strain, in which its phenotype is caused by mutation of eRF3, showed increased −1 frameshifting at a UUU UUU A slippery site. In both publications, the reporter constructs did not contain a defined downstream stimulatory signal as in the case of the stem-loop within the HIV-1 PRF signal, and a proximal stop codon was placed ∼9 bases downstream of the slippery site. In this report, the constructs contained the entire HIV-1 PRF signal, including the slippery site as well as the downstream stimulatory signal. Thus, the results obtained here are more broadly applicable to HIV-1 gene expression. Furthermore, the in vitro translation assays presented here indicate that the phenomenon is a direct effect of eRF1 depletion.
It has been reported that ribosomal pausing over the slippery site is a key factor influencing −1 frameshifting (43, 44). Thus, we utilized toeprinting to examine if inhibiting eRF1 resulted in increased ribosomal pausing at the slippery site. The toeprint indicated that ribosomal pausing at the slippery site was not augmented in the presence of a trans-dominant eRF1 mutant in rabbit reticulocyte lysates (Fig. 6). Moreover, enhanced frameshifting was independent of a proximal termination codon, suggesting that there is another mechanism that accounts for these observations. It is possible that eRF1 forms a yet to be identified complex with other factors, and this complex can augment −1 frameshifting. There is ample evidence that eRF1 binds to other factors. Multiple methods indicate that eRF1 has at least 32 cellular binding partners (Saccharomyces Genome Data base). eRF1 might also interact with HIV-1 viral proteins because it was previously reported that eRF1 binds to Moloney murine leukemia virus RT and plays a role in suppression of the termination codon separating the gag and pol genes in Moloney murine leukemia virus (49). Thus, it will be interesting to determine whether eRF1 binds to any HIV-1 viral proteins or whether down-regulation of already identified binding partners also perturbs HIV-1 PRF. In any event, the work described is the first example of a cellular factor which when depleted promotes HIV-1 PRF, and it suggests that it should be possible to identify other factors affecting this essential aspect of HIV-1 replication. Thus, these findings highlight that HIV-1 PRF is a target that can be exploited in the future for antiviral drug development.
Acknowledgments
We thank Drs. Tobias von der Haar and Gloria Merritt for supplying the yeast strain harboring the sup45/eRF1 (I222S) plasmid and Dr. Leonard Edelstein and Leia Miller for critical reading of the manuscript as well as laboratory members for helpful suggestions. We thank Dr. Lee Ann Schein and Regina Gibbions for assistance in carrying out DNA fragment analysis for toeprinting and Dr. Victor Stollar for allowing extensive utilization of equipment in his laboratory.
This work was supported by National Institutes of Health Grant P01AI057596.
- ORF
- open reading frame
- PRF
- programmed-1 ribosomal frameshifting
- HIV-1
- human immunodeficiency virus type 1
- eRF1
- eukaryotic release factor 1
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- ELISA
- enzyme-linked immunosorbent assay
- RT
- reverse transcriptase
- FAM
- carboxyfluorescein.
REFERENCES
- 1.Hayes S., Bull H. J. (1999) Acta Biochim. Pol. 46, 879–884 [PubMed] [Google Scholar]
- 2.Hung M., Patel P., Davis S., Green S. R. (1998) J. Virol. 72, 4819–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shigemoto K., Brennan J., Walls E., Watson C. J., Stott D., Rigby P. W., Reith A. D. (2001) Nucleic Acids Res. 29, 4079–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan M., Liang A., Brünen-Nieweler C., Heckmann K. (2001) J. Eukaryot. Microbiol. 48, 575–582 [DOI] [PubMed] [Google Scholar]
- 5.Vimaladithan A., Farabaugh P. J. (1998) Methods Mol. Biol. 77, 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed E. O. (2001) Somatic Cell Mol. Genet. 26, 13–33 [DOI] [PubMed] [Google Scholar]
- 7.Frankel A. D., Young J. A. (1998) Annu. Rev. Biochem. 67, 1–25 [DOI] [PubMed] [Google Scholar]
- 8.Bidou L., Stahl G., Grima B., Liu H., Cassan M., Rousset J. P. (1997) RNA 3, 1153–1158 [PMC free article] [PubMed] [Google Scholar]
- 9.Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. (1988) Nature 331, 280–283 [DOI] [PubMed] [Google Scholar]
- 10.Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. (1988) Cell 55, 1159–1169 [DOI] [PubMed] [Google Scholar]
- 11.Dulude D., Baril M., Brakier-Gingras L. (2002) Nucleic Acids Res. 30, 5094–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kollmus H., Honigman A., Panet A., Hauser H. (1994) J. Virol. 68, 6087–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulus C., Ludwig C., Wagner R. (2004) Virology 330, 271–283 [DOI] [PubMed] [Google Scholar]
- 14.Park J., Morrow C. D. (1991) J. Virol. 65, 5111–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shehu-Xhilaga M., Crowe S. M., Mak J. (2001) J. Virol. 75, 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein K. M., Goff S. P. (1988) J. Virol. 62, 2179–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I. (1995) J. Gen. Virol. 76, 1885–1892 [DOI] [PubMed] [Google Scholar]
- 18.Jacks T. (1990) Curr. Top. Microbiol. Immunol. 157, 93–124 [DOI] [PubMed] [Google Scholar]
- 19.Baril M., Dulude D., Gendron K., Lemay G., Brakier-Gingras L. (2003) RNA 9, 1246–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baril M., Dulude D., Steinberg S. V., Brakier-Gingras L. (2003) J. Mol. Biol. 331, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brierley I., Jenner A. J., Inglis S. C. (1992) J. Mol. Biol. 227, 463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinman J. D., Icho T., Wickner R. B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinman J. D., Wickner R. B. (1992) J. Virol. 66, 3669–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. (1988) Cell 55, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudin C., Mazauric M. H., Traïkia M., Guittet E., Yoshizawa S., Fourmy D. (2005) J. Mol. Biol. 349, 1024–1035 [DOI] [PubMed] [Google Scholar]
- 26.Morikawa S., Bishop D. H. (1992) Virology 186, 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staple D. W., Butcher S. E. (2003) Nucleic Acids Res. 31, 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staple D. W., Butcher S. E. (2005) J. Mol. Biol. 349, 1011–1023 [DOI] [PubMed] [Google Scholar]
- 29.Telenti A., Martinez R., Munoz M., Bleiber G., Greub G., Sanglard D., Peters S. (2002) J. Virol. 76, 7868–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinman J. D., Berry M. J. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 625–654, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31.Stahl G., Bidou L., Rousset J. P., Cassan M. (1995) Nucleic Acids Res. 23, 1557–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inge-Vechtomov S., Zhouravleva G., Philippe M. (2003) Biol. Cell 95, 195–209 [DOI] [PubMed] [Google Scholar]
- 33.Ito K., Ebihara K., Uno M., Nakamura Y. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 35.Grentzmann G., Ingram J. A., Kelly P. J., Gesteland R. F., Atkins J. F. (1998) RNA 4, 479–486 [PMC free article] [PubMed] [Google Scholar]
- 36.Frolova L. Y., Tsivkovskii R. Y., Sivolobova G. F., Oparina N. Y., Serpinsky O. I., Blinov V. M., Tatkov S. I., Kisselev L. L. (1999) RNA 5, 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mumberg D., Müller R., Funk M. (1995) Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 38.Stansfield I., Kushnirov V. V., Jones K. M., Tuite M. F. (1997) Eur. J. Biochem. 245, 557–563 [DOI] [PubMed] [Google Scholar]
- 39.Willey R. L., Smith D. H., Lasky L. A., Theodore T. S., Earl P. L., Moss B., Capon D. J., Martin M. A. (1988) J. Virol. 62, 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould P. S., Bird H., Easton A. J. (2005) BioTechniques 38, 397–400 [DOI] [PubMed] [Google Scholar]
- 41.Kushnirov V. V. (2000) Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 42.Karacostas V., Wolffe E. J., Nagashima K., Gonda M. A., Moss B. (1993) Virology 193, 661–671 [DOI] [PubMed] [Google Scholar]
- 43.Tu C., Tzeng T. H., Bruenn J. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8636–8640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somogyi P., Jenner A. J., Brierley I., Inglis S. C. (1993) Mol. Cell. Biol. 13, 6931–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartz D., McPheeters D. S., Traut R., Gold L. (1988) Methods Enzymol. 164, 419–425 [DOI] [PubMed] [Google Scholar]
- 46.Sachs M. S., Wang Z., Gaba A., Fang P., Belk J., Ganesan R., Amrani N., Jacobson A. (2002) Methods 26, 105–114 [DOI] [PubMed] [Google Scholar]
- 47.Horsfield J. A., Wilson D. N., Mannering S. A., Adamski F. M., Tate W. P. (1995) Nucleic Acids Res. 23, 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park H. J., Park S. J., Oh D. B., Lee S., Kim Y. G. (2009) FEBS Lett. 583, 665–669 [DOI] [PubMed] [Google Scholar]
- 49.Orlova M., Yueh A., Leung J., Goff S. P. (2003) Cell 115, 319–331 [DOI] [PubMed] [Google Scholar]