Abstract
Prion diseases are associated with the misfolding of the endogenously expressed prion protein (designated PrPC) into an abnormal isoform (PrPSc) that has infectious properties. The hydrophobic domain of PrPC is highly conserved and contains a series of glycine residues that show perfect conservation among all species, strongly suggesting it has functional and evolutionary significance. These glycine residues appear to form repeats of the GXXXG protein-protein interaction motif (two glycines separated by any three residues); the retention of these residues is significant and presumably relates to the functionality of PrPC. Mutagenesis studies demonstrate that minor alterations to this highly conserved region of PrPC drastically affect the ability of cells to uptake and replicate prion infection in both cell and animal bioassay. The localization and processing of mutant PrPC are not affected, although in vitro and in vivo studies demonstrate that this region is not essential for interaction with PrPSc, suggesting these residues provide conformational flexibility. These data suggest that this region of PrPC is critical in the misfolding process and could serve as a novel, species-independent target for prion disease therapeutics.
Keywords: Amyloid, Lipid Raft, Neurodegeneration, Neurological Diseases, Prions, Protein Motifs, GXXXG Motif, Hydrophobic Domain, Transmissible Spongiform Encephalopathy
Introduction
Transmissible spongiform encephalopathies, also known as prion diseases, are a family of fatal neurodegenerative disorders affecting both humans and animals. In humans, they usually present as rapidly developing dementia in the 4th to 6th decade of life with neuropathology, including astrocytosis, spongiform degeneration, and deposition of amyloid plaques. According to the protein-only model of prion propagation, these diseases are associated with the conformational conversion of the host-encoded cellular prion protein (PrPC),6 into an abnormal pathogenic isoform (PrPSc) (1). Mouse PrPC contains a flexible N-terminal region, with C-terminal residues 122–231 forming a structured globular domain (2), whereas in PrPSc, the residues 90–230 form a structured, protease-resistant core (Fig. 1A) (3). The molecular mechanisms surrounding the conformational change from PrPC to PrPSc are unknown; but prion infection would require at the very least an interaction between PrPC and PrPSc, although the infectious particle is presumed to be more complex.
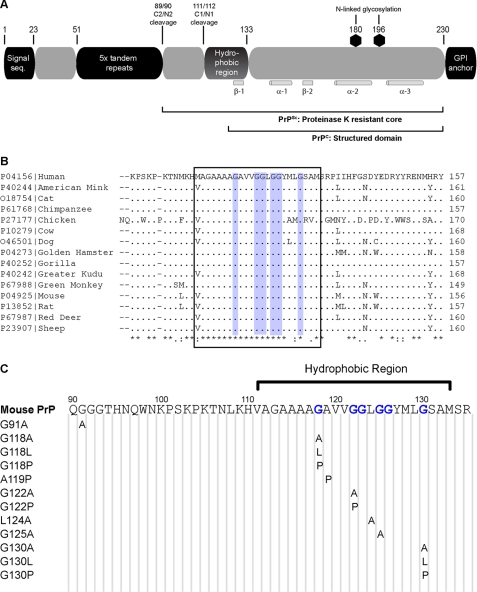
FIGURE 1.
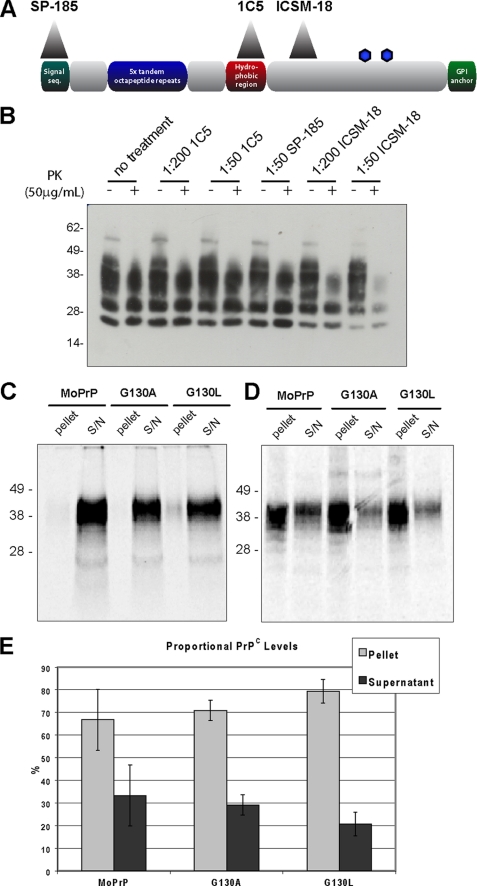
Hydrophobic region of PrP. A, overview of the prion protein and its important features. The extent to which the protein is structured in both PrPC and PrPSc is noted, as are the locations of the α-helix and β-strands in PrPC. GPI, glycosylphosphatidylinositol; seq, sequence. B, alignment of the hydrophobic region (boxed) across a sample of species with relevance to prion disease shows high levels of conservation, with perfect conservation of the glycine residues that include the GRR (blue). Alignment of PrP from all known species is available in supplemental Fig. S1. C, series of point mutations were produced, which changed several flexible glycine residues to less flexible alanine (G118A, G122A, G125A, and G130A), highly hydrophobic leucine (G118L and G130L), and structurally rigid proline (G118P, G122P, and G130P) residues. Non-glycine residues in the GRR were also mutated (A119P and L124A) as was a nonhydrophobic region amino acid (G91A).
The hydrophobic region of PrP, stretching from residues 111 to 133 in mouse PrP, may play a role in modulating prion toxicity and infectivity; synthetic peptides of this region interfere with prion propagation in vitro (4), whereas large deletions of this region prevent formation of protease-resistant PrP in cell culture models (5). The hydrophobic region is believed to undergo significant structural change following prion infection, as antibodies directed toward PrP(90–120) can detect PrPC but not PrPSc (6).
The hydrophobic region displays exceptionally high conservation across a wide range of species (Fig. 1B and supplemental Fig. S1) (7), implying that it plays a vital role in the endogenous function of PrPC, and it has recently been observed to contain several apparent repeats of a GXXXG motif (8). One of the most common motifs in transmembrane α-helices is the GXXXG motif, which is two glycines separated by any three residues, which acts by forming a shallow groove on one side of the helix and allowing close packing of transmembrane domains (9). GXXXG motifs are found as protein-protein interaction sites in a wide variety of proteins, including the region of the amyloid precursor protein that contains the Alzheimer disease-associated Aβ peptide (8) where mutation of these glycine residues alters the toxic potential of Aβ (10). GXXXG motifs are predominantly found in transmembrane regions of proteins; however, unlike amyloid precursor protein, the hydrophobic domain of PrP is only present as a transmembrane region in certain artificial systems of unknown physiological relevance (11). Furthermore, this region contains more glycine residues than required in a classical GXXXG motif. To allow for the possibility that this region is similar but functionally distinct to a GXXXG motif, we instead refer to the region as a glycine-rich region (GRR).
As the GRR contains a series of flexible residues within a region known to undergo a significant structural change in prion disease, from flexible disorder in PrPC to protease-resistant structure in PrPSc, we hypothesized that it may play a role in the process of prion infection. We created single amino acid substitutions to alter the hydrophobicity and flexibility of GRR residues and to demonstrate that even relatively conservative mutations within the GRR have a drastic effect on the propagation of prion infectivity, providing evidence that this highly conserved region of PrP is a dominant modulating factor on prion infectivity.
EXPERIMENTAL PROCEDURES
Sequence Alignments
Sequences of PrP from various species were retrieved from the Swiss Protein Database and aligned using ClustalW (European Bioinformatics Institute).
Chemicals
Unless specified otherwise, all chemicals were of molecular grade and obtained from Sigma.
Mutagenesis
The mouse PrP open reading frame was cloned into the pIRESpuro2 vector (Clontech) as described previously (12). Oligonucleotide primers (Sigma) (supplemental Table S1) were designed to introduce the single mutations detailed in Fig. 1C using QuikChange® site-directed mutagenesis kit (Stratagene).
Maintenance of Cultured Cell Lines
RK13 cells (13) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 5% fetal bovine serum, 1 mm glutamine, and 100 μg/ml penicillin/streptomycin. pIRES-MoPrP constructs were transfected with Lipofectamine 2000 (Invitrogen) and stable populations maintained in 2 μg/ml puromycin. Cell infections were performed as described previously (12) by incubating cells with 1% (w/v) M1000 infected mouse brain homogenate (14, 15) or 5% (v/v) infected RK13 cell lysate in OptiMEM + supplements and incubated 3 days, washed four times with phosphate-buffered saline (Amresco), and passaged as normal.
Confocal Microscopy
Transfected cells were plated in μ-slides (ibidi) to reach a final confluence of 40% after 2 days of incubation in standard tissue culture conditions. Cells were fixed with 3.2% paraformaldehyde and permeabilized in 0.2% Triton X-100 solution. Cells were blocked with 10% goat serum (Invitrogen), 2% bovine serum albumin in PBS. Primary antibody was 1:250 ICSM-18 (D-Gen, mouse monoclonal raised against PrP(143–153)) in blocking solution, and secondary antibody was 1:500 Alexa-488-conjugated anti-mouse (Molecular Probes), with 1:1000 4′,6-diamidino-2-phenylindole (Molecular Probes) for nuclear staining. Chamber slides were viewed on a Leica DMIRE2 confocal microscope.
Isolation of Detergent-rich Microdomains
Isolation of detergent-rich microdomains was performed using a method published previously (16). Briefly, cells were resuspended in Dulbecco's PBS on ice and lysed by sequential passage through 18-, 23-, and 27-gauge needles; lysate was rotated in 1% Triton X-100 at 4 °C for 1 h and then mixed with an equal volume of 80% sucrose. A discontinuous sucrose gradient of 30 and 5% was overlaid in 14 × 89-mm tubes (Beckman) and spun at 100,000 × g for 18 h in an Optima L-100 XP Ultracentrifuge (Beckman), Sw41 Ti rotor. Proteins of 1-ml fractions were precipitated by addition of 1/5th volume of trichloroacetic acid. Pellets were washed in acetone and resuspended in PBS before analysis by Western blotting.
Isolation of Exosomes
Exosome isolation from RK13 cells was performed as described previously (12). Cells were cultured for 3–4 days prior to removal of media. Cellular debris was removed by centrifugation at 3000 × g for 10 min; supernatant was filtered (0.2 μm) and exosomes pelleted at 100,000 × g for 1 h at 4 °C. Pellets were washed in filtered PBS, repelleted, and then resuspended in a small volume of PBS.
Western Blotting
SDS-polyacrylamide gels were transferred to polyvinylidene difluoride membranes, blocked with 5% skim milk in PBS + 0.05% Tween 20, and probed with 03R19 (rabbit polyclonal antibody raised to PrP(89–106) (15)) or ICSM-18 anti-PrP antibodies. Blots were visualized with horseradish peroxidase-linked secondary and ECL Plus.
Cell Immunoblots
Infected cells were grown to confluence on plastic coverslips (Thermanox) and transferred to nitrocellulose membranes for in situ proteolysis and immunostaining as described previously (17).
Protease Treatment of Cell Lysate
Infected cells were harvested, pelleted, and lysed in Lysis Buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.5% sodium deoxycholate, 0.5% Triton X-100). 50-μl aliquots of post-lysis supernatant were taken and mixed with 50 μl of μg/ml proteinase K. Reactions were incubated for 1 h at 37 °C before terminating the reaction with 1 mm phenylmethylsulfonyl fluoride and then visualized by Western blot analysis.
Animal Bioassay of Prion-infected Cell Lines
Prion-infected RK13 cells were resuspended, washed in Dulbecco's PBS, and lysed by five sequential freeze-thaw cycles. The equivalent of 3 × 105 cells was inoculated into the left parietal region of Tga20 transgenic mice (18, 19). Mice were observed daily for signs of disease and sacrificed under methoxyflurane anesthesia when persistent signs consistent with prion disease were evident, such as reduced motor activity, hunched posture, hind limb paresis, and ataxia. Mice were handled according to prescribed national guidelines and under ethical approval from the University of Melbourne Animal Ethics Committee.
Immunohistochemistry
Brains were removed from culled mice and bisected along the sagittal plane. The right hemisphere was fixed in Neutral Buffered Formalin (10% formalin, 29 mm NaH2PO4·H2O, 46 mm Na2HPO4, pH 7) for 1 month, decontaminated by immersion in 100% formic acid, and immobilized in paraffin blocks by the Histology Facility in the Department of Pathology, University of Melbourne. Sagittal slices were stained with hematoxylin and eosin, α-glial fibrillary acidic protein antibody, or ICSM-18 antibody, mounted on glass slides, and scanned with a Mirax Digital Slide Scanner.
Protease Treatment of Tga20 Brain Homogenate
Tga20 left brain hemispheres were weighed, and a volume of PBS equivalent to 9× brain mass was added, and the mixture was homogenized by serial needle passage to create a final 10% homogenate. 10% homogenates were mixed with 100 μg/ml proteinase K and 0.1% SDS to assist the degradation of PrPC and then incubated at 37 °C for 1 h. Digestion was halted with 10 mm phenylmethylsulfonyl fluoride, and samples were visualized by Western blot analysis.
Antibody Treatment of Infected Cells
PrP-directed antibodies SP-185 (epitope PrP residues 1–23), 1C5 (PrP 119–130), and ICSM-18 (PrP 143–153) were diluted in Opti-MEM. Prion-infected MoPrP-RK13 cells were grown in 6-well plates and treated with Opti-MEM + antibody for 1 week, in which time cell lines were passaged twice. Cells were then lysed and digested with 50 μg/ml proteinase K.
Purified PrPC-PrPSc Binding Assays
Binding to PrPSc was assessed as described previously (20). Briefly, PrP-expressing RK13 cells were metabolically labeled using [35S]methionine (MP Biomedicals) and lysed, and PrP was immunoprecipitated using 03R19 and protein G-agarose beads (Roche Applied Science). PrP was released from beads by 0.2 m acetic acid, pH 2.75, and mixed with purified PrPSc. Mixtures were incubated overnight at 37 °C, following which PrPSc and bound PrPC was pelleted by centrifugation at 16,000 × g, and pellet and supernatant fractions were analyzed by SDS-PAGE and the PhosphorImager system.
Production of Recombinant PrP
Amino acid residues 23–230 of MoPrP and G118P were cloned into the pTrcHisA vector (Invitrogen), and purified recombinant PrP was produced as described previously (21). Circular dichroism spectra were measured on a Jasco 810 spectropolarimeter, and mean residue ellipticity was determined. Recombinant PrP at 0.1 mg/ml concentration was treated with 2.5 or 5 μg/ml proteinase K for 1 h at 37 °C. The protease digestion was terminated with 1 mm phenylmethylsulfonyl fluoride, and samples were analyzed by SDS-PAGE.
RESULTS
The hydrophobic region of PrP lies at the boundary of the structured region of PrPC (2), although its presence in the protease-resistant core implies that it is structured in PrPSc (Fig. 1A) (3). The hydrophobic region is one of the most highly conserved regions in PrP (7), and the glycine residues of this region show perfect conservation across all mammalian species identified to date (Fig. 1B and supplemental Fig. S1). To analyze the role of the GRR in PrP interactions, we modified this region by creating a series of point mutations in the open reading frame of mouse PrP (Fig. 1C). The GRR glycine residues 118, 122, 125, and 130 were chosen, and single point mutations were made to alanine (a conservative alteration), leucine (a bulky hydrophobic residue), and proline (structurally constrained). The A119P and L124A mutations were chosen to introduce changes into the GRR without affecting the conserved glycine residues. A G91A mutation was generated, and as glycine 91 is relatively well conserved, it lies outside but near the hydrophobic region, and like much of the GRR is within the unstructured N-terminal tail of PrP. The wild-type and mutant PrP constructs were stably expressed in RK13 cells, a rabbit kidney epithelial cell line, and this cell lineage was chosen because it does not express detectable levels of the endogenous prion protein but is capable of propagating prion infection when transfected with murine or ovine PrP (13, 22). Transfected RK13 cell lines were shown to stably express wild-type or mutant PrP as detected by Western blot analysis, and all mutants appeared to show similar glycosylation, although PrPC expression varied between cell lines (Fig. 2A and supplemental Fig. S2). No correlation between PrPC expression level and the type of mutation could be determined, indicating that mutations were not leading to pre-emptive degradation.
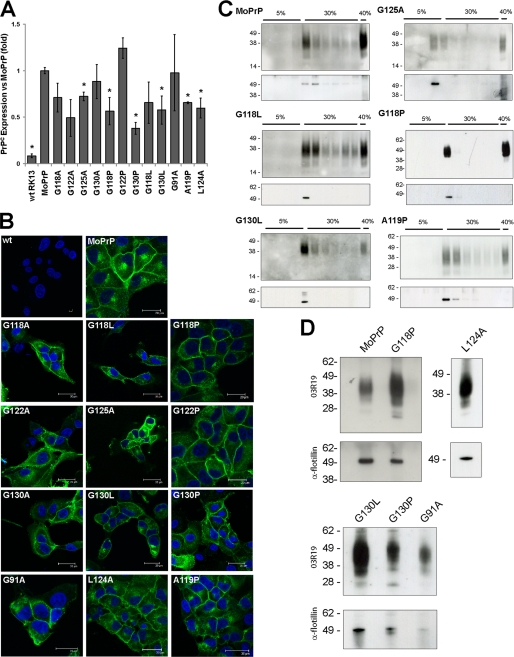
FIGURE 2.
GRR mutants are sorted normally. A, quantification of PrPC expression levels in cell lines; graph indicates normalized mean expression ± S.E. Asterisk indicates significant differences in expression (p < 0.05), and further details are available in supplemental Fig. S2. B, immunofluorescence of cell lines was performed with the PrP antibody ICSM-18 (green) and nuclear stain 4′,6-diamidino-2-phenylindole (blue). PrP was observed to localize to the plasma membrane and to an internal compartment consistent with the Golgi apparatus. This localization is similar in both MoPrP and GRR mutants, implying that mutations do not alter the subcellular expression of PrP. wt, wild type. C, cell lysates were subjected to limited Triton X-100 solubilization followed by sucrose density ultracentrifugation. Western blot analysis shows a concentration of MoPrP and GRR mutants at the interface of the 5 and 30% sucrose regions, where lipid rafts are expected to localize. The purification of lipid rafts was confirmed by probing for the raft-resident protein flotillin-2. D, exosomes were prepared from the media of transfected RK13 cells. Both prion-susceptible and prion-resistant cell lines show a concentration of PrP in exosomal pellets. Purification of exosomes was verified by probing for flotillin.
Mutant PrP Is Localized Similarly to Wild Type
Analysis of PrP-transfected cells was performed using immunofluorescence microscopy, and wild type and mutant PrP cell lines exhibit localization of PrP to the plasma membrane of transfected cells, as well as demonstrating staining of the Golgi apparatus (Fig. 2A). No indication of intracellular aggregates was detected, indicating that unlike some other mutations of PrP (23, 24) alterations to the GRR do not form inherently aggregation-prone PrP.
Although transfected cells show no gross modifications to PrP expression and trafficking, we wished to determine whether changes to specific elements of the trafficking pathway, in particular those important in prion infection, were occurring. Several cellular processes have been implicated in the process of prion infection, including the sorting of PrP into lipid rafts (25, 26). Lipid rafts are cholesterol-rich microdomains of the plasma membrane with a variety of roles in cellular biochemistry and are purified from the cell lysate by limited solubilization, followed by sucrose density ultracentrifugation (16). MoPrP is observed to localize to the density fraction corresponding to lipid rafts (Fig. 2B), which also corresponded to the known lipid-raft marker flotillin (27). All GRR mutant cell lines tested displayed localization of PrP to lipid rafts (Fig. 2B and supplemental Fig. S3), indicating that the GRR is not required for sorting of PrP to lipid rafts.
PrP is associated with exosomes, which are small vesicles formed when the multivesicular body fuses with the plasma membrane, releasing its contents into the extracellular environment (28, 29), which have previously been shown to be efficient carriers of prion infectivity (12, 30). Purification of exosomes from the MoPrP, G118P, G130L, G130P, G91A, and L124A RK13 lines demonstrates that all lines sort PrP into exosomes (Fig. 2D). These results, taken in conjunction, indicate that alterations to the GRR do not significantly alter trafficking or localization of PrP, and thus the GRR does not act as a required sorting motif for PrPC.
GRR Mutations Diminish Uptake of Prion Infection
Transfected cell lines were exposed to the M1000 prion strain, a mouse-adapted strain (31) derived from a human familial Gerstmann-Sträussler-Scheinker (GSS) case (32). Uptake of prion infection was determined by the levels of protease-resistant PrPSc in cultures after multiple passages. The cell immunoblot allows rapid detection of PrPSc, whereby cells are immobilized on nitrocellulose membranes followed by in situ lysis, proteolysis, and immunostaining, with the level of immunoreactivity corresponding to the amount of PrPSc (17). Equal numbers of cells were plated for each immunoblot allowing comparison of prion infection levels between cell lines.
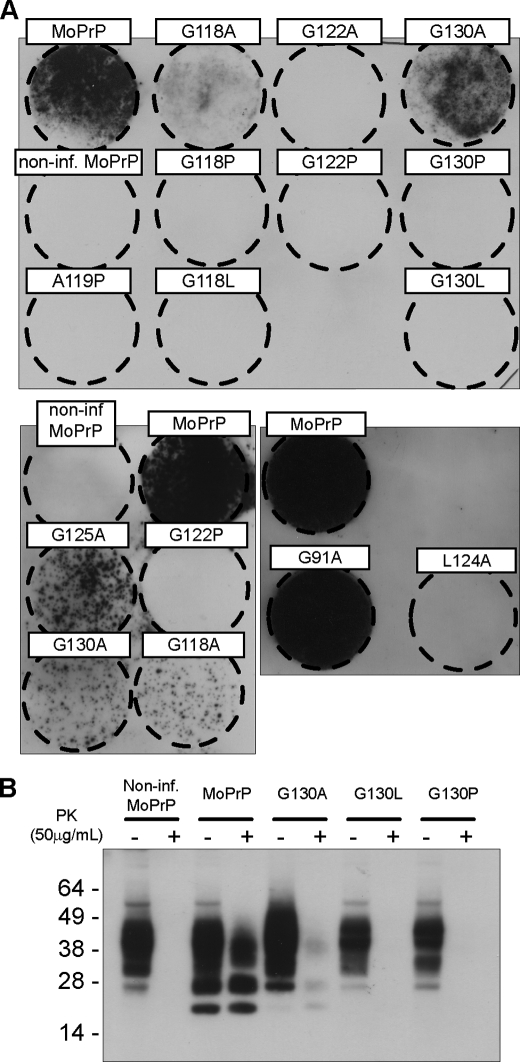
As shown previously (12), RK13 cells transfected with wild-type mouse PrP develop high levels of PrPSc following prion infection, indicating that they are propagating prion infectivity (Fig. 3A). Noninfected MoRK13 cells show no PrPSc immunoreactivity, demonstrating that the cell immunoblot can readily distinguish infected from noninfected cells. RK13 cells expressing the conservative glycine to alanine mutations exhibit lower levels of protease-resistant PrPSc; this reduction in PrPSc was observed in all cases where glycine residues were mutated to alanine (G118A, G122A, G125A, and G130A) suggesting they play an important role in modulating prion infectivity. PrPSc was undetectable in prion-exposed cells expressing the bulky leucine (G118L and G130L) and more structurally constrained proline (G118P, G122P, and G130P) mutations. No PrPSc was visible in leucine (G118L and G130L) or proline (G118P, G122P, and G130P) lines, despite repeated attempts at infection with a range of infection conditions or passage of cells to P40 post-infection (data not shown). The A119P and L124A mutations were also incapable of producing PrPSc, suggesting that conservation of the entire GRR, rather than a specific residue, is required for uptake of prion infection. In support of this, mutations outside the GRR did not alter prion propagation, cells expressing G91A-MoPrP propagate prion infection at the same level as wild-type MoPrP. Analysis for PrPSc in a subset of cell lines by Western blot (Fig. 3B) confirms the cell blot data by showing detectable levels of PrPSc in infected MoRK13 cells, decreased amounts in G130A-RK13 cells, and no detectable PrPSc in either G130P or G130L-RK13. These results strongly imply that alterations to the GRR of PrP decrease the propagation of prion infection; this decrease is not due to alterations in a specific amino acid but is proportional to the degree of alteration caused by the mutation.
FIGURE 3.
GRR mutant PrP displays lower levels of PrPSc. A, RK13 cells stably expressing wild-type or mutant PrP were infected with M1000 prions. Cell immunoblots were performed on infected cells after 10 passages and denote the presence of (PK)-resistant PrPSc as black spots of immunoreactivity (representative immunoblots of n > 3 are shown). Conservative glycine to alanine mutations reduce PrPSc levels, whereas less conservative mutations to leucine (G118L and G130L) or proline (G118P, G122P, and G130P) prevent formation of PrPSc entirely. Non-glycine mutations of the GRR (A119P and L124A) also prevent PrPSc formation, although mutations outside the GRR (G91A) have no effect. Non-inf, noninfected. B, cell blot data were verified by protease treatment of selected cell lysates, followed by Western blotting with 03R19, directed to PrP residues 89–103.
GRR Mutations Block Formation of Infectious Prions
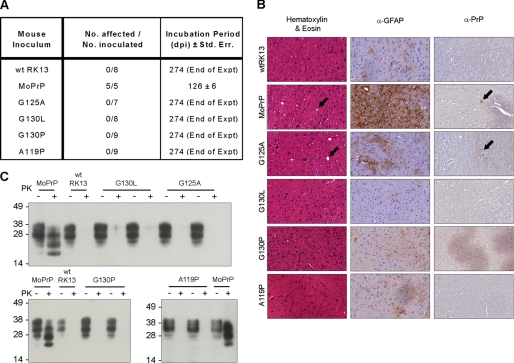
Although cell and Western immunoblotting show a decrease in PrPSc levels as the region is mutated, several lines of research have demonstrated that protease resistance and infectious potential are not necessarily concurrent (33–35). Determination of prion infectivity was obtained by animal bioassay, in which cell lysates from prion-exposed cells were inoculated into Tga20 transgenic mice, which overexpress PrP ∼5-fold and display symptoms of prion disease within a shorter time frame than wild-type mice (18). Mice inoculated with infected MoPrP-RK13 lysate developed typical signs of prion disease, such as reduced motor activity, kyphosis (hunched posture), hind limb paresis, and ataxia. Infected-MoRK13 inoculated mice progressed to terminal illness at an average of 126 days with a standard error of 6 days (Fig. 4A). Mice inoculated with wild-type RK13 (wtRK13) lysate, as well as prion-exposed G125A, G130L, G130P, or A119P-RK13 lysate, did not develop clinical signs of prion disease following inoculation, and all mice were still alive at 274 days post-inoculation, at which time the experiment was terminated. One mouse died during the experiment with an intercurrent illness, a diagnosis that was verified by the absence of PrPSc in brain tissue as detected by Western blot.
FIGURE 4.
No prion infectivity is present as detected by animal bioassay. A, lysates from infected cells were intracerebrally inoculated into Tga20 mice, which were then monitored for signs of disease and culled at terminal stages of prion infection. Infected MoRK13 inoculated mice died after an average of 126 days (S.E. of 6 days). No other mice showed signs of prion disease, although one wild-type (wt) RK13 inoculated mouse died of an intercurrent illness at 266 days. B, histology of brain slices from MoPrP-RK13 inoculated mice displays prion pathology of spongiosis, astrocytosis, and plaque deposition. Mice inoculated with wild-type, G130L, G130P, and A119P lysate did not show any signs of pathology, although limited spongiform change and plaque deposition were observed in G125A-inoculated mice. α-GFAP, antibodies directed toward glial fibrillary acidic protein. C, brain homogenates from inoculated mice were treated with 100 μg/ml proteinase K. PrPSc was observed in MoPrP-inoculated mice but not in the brains of mice inoculated with GRR mutant cell lines.
Immunohistochemistry was performed on brain slices extracted from culled mice to examine the presence of prion-related pathology (Fig. 4B). Mice infected with MoPrP lysate display spongiform change, astrocytosis, and the presence of PrP-reactive plaques. These changes were not observed in the healthy wtRK13-inoculated mice or in mice inoculated with lysate from prion-resistant G130L, G130P, and A119P RK13 cells. G125A-inoculated mice displayed spongiform change and minor plaque deposition, despite remaining outwardly healthy. It is likely that the prion titer associated with the partially prion-resistant G125A cell line is adequate to lead to infection but insufficient to cause illness within the time frame of the experiment, a condition that may be similar to previously described cases of subclinical prion infection (33, 36). Proteinase K digestion of infected brain homogenates reveals the presence of protease-resistant PrPSc in MoPrP-inoculated mice but none in the brains of mice inoculated with wild-type, G125A, G130L, G130P, or A119P lysate (Fig. 4C). Faint protease-resistant immunosignal detected in G130L-inoculated brains represents remnant undigested PrPC, as no shift in molecular weight was observed following digestion. Based on these results, we concluded that the GRR mutant cell lines were not propagating protease-sensitive infectivity, but rather that mutations were preventing the uptake of prion infection.
GRR Mutations Affect Protease Resistance of Recombinant PrP
To further examine the role of the GRR, we expressed and purified recombinant MoPrP and G118P-PrP using previously established protocols (21). Proteins were folded into their native conformation, designated α-MoPrP and α-G118P. PrP was then refolded into a β-sheet rich form known as β-PrP, which shares some attributes of PrPSc such as protease resistance and aggregation (37). Circular dichroism studies showed that no obvious differences were visible between MoPrP and G118P-PrP in both α-helical and β-sheet rich forms (supplemental Fig. S4A). However, protease treatment of recombinant PrP indicated that β-MoPrP had a higher proportion of protease-resistant material when compared with β-G118P PrP (supplemental Fig. S4, B and C). Therefore, the G118P mutant is capable of reducing the formation of aggregated material both in vivo and in vitro.
GRR Is Not Required for PrPC-PrPSc Binding
The decreased prion uptake in GRR mutant cell lines could be due to an inability of the mutant PrPC to bind PrPSc, thus preventing the infection process. Chronically infected MoPrP-RK13 cells were treated with antibodies SP-185 (epitope PrP residues 1–23), 1C5 (PrP 119–130), and ICSM-18 (PrP 143–153), to block PrPC-PrPSc interaction (Fig. 5A). The 1C5 antibody is directed toward the GRR of PrP (supplemental Fig. S5) and due to the conservation of this region presents immunoreactivity to a wide range of species (38). Following 1 week of treatment, ICSM-18 caused a reduction in PrPSc levels, as observed previously (Fig. 5B) (39). Treatment with SP-185, targeted to the signal peptide of PrP and thus not expected to have an effect on infection, did not lower the level of PrPSc present. Treatment with the GRR-directed antibody 1C5 also had no effect on PrPSc levels, indicating that blocking this region did not prevent prion replication.
FIGURE 5.
GRR is not required for binding of PrPC to PrPSc. A, overview of PrP showing the epitopes of antibodies SP-185 (directed to PrP 1–23), 1C5 (PrP 119–130), and ICSM-18 (PrP 143–153). GPI, glycosylphosphatidylinositol; seq, sequence. B, prion-infected cells were treated with antibodies, lysed, and treated with protease to isolate PrPSc. Untreated cells display the same level of PrPSc as those treated with 1C5 or SP-185, indicating these antibodies do not cure prion infection. By contrast, ICSM-18 reduces PrPSc in a dose-dependent manner. PK, proteinase K. C, radiolabeled PrPC is mixed with PrPSc and centrifuged; binding molecules will co-precipitate into the pellet fraction. When assayed in the absence of PrPSc, MoPrP, G130A, and G130L PrPC were found to primarily reside in the supernatant (S/N) fraction. D, assaying in the presence of PrPSc leads to significant levels of MoPrP, G130A, and G130L PrPC localizing to the pellet fraction, indicating that GRR mutant PrP is capable of binding to PrPSc (n = 3). E, quantification of PrPC levels in pellet and supernatant fractions following binding assay with PrPSc demonstrates no significant difference in segregation (error bars indicate 1 S.D.).
Multiple factors may affect the ability of antibodies to cure infected cell lines. To further examine the importance of the GRR in the binding of PrPC to PrPSc, we used an in vitro PrPC/PrPSc binding assay; [35S]methionine-radiolabeled MoPrP, G130A, or G130L PrPC was mixed with purified PrPSc for 24 h, followed by centrifugation. This would pellet complexes of PrPC:PrPSc (20), whereas unbound PrPC would remain in the supernatant; the proportion of PrPC in each fraction was then assessed by SDS-PAGE and autoradiography. When the assay was performed in the absence of PrPSc, all labeled PrPC was found in the supernatant fraction, indicating that PrPC is an inherently soluble, nonaggregating protein (Fig. 5C). Addition of PrPSc to the assay caused significant amounts of the MoPrP, G130A, and G130L to segregate to the pellet fraction (Fig. 5D), indicating that they retain the capability to bind to PrPSc. Quantification of PrPC levels showed no significant difference in the segregation to the pellet fraction (Fig. 5E). Therefore, the GRR is not an obligate requirement for binding of PrPC and PrPSc, and consequently the inability to infect GRR mutant cells is not due to an inability to bind infectious PrPSc.
PrPC Expression Level Does Not Affect Resistance to Prion Uptake
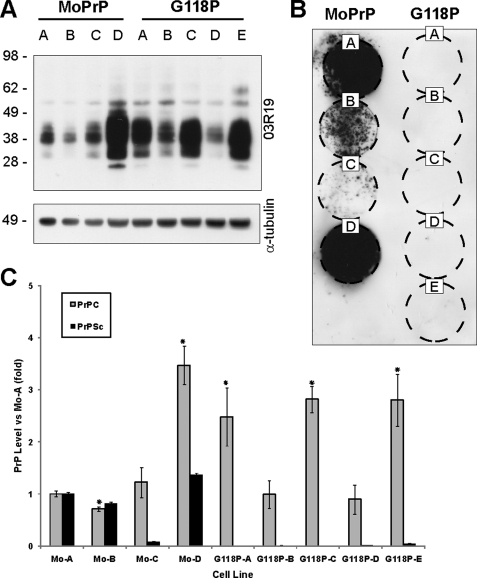
The expression of PrP is essential for the propagation of prion infectivity (40), and although previous reports have noted that the level of PrPC expressed by cultured cells does not correlate with susceptibility to prion infection (41), it remained possible that differing expression levels in GRR mutant lines are affecting prion susceptibility. To discount this, we created a series of fresh, stable cell lines expressing the prion-susceptible MoPrP or prion-resistant G118P, and these cell lines showed widely differing levels of PrPC (Fig. 6A). Cell lines were exposed to prion infection as before, and all MoPrP-expressing lines showed the development of PrPSc (Fig. 6B). PrPSc was not detected in any line expressing G118P, despite several lines showing higher expression of PrPC than susceptible MoPrP lines (Fig. 6C). Thus, we concluded that the differing levels of PrPC expression observed in GRR cell lines was not related to the susceptibility of these cell lines to prion infection.
FIGURE 6.
PrPC levels do not affect uptake of prions. A, new stable RK13 cell lines were created to express both MoPrP and the prion-resistant G118P mutant, expressing varying levels of PrPC. Tubulin probing was used to control for equal loading of cell lysate. B, cells were exposed to prions, following which cell immunoblot (passage 8) demonstrates that MoPrP cell lines develop PrPSc, whereas no PrPSc was observed in G118P lines regardless of PrPC expression levels. C, quantification of PrPC and PrPSc demonstrates that PrPC expression does not correlate with susceptibility to prion infection.
DISCUSSION
GRR Plays a Dominant Role in Prion Infection
To determine the role GRR plays in prion infection, mutations were created to cause both minor and major alterations to the hydrophobicity and bulk of the peptide structure within this region. These mutations had readily apparent effects, diminishing or abolishing propagation of prion infection as demonstrated by the lack of protease resistance or infectivity following exposure to prions. Importantly, this effect was directly related to the degree of change in the GRR, and marked effects were seen even in the relatively conservative mutations such as G118A, G122A, G125A, or G130A. Although the level of PrP expression varied in these lines, we showed that the expression of PrP does not correlate with susceptibility to prion infection. Although GRR mutant PrP showed inhibited propagation of prion infection, they exhibited approximately equivalent trafficking and processing of PrP with regard to expression, subcellular localization, sorting to lipid rafts, and release via exosomes. Constructs of PrP with deletions of β-strand 1, which contains residues 127–130 of the GRR, have previously been expressed in cells; this mutant is highly resistant to prion infection, despite having normal cellular trafficking and binding to PrPSc (42). These studies implied that changes to prion propagation are due to a physical property of the mutant PrP and that the mutations alter the binding abilities or conformational flexibility of PrP.
Our first hypothesis that the GRR was an obligate part of the PrPC-PrPSc interaction was disproved when mutants were shown to bind PrPSc in an in vitro assay. This does not rule out the possibility that the infectious prion particle includes other cofactors and that these cofactors bind through the GRR. One possibility is STI-1, a stress-inducible co-chaperone that has been shown to bind to the hydrophobic region of PrP (43) and can induce extracellular signal-regulated kinase (ERK)-dependent signaling (44). Chaperones may play an important role in prion disease; in vitro studies have suggested chaperones may accelerate aggregation of PrP (45), and increased expression of Hsp-72 chaperones has been demonstrated in Creutzfeldt-Jakob disease-affected brains (46). Binding of STI-1 to PrP may represent an important component of the formation of an infectious prion.
Structural Flexibility and the GRR
Another more likely possibility is that mutations within the GRR affect the process of conformational change. Mutations in the GRR did not appear to inhibit binding of PrPC to PrPSc, indicating that interactions within other regions such as PrP(206–223) and PrP(165–174) (47) or the N terminus (48) are sufficient to allow binding. However, the hydrophobic region is an essential determinant of prion infection; abolition of prion uptake can be achieved by removal of the AGAAAAGA palindrome (5), removal of β-strand 1, which contains part of the GRR (42), or single residue mutations within the GRR. It is known that this region undergoes significant structural rearrangement during the transition to PrPSc (6), following which the GRR is incorporated into the protease-resistant core of PrPSc (3).
PrPC contains a small β-sheet consisting of two strands comprised of residues 127–130 and 160–163 (2). This β-sheet is known to undergo slow conformational flexing when analyzed by NMR (49), and molecular dynamics simulations have indicated the transient formation of extended β-sheet structures that can incorporate the entire GRR glycines as either β-strand or inter-strand turns (50, 51). Crystal structures of PrP demonstrate a PrP-PrP dimer that involves the formations of an inter-protein β-sheet (52, 53). These results suggest that an initial step in the formation of PrPSc involves formation of a β-sheet within this area.
Glycine is the smallest and most flexible of the amino acids, whereas alanine, the next smallest residue, has comparatively limited rotational flexibility (54). Mutations that alter glycine residues to leucine and proline introduce significant changes in hydrophobicity or flexibility, and proline is known to inhibit formation of secondary structure (55). We propose that the process of conformational conversion involves the transient formation of an extended β-sheet within the hydrophobic domain, a process that is assisted by the conformational flexibility of glycine residues. Precedents exist for this, as studies of glycophorin A have demonstrated that the GXXXG-region residues provide the flexibility required to allow conversion of α-helical peptide into β-fibril amyloid, as well as providing grooves to allow close packing of adjacent β-strands (56). Alteration of glycine residues to alanine reduces the thermodynamic likelihood of this β-sheet forming, which in turn reduces the ability to convert from PrPC to PrPSc. Alteration of glycine to a leucine or proline residue introduces a large kinetic barrier to the movement of this area, which prevents formation of PrPSc. Several mutations have been described to associate with inherited prion disease that occur in or adjacent to the GRR as follows: G114V, A117V, G131V, and S132I (57–60). Interestingly, each of these mutations is to an amino acid known to be preferentially found in β-sheet structures, which may indicate a common method of action (61).
Importance of the GRR in Vivo
The role the GRR plays in prion infection can be clearly observed in the multitude of studies performed on the human codon 129 methionine/valine polymorphism, which is located within the GRR and strongly affects susceptibility to prion disease. Codon 129 homozygotes show increased susceptibility to sporadic Creutzfeldt-Jakob disease (62) and an earlier age of onset for the orally transmitted prion disease kuru (63), and almost all cases of variant Creutzfeldt-Jakob disease are methionine homozygotes (64). Dogs, one of the few mammalian species with discrepancies in the hydrophobic region, have never been reported as being susceptible to prion infection nor can canine-derived Madin-Darby canine kidney cells support prion infection (65). Finally, recent genotyping studies of members of the Fore linguistic region in Papua, New Guinea, have demonstrated a novel polymorphism that provides complete protection against kuru (66). This polymorphism, G127V, alters one of the GRR glycine residues to a hydrophobic amino acid and acts in the same manner as the glycine to leucine mutations described here. Thus, the GRR plays an important role in the transmission of prion infectivity both in vitro and in vivo. Further biophysical studies of GRR mutants are currently in progress to further understand the mechanism behind the structural alterations in the hydrophobic region, thus improving our knowledge of the conformational change of PrPC to PrPSc.
Conclusion
The glycine-rich region of PrP shows almost perfect conservation across a wide range of species. Single amino acid changes within this region dramatically reduce the ability of the mutant PrP to support prion infection, while not affecting the trafficking of PrP nor its ability to bind PrPSc. Reduction in prion infection appears proportional to the decrease in conformational flexibility in the GRR, implying that the morphological alteration from PrPC to PrPSc requires significant structural change within the hydrophobic region. Our observation that even single conservative mutations can have potent effects reinforces the importance of this region for prion propagation.
Supplementary Material
Acknowledgments
We thank Professor Charles Weissmann for the gift of the Tga20 mice and Professor John Collinge for the gift of monoclonal antibody ICSM18. We thank the Biomedical Sciences Animal Facility staff for technical assistance.
This work was supported in part by Australian Research Council Discovery Project Grant DP0987227 and National Health and Medical Research Council Program Grant 400202.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5, Table 1, and an additional reference.
- PrPC
- cellular prion protein
- PrPSc
- disease-associated conformation of PrP
- PrP
- prion protein
- GRR
- glycine-rich region
- PBS
- phosphate-buffered saline
- Mo
- mouse.
REFERENCES
- 1.Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2.Riek R., Hornemann S., Wider G., Glockshuber R., Wüthrich K. (1997) FEBS Lett. 413, 282–288 [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S. B., Groth D. F., Bolton D. C., Kent S. B., Hood L. E. (1984) Cell 38, 127–134 [DOI] [PubMed] [Google Scholar]
- 4.Chabry J., Caughey B., Chesebro B. (1998) J. Biol. Chem. 273, 13203–13207 [DOI] [PubMed] [Google Scholar]
- 5.Norstrom E. M., Mastrianni J. A. (2005) J. Biol. Chem. 280, 27236–27243 [DOI] [PubMed] [Google Scholar]
- 6.Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 7.Schätzl H. M., Da Costa M., Taylor L., Cohen F. E., Prusiner S. B. (1995) J. Mol. Biol. 245, 362–374 [DOI] [PubMed] [Google Scholar]
- 8.Barnham K. J., Cappai R., Beyreuther K., Masters C. L., Hill A. F. (2006) Trends Biochem. Sci. 31, 465–472 [DOI] [PubMed] [Google Scholar]
- 9.Russ W. P., Engelman D. M. (2000) J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 10.Hung L. W., Ciccotosto G. D., Giannakis E., Tew D. J., Perez K., Masters C. L., Cappai R., Wade J. D., Barnham K. J. (2008) J. Neurosci. 28, 11950–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison C. F., Barnham K. J., Hill A. F. (2007) J. Neurochem. 103, 1709–1720 [DOI] [PubMed] [Google Scholar]
- 12.Vella L. J., Sharples R. A., Lawson V. A., Masters C. L., Cappai R., Hill A. F. (2007) J. Pathol. 211, 582–590 [DOI] [PubMed] [Google Scholar]
- 13.Vilette D., Andreoletti O., Archer F., Madelaine M. F., Vilotte J. L., Lehmann S., Laude H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson V. A., Stewart J. D., Masters C. L. (2007) J. Gen. Virol. 88, 2905–2914 [DOI] [PubMed] [Google Scholar]
- 15.Lawson V. A., Vella L. J., Stewart J. D., Sharples R. A., Klemm H., Machalek D. M., Masters C. L., Cappai R., Collins S. J., Hill A. F. (2008) Int. J. Biochem. Cell Biol. 40, 2793–2801 [DOI] [PubMed] [Google Scholar]
- 16.Brown D. A., Rose J. K. (1992) Cell 68, 533–544 [DOI] [PubMed] [Google Scholar]
- 17.Klöhn P. C., Stoltze L., Flechsig E., Enari M., Weissmann C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11666–11671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer M., Rülicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C. (1996) EMBO J. 15, 1255–1264 [PMC free article] [PubMed] [Google Scholar]
- 19.Collins S. J., Lewis V., Brazier M. W., Hill A. F., Lawson V. A., Klug G. M., Masters C. L. (2005) Neurobiol. Dis. 20, 336–346 [DOI] [PubMed] [Google Scholar]
- 20.Priola S. A., Lawson V. A. (2001) EMBO J. 20, 6692–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman B. M., Nisbet R. M., Han S., Cappai R., Hatters D. M., Hill A. F. (2009) Biochem. Biophys. Res. Commun. 380, 564–568 [DOI] [PubMed] [Google Scholar]
- 22.Vilette D. (2008) Vet. Res. 39, 10. [DOI] [PubMed] [Google Scholar]
- 23.Grenier C., Bissonnette C., Volkov L., Roucou X. (2006) J. Neurochem. 97, 1456–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanusso G., Petersen R. B., Jin T., Jing Y., Kanoush R., Ferrari S., Gambetti P., Singh N. (1999) J. Biol. Chem. 274, 23396–23404 [DOI] [PubMed] [Google Scholar]
- 25.Vey M., Pilkuhn S., Wille H., Nixon R., DeArmond S. J., Smart E. J., Anderson R. G., Taraboulos A., Prusiner S. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14945–14949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., Avraham D. (1995) J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzer U., Prohaska R. (2001) Blood 97, 1141–1143 [DOI] [PubMed] [Google Scholar]
- 28.Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 29.Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. (1987) J. Biol. Chem. 262, 9412–9420 [PubMed] [Google Scholar]
- 30.Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9683–9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brazier M. W., Lewis V., Ciccotosto G. D., Klug G. M., Lawson V. A., Cappai R., Ironside J. W., Masters C. L., Hill A. F., White A. R., Collins S. (2006) Brain Res. Bull. 68, 346–354 [DOI] [PubMed] [Google Scholar]
- 32.Tateishi J., Ohta M., Koga M., Sato Y., Kuroiwa Y. (1979) Ann. Neurol. 5, 581–584 [DOI] [PubMed] [Google Scholar]
- 33.Hill A. F., Joiner S., Linehan J., Desbruslais M., Lantos P. L., Collinge J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10248–10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tateishi J., Kitamoto T. (1995) Brain Pathol. 5, 53–59 [DOI] [PubMed] [Google Scholar]
- 35.Chiesa R., Piccardo P., Quaglio E., Drisaldi B., Si-Hoe S. L., Takao M., Ghetti B., Harris D. A. (2003) J. Virol. 77, 7611–7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thackray A. M., Klein M. A., Aguzzi A., Bujdoso R. (2002) J. Virol. 76, 2510–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swietnicki W., Morillas M., Chen S. G., Gambetti P., Surewicz W. K. (2000) Biochemistry 39, 424–431 [DOI] [PubMed] [Google Scholar]
- 38.Choi J. K., Park S. J., Jun Y. C., Oh J. M., Jeong B. H., Lee H. P., Park S. N., Carp R. I., Kim Y. S. (2006) Hybridoma 25, 271–277 [DOI] [PubMed] [Google Scholar]
- 39.White A. R., Enever P., Tayebi M., Mushens R., Linehan J., Brandner S., Anstee D., Collinge J., Hawke S. (2003) Nature 422, 80–83 [DOI] [PubMed] [Google Scholar]
- 40.Büeler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., Weissmann C. (1993) Cell 73, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 41.Enari M., Flechsig E., Weissmann C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9295–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorberg I., Chan K., Priola S. A. (2001) J. Virol. 75, 10024–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanata S. M., Lopes M. H., Mercadante A. F., Hajj G. N., Chiarini L. B., Nomizo R., Freitas A. R., Cabral A. L., Lee K. S., Juliano M. A., de Oliveira E., Jachieri S. G., Burlingame A., Huang L., Linden R., Brentani R. R., Martins V. R. (2002) EMBO J. 21, 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Americo T. A., Chiarini L. B., Linden R. (2007) Biochem. Biophys. Res. Commun. 358, 620–625 [DOI] [PubMed] [Google Scholar]
- 45.Stöckel J., Hartl F. U. (2001) J. Mol. Biol. 313, 861–872 [DOI] [PubMed] [Google Scholar]
- 46.Kovács G. G., Kurucz I., Budka H., Adori C., Müller F., Acs P., Klöppel S., Schätzl H. M., Mayer R. J., László L. (2001) Neurobiol. Dis. 8, 881–889 [DOI] [PubMed] [Google Scholar]
- 47.Horiuchi M., Caughey B. (1999) EMBO J. 18, 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solforosi L., Bellon A., Schaller M., Cruite J. T., Abalos G. C., Williamson R. A. (2007) J. Biol. Chem. 282, 7465–7471 [DOI] [PubMed] [Google Scholar]
- 49.Viles J. H., Donne D., Kroon G., Prusiner S. B., Cohen F. E., Dyson H. J., Wright P. E. (2001) Biochemistry 40, 2743–2753 [DOI] [PubMed] [Google Scholar]
- 50.Guilbert C., Ricard F., Smith J. C. (2000) Biopolymers 54, 406–415 [DOI] [PubMed] [Google Scholar]
- 51.Alonso D. O., DeArmond S. J., Cohen F. E., Daggett V. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2985–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haire L. F., Whyte S. M., Vasisht N., Gill A. C., Verma C., Dodson E. J., Dodson G. G., Bayley P. M. (2004) J. Mol. Biol. 336, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 53.Antonyuk S. V., Trevitt C. R., Strange R. W., Jackson G. S., Sangar D., Batchelor M., Cooper S., Fraser C., Jones S., Georgiou T., Khalili-Shirazi A., Clarke A. R., Hasnain S. S., Collinge J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hovmöller S., Zhou T., Ohlson T. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 768–776 [DOI] [PubMed] [Google Scholar]
- 55.Chou P. Y., Fasman G. D. (1974) Biochemistry 13, 222–245 [DOI] [PubMed] [Google Scholar]
- 56.Liu W., Crocker E., Zhang W., Elliott J. I., Luy B., Li H., Aimoto S., Smith S. O. (2005) Biochemistry 44, 3591–3597 [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez M. M., Peoc'h K., Haïk S., Bouchet C., Vernengo L., Mañana G., Salamano R., Carrasco L., Lenne M., Beaudry P., Launay J. M., Laplanche J. L. (2005) Neurology 64, 1455–1457 [DOI] [PubMed] [Google Scholar]
- 58.Mallucci G. R., Campbell T. A., Dickinson A., Beck J., Holt M., Plant G., de Pauw K. W., Hakin R. N., Clarke C. E., Howell S., Davies-Jones G. A., Lawden M., Smith C. M., Ince P., Ironside J. W., Bridges L. R., Dean A., Weeks I., Collinge J. (1999) Brain 122, 1823–1837 [DOI] [PubMed] [Google Scholar]
- 59.Panegyres P. K., Toufexis K., Kakulas B. A., Cernevakova L., Brown P., Ghetti B., Piccardo P., Dlouhy S. R. (2001) Arch. Neurol. 58, 1899–1902 [DOI] [PubMed] [Google Scholar]
- 60.Hilton D. A., Head M. W., Singh V. K., Bishop M., Ironside J. W. (2009) Neuropathol. Appl. Neurobiol. 35, 111–115 [DOI] [PubMed] [Google Scholar]
- 61.Chou P. Y., Fasman G. D. (1974) Biochemistry 13, 211–222 [DOI] [PubMed] [Google Scholar]
- 62.Palmer M. S., Dryden A. J., Hughes J. T., Collinge J. (1991) Nature 352, 340–342 [DOI] [PubMed] [Google Scholar]
- 63.Cervenáková L., Goldfarb L. G., Garruto R., Lee H. S., Gajdusek D. C., Brown P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13239–13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mead S. (2006) Eur. J. Hum. Genet. 14, 273–281 [DOI] [PubMed] [Google Scholar]
- 65.Polymenidou M., Trusheim H., Stallmach L., Moos R., Julius C., Miele G., Lenz-Bauer C., Aguzzi A. (2008) Vaccine 26, 2601–2614 [DOI] [PubMed] [Google Scholar]
- 66.Mead S., Whitfield J., Poulter M., Shah P., Uphill J., Campbell T., Al-Dujaily H., Hummerich H., Beck J., Mein C. A., Verzilli C., Whittaker J., Alpers M. P., Collinge J. (2009) N. Engl. J. Med. 361, 2056–2065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.