Abstract
Cell surface pili are polymeric protein assemblies that enable bacteria to adhere to surfaces and to specific host tissues. The pili expressed by Gram-positive bacteria constitute a unique paradigm in which sortase-mediated covalent linkages join successive pilin subunits like beads on a string. These pili are formed from two or three distinct types of pilin subunit, typically encoded in small gene clusters, often with their cognate sortases. In Group A streptococci (GAS), a major pilin forms the polymeric backbone, whereas two minor pilins are located at the tip and the base. Here, we report the 1.9-Å resolution crystal structure of the GAS basal pilin FctB, revealing an immunoglobulin (Ig)-like N-terminal domain with an extended proline-rich tail. Unexpected structural homology between the FctB Ig-like domain and the N-terminal domain of the GAS shaft pilin helps explain the use of the same sortase for polymerization of the shaft and its attachment to FctB. It also enabled the identification, from mass spectral data, of the lysine residue involved in the covalent linkage of FctB to the shaft. The proline-rich tail forms a polyproline-II helix that appears to be a common feature of the basal (cell wall-anchoring) pilins. Together, our results indicate distinct structural elements in the pilin proteins that play a role in selecting for the appropriate sortases and thereby help orchestrate the ordered assembly of the pilus.
Keywords: Cell Surface Protein, Crystal Structure, Mass Spectrometry (MS), Protein Assembly, Protein Structure, Bacterial Pili, Isopeptide Bonds, Sortase Action, Streptococcus pyogenes
Introduction
Pili (or fimbriae) are hair-like protein appendages common in Gram-negative and Gram-positive bacteria. In many instances, pili are crucial for pathogenesis, because they mediate adhesion and enable colonization of a host (1). In Gram-positive pathogens such as Corynebacterium diphtheriae or Streptococcus pyogenes, the genes for the pilus assembly are arranged in pathogenic islets that encode one major pilin, one or two minor (ancillary) pilins, and pilus-specific sortases (2–5). The latter catalyze covalent polymerization of the major pilins by the formation of intermolecular amide bonds between the C terminus of one subunit and a lysine residue on the next (2, 4, 5). This covalent shaft assembly is a hallmark of the Gram-positive pilus structure. Another striking feature of the pilus shaft is the occurrence of intramolecular isopeptide (amide) bonds in the major pilins that confer stability on these subunits and on the pilus assembly (6–8).
The minor pilins, in contrast, are less well characterized, and questions arise as to their roles and modes of incorporation into the pili, due to the fact that some pili (e.g. those for Bacillus cereus) have only one minor pilin, whereas most others have two (9). The best characterized of the three-component pili are those from C. diphtheriae. In the prototypical pili from this organism, the pilus-specific sortase SrtA polymerizes the major pilin SpaA to form a pilus shaft, which carries the minor pilin SpaC on its tip (2). Another minor pilin, SpaB, is incorporated at the base of the pilus and is tethered to the peptidoglycan by the so-called housekeeping sortase SrtF (10). Thus, SpaB anchors the pilus structure to the cell wall.
This pattern, in which a major pilin forms the polymeric pilus shaft, with minor pilins as the cap and the cell wall anchor, has emerged as a consistent paradigm for the three-component pili of other Gram-positive bacteria such as Streptococcus agalactiae (11, 12), S. pneumoniae (13), and most recently, S. pyogenes.4
At the structural level, three-dimensional structures are available for the shaft-forming major pilins of S. pyogenes, C. diphtheriae, and Bacillus cereus (6, 8, 15), and the amide bonds that join these major pilin subunits have in each case been characterized by mass spectrometry. In contrast, much less is known so far of the structural determinants that underlie the incorporation of minor pilins at the tip and the base.
Here, we focus on the important human pathogen S. pyogenes (group A streptococcus (GAS)).5 GAS colonizes host epithelia, causing benign but very common diseases such as acute tonsillitis and pharyngitis, but can also invade underlying soft tissues to cause lethal invasive disease such as streptococcal toxic shock syndrome or necrotizing fasciitis (16). The search for GAS vaccine candidates led to the discovery of pilus structures on this bacterium (3). These pili are essential for adhesion of S. pyogenes to pharyngeal cell lines, human tonsil explants, and primary keratinocytes and appear to be involved in biofilm formation (17, 18).
GAS pilin proteins are encoded by genes of the highly variable FCT region, of which nine types have been described so far (19). Except for FCT-1, these encode three pilin proteins, of which the major pilin (usually designated FctA or backbone pilin (BP)) forms the shaft, with the two minor pilins at the tip and the base; Cpa or ancillary pilin 1 (AP1) forms the cap and FctB or ancillary pilin 2 (AP2) acts as the cell wall anchor of the pilus (19, 20).4 The crystal structure of Spy0128, the BP from GAS serotype M1, strain SF370, revealed a conserved lysine (Lys-161) to which the next BP subunit is covalently attached during polymerization of the backbone (6). The same lysine residue of the BP is also used to incorporate Cpa/AP1 at the tip of the pilus, as has been shown for several strains (21).4 The second minor pilin, FctB/AP2 (Spy0130 in GAS M1 strain SF370), has also been reported to be covalently linked to the pilus structure (3, 18) and has been located in the cell wall by immunoelectron microscopy (3). Importantly, it has been shown recently that a Δspy0130 knock-out mutant still forms pili, but that these are not attached to the cell wall and are instead released into the culture medium.4 A ΔfctB knock-out in a GAS serotype M49 strain similarly results in loss of pili from the cell wall (22). These experiments identify FctB/AP2 as the cell wall anchor of GAS pili. Neither the structure of AP2, nor its mode of attachment to the shaft of the pilus, has as yet been determined, however.
Our long term goal is to obtain a complete atomic description of the GAS pilus assembly. Building on our crystal structure of the GAS backbone pilin (6) we here address the structure of the basal pilin AP2, which anchors the GAS pilus to the cell wall. After unsuccessful attempts to crystallize the AP2 subunit from the M1 strain (Spy0130), we have solved the high resolution structure of its FctB homolog from GAS strain 90/306S. The structure reveals an Ig-like domain with an extended proline-rich tail. Structural homology with the N-terminal domain of Spy0128, the GAS BP subunit from the M1 strain, points to a conserved lysine on AP2 as the site of the covalent linkage between AP2 and BP, a linkage that we have also confirmed by mass spectral analysis of native M1 pili. The FctB structure further identifies the polyproline tail as an important structural determinant in anchoring the pilus to the cell wall.
EXPERIMENTAL PROCEDURES
Expression and Purification of FctB
The fctB gene without the predicted signal peptide and the C-terminal membrane association helix was cloned by PCR from the GAS strain 90/306S. This strain is a T9 serotype (from its fctA sequence; data not shown) but is of unknown M serotype. The FctB protein was overexpressed in Escherichia coli and purified by immobilized metal ion affinity chromatography and gel filtration as described before (23). FctB substituted with selenomethionine (SeMet-FctB) was expressed according to a protocol described by Van Duyne et al. (24). SeMet-FctB was purified as described for native FctB (23), except that 1 mm β-mercaptoethanol was added to lysis, wash, and elution buffer, 2 mm β-mercaptoethanol to Tris-buffered saline and 10 mm dithiothreitol to the standard storage buffer (10 mm Tris/HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.3 mm NaN3).
Crystallization of FctB and X-ray Data Collection
Native FctB crystals were grown by mixing 1–2 μl of protein solution (27 mg/ml) with 1–2 μl of precipitant (100 mm sodium cacodylate, pH 6.1–6.5, 0.9–1 m sodium citrate) at 18 °C (23). SeMet-FctB crystals were grown under the same conditions by streak-seeding from crystals of native FctB. Crystals of native FctB and SeMet-FctB were transferred to cryoprotectant (100 mm sodium cacodylate, pH 6.5, 1 m sodium citrate, 20% (v/v) glycerol) prior to flash-freezing in liquid nitrogen. X-ray diffraction data were collected in-house (23). Further native FctB diffraction data were collected at the Australian Synchrotron on beamline PX-2. The small size of the beam allowed data to be collected from different regions within a crystal, which were found to have quite variable diffraction properties. All data sets (Table 1) were processed as described before (23).
TABLE 1.
Data collection, processing, and phasing statistics
| Native FctB 1 | SeMet-FctB | Native FctB 2 | |
|---|---|---|---|
| X-ray source | Rotating anode | Rotating anode | Australian Synchrotron |
| Wavelength (Å) | 1.5418 | 1.5418 | 0.979463 |
| Resolution rangea (Å) | 31.15–2.90 (3.05–2.90) | 31.15–2.90 (3.05–2.90) | 19.56–1.90 (2.00–1.90) |
| Space group | P65 | P65 | P65 |
| Unit cell axes (Å) | a = b = 95.15, c = 100.25 | a = b = 95.09, c = 100.20 | a = b = 95.19, c = 100.46 |
| Angles (°) | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = 90, γ = 120 |
| Total observationsa | 143,360 (19,699) | 52,394 (2351) | 307,803 (44,910) |
| Unique reflectionsa | 11,506 (1,636) | 10,316 (490) | 40,679 (5,934) |
| Redundancy | 12.5 | 5.1 | 7.6 |
| Completenessa (%) | 99.7 (98.1) | 99.4 (96.7) | 99.9 (100) |
| Mean I/σ(I)a | 27.5 (7.6) | 17.2 (4.9) | 19.2 (4.5) |
| Rmerge (%)a,b | 9.5 (36.5) | 8.1 (30.4) | 6.6 (53.1) |
| Phasing power centric/acentric | 0.735/0.769 | ||
| Cullis R-factor centric/acentric | 0.881/0.755 | ||
| Figure-of-merit centric/acentric | 0.274/0.210 | ||
| Number of sites | 3 |
a Values in parentheses are for the outermost resolution shell.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl).
Phase Determination and Model Building
In-house datasets for native and SeMet-FctB to 3.0-Å resolution were combined using POINTLESS and scaled using SCALA (25). In a single isomorphous replacement approach, AutoSHARP (26) was used to find and refine the selenomethionine sites in SeMet-FctB (1 per molecule). Three sites were found, two of which were very close together and had occupancies of 0.39 each (0.87 for the third site). Initial phases were derived from these sites using AutoSHARP and were improved by density modification on the initial electron density map. The resulting electron density was suitable for automated model-building with BUCCANEER (27) in combination with REFMAC (28). This model was used for molecular replacement with the native data to 1.9-Å resolution using PHASER (29). The structure was then refined through cycles of manual building in COOT (30) and maximum-likelihood refinement in PHENIX (31). For the final rounds of refinement, hydrogens were added in their riding positions and two translation/libration/screw (TLS) groups per molecule were defined based on an assessment provided by the TLSMD server (32). The latter significantly improved the quality of the electron density maps. The model was validated using MOLPROBITY (33). Structure comparisons were performed using the SSM algorithm (34) in COOT. The figures were prepared using PyMOL (35). Refinement statistics are in Table 2. Atomic coordinates and structure factors have been deposited into the Protein Data Bank (www.pdb.org) with accession code 3KLQ.
TABLE 2.
Refinement statistics
| Resolution rangea | 19.55–1.90 (1.95–1.90) |
| R/Rfree (%)a,b | 15.9/18.2 (21.6/22.6) |
| Protein atoms | 2,408 |
| Water molecules | 231 |
| Average B-factors (Å2) for | |
| Main-chain atoms | 37.0 |
| Side-chain atoms | 45.7 |
| Water molecules | 45.3 |
| r.m.s. deviations from ideal geometry | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.77 |
| Residues in favored regions (33) | 100% |
a Values in parentheses are for the outermost resolution shell.
b R and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Rfree was calculated over 5% of amplitudes that were chosen at random and not used in refinement.
Model of the Pilus of S. pyogenes Serotype M1
Models of the M1 basal pilin Spy0130 and the C-terminal domain of the adhesin Spy0125 were built from the coordinates of FctB and the C-terminal domain of Spy0128 (6), respectively, using MODELLER (36). The models of Spy0130 and Spy0125 were then docked onto the structure of the backbone pilin Spy0128 (PDB code 3B2M), based on the pilus-like molecular association seen in crystals of Spy0128 (6). The Spy0130 model was superimposed on the N-terminal domain of Spy0128 where it abuts the C terminus of the next molecule in the crystal, and the model of the C-terminal domain of Spy0125 was similarly superimposed on the C-terminal domain of Spy0128 to dock against the N-terminal domain of the next molecule.
CD Spectroscopy
FctB and Spy0130 from S. pyogenes strain SF370 were purified as described previously (23). Both were dialyzed thoroughly against 5 mm phosphate buffer, pH 7.5, and diluted to 3–4 μm. CD spectra were recorded on a PiStar-180 (Applied Photophysics) spectrometer at ambient temperature.
Proteolytic Digestion and Mass Spectral Analysis of Pili
Pili were purified from S. pyogenes strain SF370 and analyzed as described before (6). Briefly, cell wall extracts were separated by SDS-PAGE, and bands containing pili were cut out and digested using trypsin (Promega, Madison, WI). The fragments were analyzed using a Q-STAR XL Hybrid MS/MS system (Applied Biosystems). Peptides were identified manually or with a Mascot search engine version 2.0.05 (Matrix Science).
RESULTS
Structure Determination
Our original goal was to determine the structure of Spy0130, the basal pilin from the M1 strain SF370, for which we had mass spectral data from the native pili. When all attempts to crystallize Spy0130 failed, however, we turned to its homologue from a local isolate of GAS strain 90/360S. The latter is of unknown M serotype but was typed as T9 from its fctA sequence, and PCR analysis of the FCT islet showed it to have the same gene order as FCT-2, FCT-3, and FCT-4 strains. Its fctA and fctB genes are >97% identical to those from the M9 strain 2720, which is an FCT-4-containing strain (19). Constructs for Spy0130 and for FctB from strain 90/306S were expressed in E. coli and purified, with high quality crystals being obtained for FctB (23). The construct used encompasses the mature protein from the signal peptidase cleavage site through to the LPLAG sortase cleavage motif. The crystal structure of FctB was solved by single isomorphous replacement using in-house diffraction data from SeMet-substituted crystals of FctB. The solvent content of the crystals was 68% and enabled successful density modification despite low phasing power (Table 1). The structure was refined at 1.9-Å resolution to an R factor of 15.9% (Rfree = 18.2%) (Table 2). The asymmetric unit contains two molecules, A and B, of which molecule A lacks only the first two N-terminal residues, whereas molecule B lacks interpretable electron density for one N-terminal and the 7 C-terminal residues 133–139. Here, we take molecule A as the representative model for FctB.
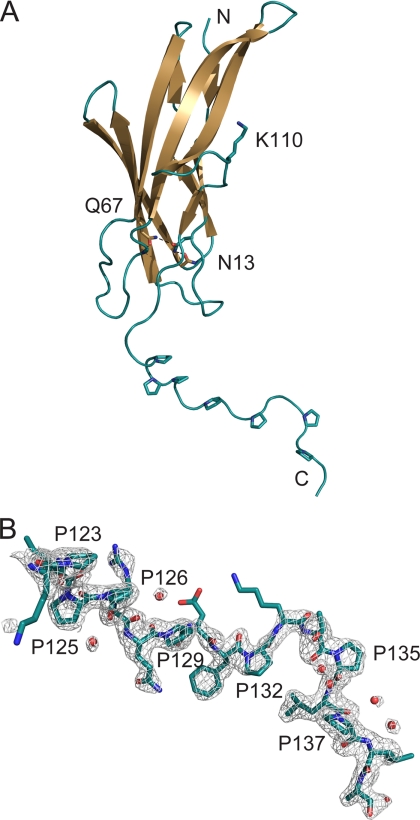
Structure of FctB
The structure of FctB consists of a compact, all-β domain (residues 3–120) with an immunoglobulin-like (Ig-like) fold, followed by an extended proline-rich tail that forms a polyproline-II (PPII)-like helix (Fig. 1A). The Ig-like domain is formed by a 5-stranded β-sheet (C-B-E-F-G1) and a 3-stranded β-sheet (D-A-G2) that pack to give a β-sandwich. Strand G is interrupted by an omega (Ω)-loop (residues 102–111) that enables this strand to transfer its hydrogen bonding from one sheet to the other. Beyond Leu-120, the C-terminal 19 residues form an extended structure that projects from the globular Ig-like domain (Fig. 1). This C-terminal tail has a high frequency of proline residues, which occur with significant regularity; residues 123–137 have the sequence PXPPXXPXXPXXPXXP. For the most part, the electron density for this region is surprisingly good, especially in molecule A (Fig. 1B), and the structure is well defined.
FIGURE 1.
Three-dimensional structure of FctB. A, the fold of FctB, drawn as a schematic diagram. Side chains of important residues discussed in the text and of prolines in the proline-rich tail are shown in stick mode, where red indicates oxygen and blue nitrogen atoms. Hydrogen bonds are depicted as dashed lines. B, the final 2Fobs − Fc electron density map at 1σ contour level with the model for the proline-rich tail of FctB. Water molecules are shown as red spheres. Some side chains are disordered but were included for clarity.
The overall shape of the FctB molecule strongly resembles the low resolution model derived for Spy0130, based on SAXS data (37), This model has approximate dimensions of 105 × 25 × 14 Å, agreeing well with those of FctB (80 × 30 × 20 Å). It also displays a very similar division into two elongated domains of different width (37), consistent with a globular IgG-like domain and a tail-domain. These data, coupled with the 33% sequence identity for the two proteins (supplemental Fig. S3) suggest that FctB and Spy0130 share the same structure, an IgG-like domain linked to a proline-rich tail domain.
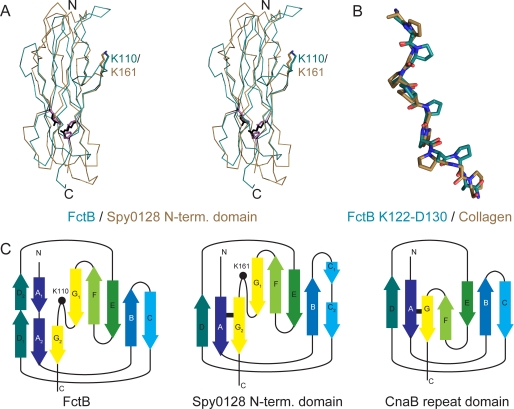
Searches of the Protein Data Bank with SSM (34) show that the closest structural homolog of FctB is the N-terminal domain of the major pilin Spy0128 from S. pyogenes serotype M1 (PDB identifier 3B2M) (6). FctB aligns with this domain with a root-mean-square (r.m.s.) difference in Cα atom positions of 1.78 Å over 96 residues (Fig. 2A). Like the N-terminal domain of Spy0128, the overall fold and the topology of FctB are related to the inverse IgG fold of the CnaB repeat domains of the staphylococcal collagen-binding protein Cna (38) (Fig. 2C).
FIGURE 2.
Comparison of FctB and structurally related proteins. A, structural alignment of FctB (residues 3–123, turquoise) and the N-domain of Spy0128 (6) (brown) shown as a Cα-trace. Residues involved in the isopeptide bond of Spy0128 are in black, and their homologs are in FctB in magenta. B, superposition of residues Lys-122 through Asp-130 of the C-terminal polyproline-II-like helix of FctB (turquoise) with a collagen peptide (PDB code 2DRX, brown) (14). C, topology diagrams for FctB (left), the N-domain of Spy0128 (middle), and a CnaB domain (right). The positions of Lys-110 in FctB and Lys-161 in Spy0128 are indicated by a dot, and the isopeptide bonds of Spy0128 and CnaB are indicated by a black bar. Lettering is according to Kang et al. (6).
The two β-sheets of the Ig-like domain of FctB are linked by a 2.8-Å hydrogen bond between Asn-13 Oδ1 and Gln-67 Nϵ2 in the hydrophobic core. The side chains of Asn-13 and Gln-67 make further hydrogen bonds with the peptide oxygen of Gly-44 and peptide nitrogen of Asp-78, respectively, creating a hydrogen network through the hydrophobic core that bridges four strands of the two β-sheets. This hydrogen bond network in FctB takes the place of the intramolecular isopeptide bond that stabilizes the N-terminal domain of Spy0128 (6, 39) and which is found also in other CnaB domains (6) (Fig. 2C). FctB Asn-13 aligns precisely with Lys-36 of the Spy0128 isopeptide bond and Gln-67 in FctB replaces Glu-117, which catalyzes the formation of the isopeptide bond in Spy0128. However, Asn-168, which is covalently bound to Lys-36 in Spy0128, is replaced by Pro-117 in FctB. Thus, although FctB does not contain an intramolecular isopeptide bond, there is an internal stabilizing hydrogen bond network in an identical region of the largely conserved hydrophobic core of FctB (Figs. 2 and supplemental Fig. S1).
A major stretch of the proline-rich tail of FctB, encompassing residues 123–135, displays significant regularity in sequence and structure (Fig. 1). Four of the six prolines in this sequence PXPPXXPXXPXXP, which immediately precedes the sortase motif LPLA (residues 136–139), are orientated to the same side of the strand, with residues Pro-123 to Pro-132 forming a PPII-like helix, based on ϕ/ψ angles (40). The same PPII-like helix is seen for the proline-rich tails of both molecules of the asymmetric unit, differing only in their overall orientation. A hinge at Leu-120 at the start of the tail enables the rigid proline-rich region to flex like a lever arm relative to the IgG-like domain. CD spectra for both FctB and Spy0130 (supplemental Fig. S2) contain a peak at ∼220–240 nm that is characteristic of the PPII helix, confirming its occurrence in solution. The SAXS data of Solovyova et al. (37) is also consistent with this interpretation.
Incorporation of AP2 into the Pilus
Structural superpositions of FctB onto the N-terminal domain of Spy0128 and the B domains of the Staphylococcus aureus Cna protein show that both FctB and the Spy0128 N-terminal domain share a common loop not present in the archetypal CnaB domains. This Ω-loop interrupts β-strand G and contains a conserved lysine residue (FctB, Lys-110) that is in an identical position to Lys-161 of Spy0128 (Fig. 2A). Lys-161 is the essential lysine used in formation of the polymeric shaft of the GAS pilus; its Nζ atom forms an intermolecular isopeptide bond with the C-terminal Thr-311 of the LPXTG sortase motif of the next BP subunit (6). Based on the structural match of FctB Lys-110 to Spy0128 Lys-161, we hypothesized that Lys-110 of FctB, and the equivalent lysine Lys-120 of Spy0130, is the lysine that covalently links AP2, the basal minor pilin, to the LPXTG motif of the BP.
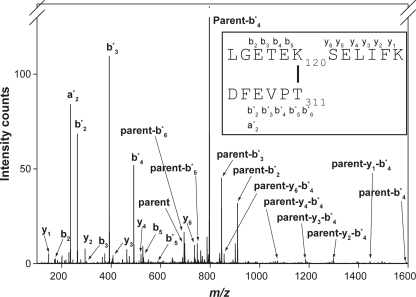
We used mass spectrometry to confirm the presence of this intermolecular isopeptide bond in native GAS pili. We had previously purified pilus polymers from GAS strain SF370 (6), but in the absence of a recognizable sequence motif we could not identify peptides incorporating the Spy0128-Spy0130 linkage. With the knowledge that Lys-120 of Spy0130 was a likely candidate for the essential lysine, we were able to sort through unassigned peptides from the pilus digest and identify a peptide containing both Spy0130 Lys-120 and the C-terminal Thr-311 of Spy0128 (Fig. 3). This peptide, including the amide bond between Spy0130 and Spy0128, had an m/z 694.683+. The observed mass of this peptide corresponds exactly to the expected sum for the peptide surrounding Spy0130 Lys-120 and the C-terminal sortase motif of Spy0128, less the mass of a water molecule released by the formation of the amide bond (Fig. 3 and supplemental Table S1). The identity of this peptide confirms that the covalent linkage between AP2 and BP in GAS pili involves a conserved lysine in the AP2 proteins (Lys-120 of Spy0130 and Lys-110 of FctB, supplemental Fig. S3) that is joined to Thr-311 from the C-terminal sortase motif of the BP Spy0128.
FIGURE 3.
Identification of intermolecular amide bond by mass spectrometry. The fragmentation pattern of a parent ion (inset) of m/z 694.683+ derived from proteolysis of native GAS pili reveals the sequences flanking the amide bond that joins the C terminus of the major pilin Spy0128 to Lys-120 in the basal pilin Spy0130. Ion types are indicated, and the proposed structure of the parent ion is presented in the inset. A full list of daughter ions is in supplemental Table S1.
DISCUSSION
Molecular studies of the pili expressed by Gram-positive pathogens such as S. pyogenes and C. diphtheriae reveal a mechanism of pilus assembly completely different from that of Gram-negative bacteria (2–6, 8). They are covalent polymers, in which the pilin subunits are joined by amide bonds, whose formation is catalyzed by sortase transpeptidases. Moreover, the sortase reactions appear to be substrate-specific, in that polymerization of the major pilin to form the shaft depends on a pilus-specific sortase, encoded in the same gene cluster as the pilin subunits, whereas attachment of the pilus to the cell wall is mediated by a general (housekeeping) sortase (1, 4, 5).
Fundamental to understanding Gram-positive pilus assembly are the questions of what structural elements in the pilin subunits determine their role in the pilus, where they are incorporated into the overall assembly and by which sortase. Sortase-mediated polymerization takes place in two steps: sortase cleavage of the LPXTG motif and formation of a covalent adduct, followed by resolution of this adduct by the attack of an appropriate nucleophilic amino group (2, 4, 5). The first step has been shown in numerous studies to depend on the specific sortase motif. Thus for the GAS M1 pilins these sequences are VVPTG for Cpa, EVPTG for Spy0128, and LPSTG for Spy0130, and for the SpaA pili of C. diphtheriae they are LPLTG for SpaA and SpaC, but LAFTG for SpaB (supplemental Table S2). The similarity of the motifs displayed by the tip and backbone pilins is consistent with their recognition by the same sortase, whereas the basal pilins share a more generic LPXTG-type motif, consistent with attachment by a housekeeping sortase.
The structural determinants that select for a particular nucleophile to resolve the sortase-pilin adduct in the second step are clearly more subtle, however. For polymerization, only one lysine out of many on the pilin surface is selected. Many Gram-positive major pilins share a recognizable YPKN pilin motif that includes the essential lysine (2). Such motifs are not apparent in minor pilins, however, nor in some major pilins. For the basal pilin SpaB, for example, a lysine, Lys-139, which is essential for incorporation into the pilus, has been identified but is not part of any recognizable motif (10) and in the GAS major pilins such as Spy0128 no conserved pilin sequence motif is present (6). For anchoring to the cell wall only the adduct of the pilin with the housekeeping sortase can be resolved by the peptidoglycan amino group (10–12), again implying more extensive structural recognition between these components.
Three-dimensional structural data are mostly limited to the major pilin subunits, Spy0128 from GAS (6), SpaA from C. diphtheriae (8), and BcpA from B. cereus (15). These analyses have provided structural models for the pilus shaft, identified the intermolecular linkages between the major pilin subunits and the structural context of the lysines involved, and revealed the importance of internal isopeptide bonds for pilus stability (6, 8, 39). In contrast, the structure of only two ancillary pilins is known, RrgA from S. pneumoniae (41) and GBS52 from S. agalactiae (42). RrgA is the adhesin at the tip of the pneumococcal pilus (13, 41). The situation is less clear for GBS52. Although it was initially described as an adhesin (42), it is more likely that GBS104 from the GBS52 pilus islet forms the adhesin at the pilus tip, because it shares >50% sequence identity with RrgA (41). GBS52 on the other hand has characteristics more in keeping with basal pilins, such as a C-terminal proline-rich region.
Three-dimensional structural determinants that underlie the incorporation of the APs in GAS pili have until now largely remained unknown. There is now ample evidence as to the relative roles of the two GAS APs. AP1/Cpa has been shown to be the adhesin at the tip of the pilus, for both M3 and M1 serotypes of GAS (21).4 The Δspy0130 knock-out mutant of GAS M1 serotype shows conclusively the role of AP2/Spy0130 in anchoring the M1 pili to the cell wall,4 and there is strong evidence also from a ΔfctB knock-out in an M49 strain (22). We have not carried out similar knock-out studies on fctB of the 90/306S strain. However, the gene organization for this strain is the same as for strains bearing the FCT-2, FCT-3, or FCT-4 islets (M1, M3, M5, M9, M49, and many others (19)), i.e. cpa/ap1-lepA/sipA-fctA/bp-srtC1/C2-fctB/ap2. The AP1, BP, and AP2 proteins are clearly differentiated by sequence (19), and the AP2 proteins share high sequence identity with each other (FctB from strain 90/306S has 98% sequence identity with M49 FctB). These data indicate that these pili are assembled in an analogous manner. Functionally, the GAS AP2 pilins such as Spy0130, and by homology FctB, can thus be grouped with the pilins shown to act as the pilus cell wall anchor in other Gram-positive bacteria: SpaB from C. diphtheriae (10), GBS150 from S. agalactiae strain 2603V/R (11), PilC from S. agalactiae strain NEM316 (12) and RrgC from S. pneumoniae (13).
The crystal structure of AP2/FctB, described here, identifies several key structural features that we propose are critical to its role in the pilus assembly. Firstly, it shows that the N-terminal Ig-like domain of FctB is homologous with the N-terminal domain of the GAS major pilin Spy0128, despite minimal sequence identity (<15%). Importantly, this AP2 protein also presents a lysine residue (Lys-110 in FctB, and by homology Lys-120 in Spy0130) in exactly the same structural context as the essential lysine (Lys-161) of Spy0128. We hypothesized that, just as Lys-161 forms the amide bond to the C terminus of the next Spy0128 subunit to form the polymeric shaft, so Lys-110 of FctB (or Lys-120 of Spy0130) forms an amide bond to the C terminus of the Spy0128 subunit at the base of the pilus. This is confirmed by our mass spectral analysis of native GAS M1 pili, showing that Lys-120 is indeed joined to the C-terminal threonine from the sortase recognition motif of Spy0128 and hence to the pilus shaft. Sequence comparisons show that these lysine residues are not part of any recognizable motif analogous to the YPKN motif.
We conclude that the conserved spatial environment of this lysine in the basal pilin AP2 (FctB/Spy0130), and of the essential lysine in the BP (FctA/Spy0128) is likely to be a key structural determinant in its linkage to the proximal subunit of the pilus shaft. The same sortase, Spy0129 (SrtC2), catalyzes both the polymerization of the BP Spy0128 and the attachment of AP2 Spy0130 to the last BP subunit of the shaft. Sortase recognition must depend both on the specific C-terminal sequence motif (LPXTG or a variant) and on the structural context of the lysine that resolves the thioester intermediate. The structural homology between the N-terminal domains of the BP and AP2 pilins explains why the same sortase can act on both; in both proteins the lysine side chain projects from an Ω-loop that interrupts β-strand G (Fig. 2) and is supported by contact with a conserved Val/Ile residue (Ile-101 in FctB and Val-154 in Spy0128). This environment enables the linkage of AP2 to the extended BP shaft and so anchors the pili to the cell wall.
A second striking feature of the structure of FctB is the C-terminal PPII-type helix that extends from the N-terminal Ig-like domain. The sequences of the basal pilins Spy0130 from S. pyogenes strain SF370, SpaB from C. diphtheriae, GBS150 from S. agalactiae strain 2603V/R, and PilC from S. agalactiae strain NEM316 all possess proline-rich C-terminal domains prior to the LPXTG sortase motif (Table 3 and supplemental Table S2). The presence of a proline-rich C-terminal tail is not unique to pilin proteins and can be found in other cell wall-anchored proteins (37), although it is by no means universal. Because only one pilin protein per pilus operon appears to have a proline-rich tail, we further propose that such domains prior to a sortase motif provide a common structural motif involved in cell wall anchoring. This would identify FctB, SpaE, and SpaI as the cell wall anchors of their respective pilus structures (Table 3 and supplemental Table S2) and further suggests that the minor pilin GBS52 from S. agalactiae probably serves as a cell wall-anchored subunit rather than an adhesin at the tip of the pilus. The structure of GBS52 (PDB identifier 2PZ4) (42) indeed shows a shorter but similar PPII-helix at its C terminus.
TABLE 3.
C-terminal sequences preceding the sortase motif of proven and putative cell wall anchors of pili from S. pyogenes, S. agalactiae, and C. diphtheriae
The sortase motifs are underlined, and proline residues are highlighted in boldface. Note that S. agalactiae strain 2603V/R and C. diphtheriae have several pilus clusters. A full table, including the C-terminal sequences of other pilins within these pilin clusters, can be found in the supplemental material (supplemental Table S2). The NCBI accession numbers are in parentheses.
| S. pyogenes M1 strain SF370 | ||
|---|---|---|
| Spy0130 (NP_268519.1) | PEPHQPDTTEKEKPQKKRNGILPSTG | Cell wall anchor |
| S. pyogenes serotype M3 MGAS315 or M5 strain Manfredo | ||
| FctB (NP_663906.1) | VKPIPPRQPNIPKTPLPLAG | Putative cell wall anchor |
| S. agalactiae strain NEM316 | ||
| PilC (NP_735911.1) | ETPPPTNPKPSQPLFPQSFLPKTG | Cell wall anchor |
| S. agalactiae strain 2603V/R | ||
| GBS52 (NP_687666.1) | VPTPKVPSRGGLIPKTG | Putative cell wall anchor |
| GBS150 (NP_688402.1) | ETPPPTNPKPSQPLFPQSFLPKTG | Cell wall anchor |
| C. diphtheriae strain NCTC 13129 | ||
| SpaB (NP_940342.1) | PGAPNVPSVPSPPSVTSPAPKKTPPRLAFTG | Cell wall anchor |
| SpaE (NP_938628.1) | PSTPPPGHTPPLRETPGSGDEKEREQGDLALTG | Putative cell wall anchor |
| SpaI (NP_940530.1) | VPGTPKTPGKPDLPEKFRKEVTDRLGNTG | Putative cell wall anchor |
What might be the function of this PPII helix and extended tail prior to the sortase motif? Pro-rich stretches are often pivotal in protein-protein interactions (43), and PPII helices at the C terminus of cell wall-anchored proteins could play a role in interaction with other proteins such as chaperones or complexes involved in export or assembly. It is also possible that this feature is important in the interaction with the housekeeping sortase that anchors these proteins to the cell wall. Most likely, however, the PPII helix may simply ensure that a cell wall-anchored protein actually protrudes from the thick bacterial peptidoglycan. It is notable that, whereas the sortase motif of AP2 is well separated from its Ig-like domain, by this C-terminal tail, that of the BP subunits is located only a few residues after the end of their (globular) C-terminal domain.
A mixture of charged groups and prolines, which are the only common feature of PPII helices at the sequence level (44), may also be suitable for the environment of the peptidoglycan consisting of a mixture of sugar rings with charged side chains and amino acids. In a similar context, N- and C-terminal PPII helices have been described previously as “sticky arms” (45), and we hypothesize that this might be their role in the case of the basal minor pilins. Although there is some regularity in the positioning of prolines in the PPII helix of FctB, superposition onto a collagen PPII helix (Fig. 2B) shows that the exact location of the prolines does not affect the conformation of the helix.
Gene knock-out studies on the GAS M1 strain SF370 have shown that a ΔsrtA knock-out leads to the same phenotype as a Δspy0130 mutant, i.e. the release of pili into the culture medium.4 A ΔfctB knock-out similarly results in loss of pili from the cell wall (22). These data indicate that Spy0130 and other AP2 proteins are the basal pilins for GAS pili and should then be tethered to the cell wall by the housekeeping sortase SrtA (46). This is expected to require an LPXTG-type sortase motif, such as is seen for other basal pilins such as SpaB (10), GBS150 (11), and PilB (12) (Table 3). In contrast, the deviating sortase motifs of the major pilins and AP1/Cpa tip pilin (supplemental Table S2) are recognized and processed by pilin-specific sortases for polymerization into the pilus (3, 21, 47, 48).
Spy0130 does indeed have an LPXTG-type motif (Table 3). The LPLAG motif present in FctB differs slightly, in the substitution of Ala for Thr, but this T/A substitution is found in almost all sequenced AP2/FctB genes from a variety of GAS serotypes. Given the strong conservation in sequence and in the location of the ap2 gene in the FCT islets of different strains, it is reasonable to conclude that the AP2 proteins act as the basal pilins in all GAS strains, and that LPLAG is a common variant for SrtA recognition in these strains. LPXAG is found as a variant for LPXTG in S. aureus (49, 50), and the interleukin-8-protease in S. pyogenes similarly has an LPXAG motif in many strains. Moreover, structural analyses of S. aureus SrtA in complex with an LPXTG peptide analog (51) and of S. pyogenes SrtA with modeled peptide (52) both indicate that recognition is primarily dependent on the Leu-Pro dipeptide and that the Thr makes no specific interaction.
Intriguingly, FctB has no intramolecular isopeptide bond such as is found in the major pilins and in GBS52, but this is unlikely to affect the mechanical strength of the GAS pili. In Spy0128 a linear arrangement of isopeptide bonds connects the β-strands in both N- and C-terminal domains (6), and this arrangement may be crucial for resisting tensile forces exercised on the pilus structure (39, 53, 54). In FctB, however, the essential lysine that bonds to the pilus shaft resides on the final β-strand of the Ig-like domain, which is followed directly by the proline-rich domain and the LPXTG linkage to the peptidoglycan. Additional stabilization is likely to be given by its partial burial in the cell wall.
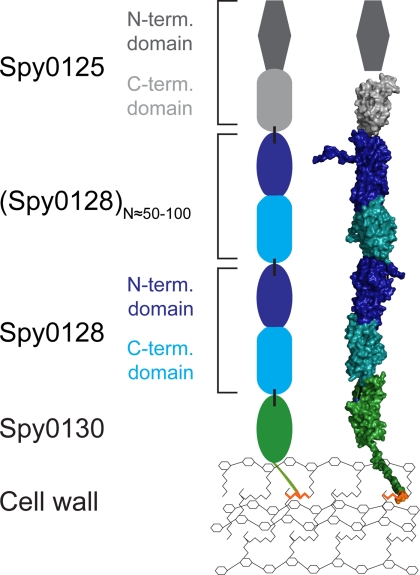
Based on these data, we constructed a model of the pilus structure and its anchoring to the cell wall (Fig. 4). Using MODELLER (36) and the coordinates of FctB, we built a homology model for Spy0130, the serotype M1 AP2 protein. We then docked the N-terminal Ig-like domain of this basal pilin on to the C-terminal domain of the BP Spy0128, based on the pilus-like docking of the N- and C-terminal domains of Spy0128 in crystals of Spy0128 (6). The four C-terminal residues of Spy0128, which were missing from the crystal structure and would terminate in the intermolecular amide linkage to Lys-120, were not modeled. Given the high sequence identity between the C-terminal domains of Spy0128 and Spy0125/Cpa (6), the latter was also modeled and docked onto the N-terminal domain of Spy0128. In this working model, Spy0128 monomers are polymerized into the pilus shaft, Spy0125 forms the tip of the pilus (21),4 and Spy0130 is at the base with its proline-rich tail likely to be buried in the cell wall and covalently linked to the peptidoglycan.
FIGURE 4.
Model for the full structure and anchoring of the pilus from S. pyogenes. The surface model (right) of the pilus was scaffolded on the crystal structure of the major pilin Spy0128 from S. pyogenes M1 (6). Models for Spy0130 and the C-terminal domain of Spy0125 were generated based on the structures of FctB and the C-terminal domain of Spy0128, respectively. The LPST sortase motif of the model of Spy0130 is highlighted in orange. Dashed lines indicate the location of intermolecular isopeptide bonds. The schematic model (left) includes data from Ref. 21 and Footnote 4. Intermolecular isopeptide bonds are indicated as black bars. Domains of similar structure are drawn as the same shape. The number of Spy0128 monomers is a rough estimate.
CONCLUSIONS
The currently available sequence and structural data make it increasingly clear that these Gram-positive pili are assembled in a modular fashion, using pilin subunits with Ig-like domains of shared evolutionary origin, arranged like beads on a string. For the GAS pili, we can build a plausible model, in which the polymeric shaft is generated by the action of a pilus-specific sortase that links a lysine residue from the N-terminal domain of one major pilin subunit to the C terminus of the next. The minor pilin at the base mimics the N-terminal domain of the major pilin, allowing it to be joined to C-terminal end of the shaft using the same recognition processes as are involved in the shaft polymerization. This minor pilin also has a proline-rich domain as a “spacer” between its Ig-like domain and the LPXTG motif that targets it to cell wall attachment. Although no structure is yet available for Cpa, it is known to have a C-terminal domain homologous with the C-terminal domain of Spy0128 (6) and to be attached to the N-terminal domain of the major pilin using the same lysine that is involved in shaft polymerization (21).4 We anticipate that similar modular principles will apply to other Gram-positive bacterial pili.
Supplementary Material
This work was supported in part by the Marsden Fund of New Zealand, the Health Research Council of New Zealand, and the Tertiary Education Commission through funding of the Maurice Wilkins Centre for Molecular Biodiscovery.
The atomic coordinates and structure factors (code 3KLQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
W. D. Smith, J. A. Pointon, E. Abbot, H. J. Kang, E. N. Baker, B. H. Hirst, J. A. Wilson, M. J. Banfield, and M. A. Kehoe, submitted for publication.
- GAS
- Group A streptococci
- FCT
- fibronectin- and collagen-binding protein and T-antigen
- AP1
- -2, ancillary pilins 1 and 2
- BP
- backbone pilin
- SeMet
- selenomethionine
- PPII
- polyproline-II
- r.m.s.
- root mean square.
REFERENCES
- 1.Proft T., Baker E. N. (2009) Cell. Mol. Life Sci. 66, 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ton-That H., Schneewind O. (2003) Mol. Microbiol. 50, 1429–1438 [DOI] [PubMed] [Google Scholar]
- 3.Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006) Nat. Rev. Microbiol. 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 5.Scott J. R., Zähner D. (2006) Mol. Microbiol. 62, 320–330 [DOI] [PubMed] [Google Scholar]
- 6.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. (2007) Science 318, 1625–1628 [DOI] [PubMed] [Google Scholar]
- 7.Budzik J. M., Marraffini L. A., Souda P., Whitelegge J. P., Faull K. F., Schneewind O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H. J., Paterson N. G., Gaspar A. H., Ton-That H., Baker E. N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16967–16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh S. Y., Budzik J. M., Schneewind O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13703–13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandlik A., Das A., Ton-That H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobbs A. H., Rosini R., Rinaudo C. D., Maione D., Grandi G., Telford J. L. (2008) Infect. Immun. 76, 3550–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konto-Ghiorghi Y., Mairey E., Mallet A., Duménil G., Caliot E., Trieu-Cuot P., Dramsi S. (2009) PLoS Pathogens. 5, e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilleringmann M., Ringler P., Müller S. A., De Angelis G., Rappuoli R., Ferlenghi I., Engel A. (2009) EMBO J. 28, 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuyama K., Narita H., Kawaguchi T., Noguchi K., Tanaka Y., Nishino N. (2007) Biopolymers 86, 212–221 [DOI] [PubMed] [Google Scholar]
- 15.Budzik J. M., Poor C. B., Faull K. F., Whitelegge J. P., He C., Schneewind O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19992–19997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., Kehoe M. A. (2007) Cell. Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 18.Manetti A. G., Zingaretti C., Falugi F., Capo S., Bombaci M., Bagnoli F., Gambellini G., Bensi G., Mora M., Edwards A. M., Musser J. M., Graviss E. A., Telford J. L., Grandi G., Margarit I. (2007) Mol. Microbiol. 64, 968–983 [DOI] [PubMed] [Google Scholar]
- 19.Falugi F., Zingaretti C., Pinto V., Mariani M., Amodeo L., Manetti A. G., Capo S., Musser J. M., Orefici G., Margarit I., Telford J. L., Grandi G., Mora M. (2008) J. Infect. Dis. 198, 1834–1841 [DOI] [PubMed] [Google Scholar]
- 20.Kratovac Z., Manoharan A., Luo F., Lizano S., Bessen D. E. (2007) J. Bacteriol. 189, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quigley B. R., Zähner D., Hatkoff M., Thanassi D. G., Scott J. R. (2009) Mol. Microbiol. 72, 1379–1394 [DOI] [PubMed] [Google Scholar]
- 22.Nakata M., Köller T., Moritz K., Ribardo D., Jonas L., McIver K. S., Sumitomo T., Terao Y., Kawabata S., Podbielski A., Kreikemeyer B. (2009) Infect. Immun. 77, 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linke C., Young P. G., Kang H. J., Proft T., Baker E. N. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 177–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 25.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 27.Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 33.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2002) The PyMOL molecular graphics system, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 36.Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., Stuart A. C., Marti-Renom M. A., Madhusudhan M. S., Yerkovich B., Sali A. (2003) Nucleic Acids Res. 31, 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solovyova A. S., Pointon J. A., Race P. R., Smith W. D., Kehoe M. A., Banfield M. J. (2010) Eur. Biophys. J. 39, 469–480 [DOI] [PubMed] [Google Scholar]
- 38.Deivanayagam C. C., Rich R. L., Carson M., Owens R. T., Danthuluri S., Bice T., Höök M., Narayana S. V. (2000) Structure 8, 67–78 [DOI] [PubMed] [Google Scholar]
- 39.Kang H. J., Baker E. N. (2009) J. Biol. Chem. 284, 20729–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollingsworth S. A., Berkholz D. S., Karplus P. A. (2009) Protein Sci. 18, 1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izoré T., Contreras-Martel C., El Mortaji L., Manzano C., Terrasse R., Vernet T., Di Guilmi A. M., Dessen A. (2010) Structure 18, 106–115 [DOI] [PubMed] [Google Scholar]
- 42.Krishnan V., Gaspar A. H., Ye N., Mandlik A., Ton-That H., Narayana S. V. L. (2007) Structure 15, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay B. K., Williamson M. P., Sudol M. (2000) FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 44.Stapley B. J., Creamer T. P. (1999) Protein Sci. 8, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williamson M. P. (1994) Biochem. J. 297, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott J. R., Barnett T. C. (2006) Annu. Rev. Microbiol. 60, 397–423 [DOI] [PubMed] [Google Scholar]
- 47.Barnett T. C., Patel A. R., Scott J. R. (2004) J. Bacteriol. 186, 5865–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnett T. C., Scott J. R. (2002) J. Bacteriol. 184, 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roche F. M., Massey R., Peacock S. J., Day N. P., Visai L., Speziale P., Lam A., Pallen M., Foster T. J. (2003) Microbiology 149, 643–654 [DOI] [PubMed] [Google Scholar]
- 50.Comfort D., Clubb R. T. (2004) Infect. Immun. 72, 2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suree N., Liew C. K., Villareal V. A., Thieu W., Fadeev E. A., Clemens J. J., Jung M. E., Clubb R. T. (2009) J. Biol. Chem. 284, 24465–24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Race P. R., Bentley M. L., Melvin J. A., Crow A., Hughes R. K., Smith W. D., Sessions R. B., Kehoe M. A., McCafferty D. G., Banfield M. J. (2009) J. Biol. Chem. 284, 6924–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeates T. O., Clubb R. T. (2007) Science 318, 1558–1559 [DOI] [PubMed] [Google Scholar]
- 54.Alegre-Cebollada J., Badilla C. L., Fernández J. M. (2010) J. Biol. Chem. 285, 11235–11242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.