Abstract
The transport of biomolecules across cell boundaries is central to cellular function. While structures of many membrane channels are known, the permeation mechanism is known only for a select few. Molecular Dynamics (MD) is a computational method that can provide an accurate description of permeation events at the atomic level, which is required for understanding the transport mechanism. However, due to the relatively short timescales accessible to this method, it is of limited utility. Here, we present a method for all-atom simulation of electric field-driven transport of large solutes through membrane channels, which in tens of nanoseconds can provide a realistic account of a permeation event that would require a millisecond simulation using conventional MD. In this method, the average distribution of the electrostatic potential in a membrane channel under a transmembrane bias of interest is determined first from an all-atom MD simulation. This electrostatic potential, defined on a grid, is subsequently applied to a charged solute to steer its permeation through the membrane channel. We apply this method to investigate permeation of DNA strands, DNA hairpins, and alpha-helical peptides through alpha-hemolysin. To test the accuracy of the method, we computed the relative permeation rates of DNA strands having different sequences and global orientations. The results of the G-SMD simulations were found to be in good agreement in experiment.
I. INTRODUCTION
Alpha-hemolysin is a bacterial toxin that self-assembles in a lipid membrane to form a water-filled transmembrane pore1. Applying a transmembrane electric potential, solutes of various molecular weights can be admitted into the channel, including rather long (up to 1300 nucleotides) single DNA and RNA strands2,3. The presence of a solute in the channel reduces the transmembrane ionic current below the open pore level, which can be used to identify the solute’s type and concentration4. Such ionic current blockades in α-hemolysin have been used to detect small organic analytes5, metal ions6, drugs7, peptides8, DNA hairpins9, modified DNA strands10, proteins11,12, and to detect rupture events in nanopore force spectroscopy experiments13,14. In the case of DNA and RNA strands, the level of the ionic current blockades depends on the sequence of the nucleotide fragment confined in the pore constriction2,15–17, which in principle can be used to create a device for high-throughput DNA sequencing18,19.
Although α-hemolysin has been successfully employed as a stochastic sensor in a number of studies, the ionic current recordings have limited utility, as the level of the ionic current blockade is determined not only by the atomic-detail structure of the solute, but also by the solute’s microscopic conformation. Hence, to unambiguously interpret the ionic current recordings, the microscopic conformation of the solute in α-hemolysin has to be determined to atomic resolution. Direct experimental visualization of solute conformation in α-hemolysin is not currently possible.
Molecular Dynamics (MD) is a computational method that can provide a realistic account of a transmembrane permeation event20–30. In this method, a biomolecular system is represented by an ensemble of particles that move and interact according to the laws of classical mechanics31. Recently, this method was employed to relate the microscopic conformations of DNA in α-hemolysin to the measured ionic current blockades, revealing that DNA’s global orientation is a significant factor determining the current blockade32. Due to the recent dramatic advances in computational technology33, the ionic conductance of α-hemolysin can now be accurately computed from all-atom MD simulations34. Nevertheless, it is still not feasible to simulate using the “brute-force” approach to the permeation of larger solutes, such as DNA and proteins, since typical permeation times of such solutes range from tens of microseconds to milliseconds. In this manuscript, we present a computational method that, in tens of nanoseconds, can provide a realistic account of a permeation event that would require a millisecond simulation using conventional MD.
One of the methods developed to overcome the timescale limitation of MD is Steered Molecular Dynamics (SMD)35,36. In this method, an external force is applied to a solute to facilitate the crossing of free-energy barriers in the channel, thus reducing the permeation timescale. The applied force is usually implemented by attaching a moving harmonic (spring-like) restraint to one or more atoms in the system35–37. The outcome of many repetitive SMD simulations can be related to the profile of the Potential of Mean Force in the channel through Jarzinsky’s identity38,39. Coarser-scale simulations of membrane transport, although able to describe the general features of the permeation process40–44, by their very design cannot reveal a permeation mechanism that is sensitive to atomic-scale details.
Although SMD is a popular method to investigate permeation of small solutes through membrane channels 30,45,46, the application of conventional SMD is troublesome in the case of long linear biopolymers, such as proteins, nucleic acids, or any other biomolecules that are or may transiently become structurally disordered during the permeation process. If a steering force is applied to a terminal of such a solute, this force is likely to distort the solute’s conformation, e.g. overstretch DNA or unfold a protein. An example of such an SMD simulation is shown in Fig. 1(a–d). Because the timescale of the SMD simulation is too short for relaxation forces to act in response to the strain produced by the steering force, the solute enters the channel in a distorted, physiologically unfeasible conformation. Applying the SMD force to the center of mass of the solute produces similar distortions, as shown in Fig. 1(e–h).
Figure 1.
SMD simulations of DNA translocation through α-hemolysin. (a–d) SMD force applied to the phosphorous atom of the first nucleotide of the DNA strand. The DNA stretches as it traverses through the pore constriction. This 3 ns simulation was done using an SMD pulling velocity of 43Å/ns and a spring constant of 500 kcal/mol·Å2. (e–h) SMD force applied to the center of mass of the DNA strand. (i–l) DNA permeation driven by a uniform electric field. This simulation was performed by applying constant forces to individual DNA atoms. The magnitude of each force was computed as a product of the atomic charge and the electric field equivalent to a 12 V transmembrane bias. The total simulation time is 4 ns. (m–p) G-SMD simulation of DNA translocation. The transmembrane bias was set to 1.2 V with the scaling factor of N = 10 (see Methods). The total simulation time is 5 ns. Animations of these trajectories are available in the supplementary information58.
An alternative method for accelerating the permeation processes is to scale up the transmembrane potential that drives the solutes across the cell membrane. This method was recently employed to simulate the permeation of ions and DNA through biological and artificial membranes32,34,47. However, the maximum transmembrane bias that can be applied in such simulations is limited by the stability of the membrane, as a lipid bilayer in an MD simulation is prone to breaking when the transmembrane bias exceeds ~2 V. This leads to ion leakage, which subsequently distorts the driving potential and thereby results in a simulation of an unrealistic permeation event. Applying a uniform external electrostatic field only to the solute’s atoms fails to produce a successful translocation, as illustrated in Fig. 1(i–l).
Here we present a generalization of the SMD methodology tailored to simulations of membrane potential-driven transport of large solutes. In order to simulate a permeation event, we amplify the three-dimensional (3-D) electrostatic potential derived from an all-atom MD simulation34 and apply it to the permeating solute only. By doing so we facilitate faster translocation of solutes without straining the structure of the channel or the membrane. Moreover, as the shape of the potential applied to the solutes in our method exactly matches the transmembrane potential driving the solutes in experiment, the field of forces is very realistic, and because the forces are distributed over the entire solute, the strain introduced into the solute’s conformation is much smaller than that introduced by conventional SMD. An example of such a simulation is shown in Fig. 1(m–p).
In this study, we apply the Grid-SMD (G-SMD) method to simulate the translocation of DNA through α-hemolysin. First, we investigate how the translocation rate of a DNA strand depends on the magnitude of the applied potential and examine the strain introduced into the DNA conformation. Next, we compare the results of G-SMD simulations to experiment by measuring the translocation velocity of DNA strands of different sequences and global orientations. Finally, we apply our method to simulate the permeation of a DNA hairpin and of an α-helical peptide through α-hemolysin, and discuss the limitations and possible artifacts of the G-SMD method.
II. METHODS
A. Microscopic model of α-hemolysin
An all-atom model of the α-hemolysin channel assembled with a lipid bilayer was constructed, simulated and tested extensively as described in Ref. 34. Atomic coordinates of α-hemolysin were taken from the Protein Data Bank (entry 7AHL). Coordinates of the atoms missing from the crystallographic structure were reconstructed using the psfgen structure building module of NAMD33. All histidine residues were assigned the HSE protonation state (pH 8.0 conditions)34. Water molecules were placed in the internal cavities of the protein using the Dowser program48. Following that, a 3 Å layer of water was created around the entire protein using the Solvate program36, which also populated the transmembrane pore and the seven side channels with water. The resulting structure was oriented in space to align the symmetry axis of the transmembrane pore with the z-axis. Next, the protein was embedded in a patch of a pre-equilibrated and solvated DPPC lipid bilayer. All lipid molecules that overlapped with the protein stem were removed, along with all water molecules around the stem of the protein that overlapped with the lipid bilayer. The protein-lipid complex was solvated in a rectangular volume of pre-equilibrated TIP3P49 water molecules. Corresponding to a solution concentration of 1 M, K+ and Cl− ions were added at random positions located at least 4 Å away from the protein, DNA and membrane, and 3 Å away from each other. The resulting system measured 135 × 137 × 148 Å3 and included 288,678 atoms. Following 2,000 steps of minimization with all protein atoms fixed, the system was equilibrated in the NpT ensemble for 1.3 ns with the backbone of the protein restrained, and for another 3.0 ns without any restraints34.
B. Microscopic models of single-stranded DNA/α-hemolysin
Single strands of DNA were threaded through the α-hemolysin pore using the phantom pore procedures described in detail in Refs. 32 and 50. A 58-base pair double-stranded DNA helix was built from individual base pairs in the geometry suggested by Quanta51. Single-stranded DNA was obtained from that structure by removing one of the strands. The remaining strand was then solvated in a pre-equilibrated volume of TIP3P water molecules; K+ and Cl− ions were added, corresponding to a 1M concentration. The resulting system was equilibrated for 12 ns. The poly(dA)58, poly(dC)58 and poly(dAdC)29 strands were constructed from the equilibrated DNA strand by replacing DNA bases with adenine or cytosine.
In order to thread a DNA strand through the α-hemolysin pore, KCl electrolyte was built around a DNA strand, conforming to the shape of the α-hemolysin pore32. The shape of the α-hemolysin pore was represented by a mathematical surface, which we refer to as a phantom pore. Initially, the phantom pore was made 2nm wider in diameter than the pore of α-hemolysin, so that the entire 58-nucleotide strand could fit into it. The pore was then gradually shrunk in a 2 ns simulation to the shape of the α-hemolysin pore. At the same time, 10 pN forces pushed all atoms laying outside of the shrinking surface toward the center of the pore. At the end of the simulation, the DNA strand adopted a straight conformation that conformed to the shape of the α-hemolysin pore. We carried out two such simulations (in the NpT ensemble) corresponding to the two global orientations of the DNA strand inside the pore. Each DNA strand, as well as the ions found in the stem region of the phantom pore, were merged with the all-atom model of the α-hemolysin channel. In the resulting structure, water and ions covered the DNA strand completely; one such system is shown in Fig. 2(a). The final systems measured 135 × 137 × 183 Å3 and included 356,065 atoms in the case of poly(dA)58, 355,952 atoms in the case of poly(dC)58 and 356,005 atoms in the case of poly(dAdC) 29. Following 2,000 steps of minimization with all DNA atoms fixed, each system was equilibrated for 2 ns in the NpT ensemble.
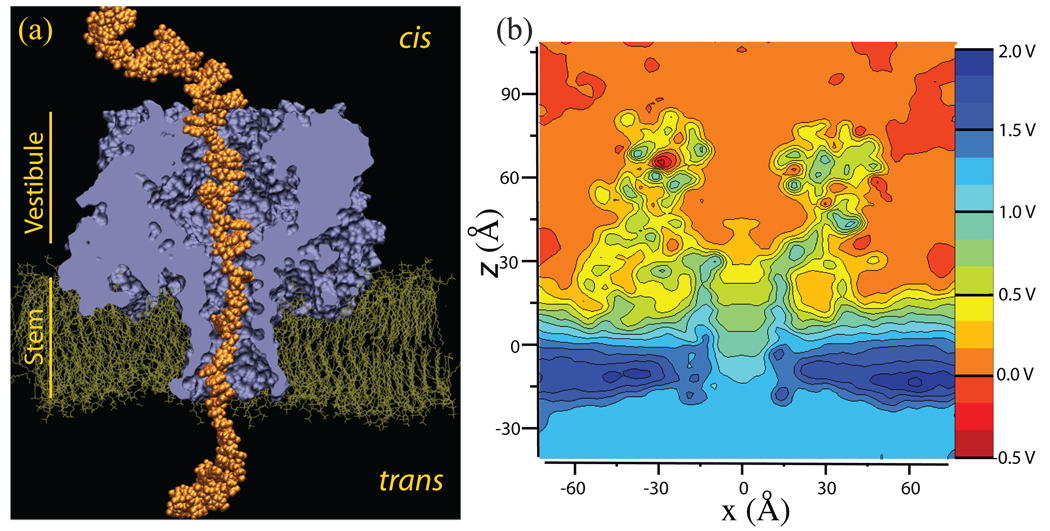
Figure 2.
The set-up of G-SMD simulations. (a) Microscopic model of the α-hemolysin/DNA system suspended in a lipid bilayer membrane. The DNA atoms are drawn as orange spheres. The channel is drawn as a molecular surface (violet) separating the protein from the membrane and water. This surface is cut by a plane perpendicular to the lipid bilayer, passing through the geometrical center of the protein. The DPPC lipid bilayer is shown in green, water and ions are not shown. The stem and vestibule sections of the protein are indicated; the constriction, the narrowest part of the channel, lies between them. The model comprises of 356,065 atoms. (b) The electrostatic potential map of α-hemolysin. The figure shows a cut through the averaged (over a 5.3 ns MD simulation and the sevenfold symmetry of α-hemolysin) electrostatic potential along the z-axis. A 1.2 V transmembrane bias was applied in this simulation. This 3-D potential is used in our G-SMD simulations to drive translocation of DNA through α-hemolysin.
C. Microscopic model of Z-DNA hairpin/α-hemolysin
The atomic coordinates of a 5’-d(CGCGCGTTTTCGCGCG)-3’ Z-DNA hairpin were taken from the Protein Data Bank (code 1D16). Two CG base pairs were added at the end of the double-stranded portion of the hairpin. The resulting structure was placed in a 0.13M aqueous solution of KCl, and, after a short minimization, was equilibrated for 5 ns at 295K in the NpT ensemble. During the equilibration, the root mean square deviation of the DNA structure from the X-ray model fluctuated within 1.75 Å. The equilibrated model of the hairpin was merged with α-hemolysin, placing the hairpin into the opening of the channel that connects the vestibule with the cis compartment (see Fig. 2(a) for the cis/trans convention used). The termini of the hairpin were located at the level of the Asp4 ring of the channel, whereas the loop of the hairpin protruded outside of the vestibule. The number of ions in the system was adjusted to ensure the system’s overall neutrality. The final system (288,618 atoms) was minimized keeping coordinates of all DNA atoms fixed, and equilibrated for 0.7 ns at 295 K. For the first 50,000 steps (50 ps) of the equilibration, harmonic restraints were applied to the protein, DNA and the lipid bilayer.
D. Microscopic model of α-helical peptide/α-hemolysin
A microscopic model of an α-helical peptide was constructed by mutating the sequence of a template α-helix (Protein Data Bank code 1HTM) into the desired sequence: MLSRQQSQRQSRQQSQRQSRYLL. The resulting structure was equilibrated at 310K in 1M KCl solution for 8 ns in the NpT ensemble. During the equilibration the peptide preserved its α-helical structure (95% of the total structure). The equlibrated peptide was aligned with the symmetry axis of α-hemolysin. The N-terminal of the peptide was placed at the level of the Lys131 ring, which is located near the trans side-entrance into the channel. All water molecules within 1.5 Å of the peptide were removed. The resulting system was neutral and comprised 288,743 atoms. The system underwent 2000 steps of minimization, followed by gradual heating from 0 to 298K in 10 ps, and subsequent 80 ps equilibration. For the first 50 ps of the equilibration coordinates of the peptide were restrained by harmonic constraints.
E. MD methods
All our MD simulations were performed using the program NAMD33, periodic boundary conditions and Particle Mesh Ewald (PME) full electrostatics52 with a dielectric constant ε = 1. PME was computed using a grid spacing of 007E;1.1 Å per grid point in each dimension. Systems comprised entirely of water, ions and nucleic acids were simulated using the AMBER9553 force field; the CHARMM2754 force field was employed for all other systems. The temperature was kept at 295K by applying Langevin forces55 to all heavy atoms; the Langevin damping constant was set to 1 ps−1. The integration timestep chosen was 1 fs. The equilibration in the NpT ensemble was performed using the Nosé-Hoover Langevin piston pressure control56 at 1 bar. Van der Waals energies were calculated using a smooth (10–12 Å) cutoff. Restraints were imposed by harmonic forces; the force constants were set to 1 kcal/mol· Å2. All simulations in an external electric field were carried out in the NVT ensemble.
F. Electrostatic potential
The procedures for determining the average distribution of an electrostatic potential from MD trajectories were described in detail elsewhere34. A 5.3 ns simulation of the α-hemolysin system containing no DNA was carried out while applying a uniform external electric field equivalent to a 1.2 V bias. The simulation produced 5,300 snapshots of the system configuration. For every configuration, an instantaneous distribution of the electrostatic potential was computed using the PME electrostatics module of NAMD33. The instantaneous potentials were averaged over the entire MD trajectory and over the sevenfold symmetry of α-hemolysin. A slice through such a potential along the xz-plane is shown in Fig. 2(b). The electrostatic potential used in our G-SMD simulation was computed over a 96 × 96 × 96 grid using gaussians of width β = 0.395 Å−1.
G. Grid-SMD simulations
To carry out G-SMD simulations, the systems were simulated in the NVT ensemble, applying an external electric field equivalent to a 1.2 V bias. Steering forces derived from the 3-D distribution of the electrostatic potential were applied to all atoms of DNA using either a tclforces script or a custom version of NAMD. These forces were computed from the finite differences of the potential and were scaled with the charge of the DNA atoms and with the scaling factor N. Only the z-components of the forces were applied. In our G-SMD simulations, we varied the scaling factor N from 0 to 10, thereby changing the effective bias from 1.2 to 13.2 V in the z-direction (effective bias is (N + 1) × 1.2 V). No steering force was applied in the xy-plane. To keep the potential aligned with the protein, the latter was restrained through harmonic forces applied to the α-carbon atoms. Future versions of NAMD will incorporate G-SMD capabilities.
H. Rate of DNA transport
To quantitatively characterize translocation of DNA through α-hemolysin, we adapted the method used for computing ionic currents34,47,57. An instantaneous current of DNA nucleotides was computed as
| (1) |
where zi and mi are the z-coordinate and the mass of atom i, respectively, L is the extension of the system along the direction of the current, and MDNA is the mass of one nucleotide. The sum in Eq. 1 runs over all DNA atoms in the volume of interest, which in our case was either the entire channel or the channel’s constriction. To compute the average translocation velocity at a given bias, instantaneous currents of DNA nucleotides IDNA(t) were first integrated with respect to time to produce a cumulative current curve; applying a linear regression fit to the cumulative current curve yielded the average translocation velocity.
III. RESULTS
After the systems comprising DNA, α-hemolysin, lipid bilayer membrane, water and ions were assembled and equilibrated (see Methods), external fields of different magnitudes were applied to the DNA, and the resulting displacements of the DNA in the channel were recorded. All simulations were carried out applying an electric field equivalent to a 1.2 V transmembrane bias to all atoms in the system34,47,57. In addition, the z-component of the forces derived from the average distribution of the electrostatic potential were multiplied by the scaling factor N and applied to DNA atoms only. The steering forces were not applied in the x- and y-directions. A table summarizing the results of all MD simulations performed is provided in the supplementary information58.
First, we investigate how the introduction of a steering potential influences the velocity of DNA translocation. In Fig. 3 we plot the cumulative number of permeated nucleotides against the simulation time for four G-SMD runs in which the steering field was applied with the scaling factor N of 1, 3, 7 and 10. In the same plot we display the results of a simulation without a steering field, i.e., N = 0 (a 1.2 V transmembrane bias was applied in all simulations). The DNA was observed to permeate the channel faster at larger steering fields. In the inset to Fig. 3 we plot the average DNA translocation velocity against the effective bias applied, i.e., (N + 1) × 1.2 V. The DNA translocation velocity was observed to scale superlinearly with the effective bias until N = 3. Note that at N = 3, the translocation velocity is about 1 nucleotide per nanosecond, approximately 1000 times faster than in experiment.
Figure 3.
Dependence of DNA translocation velocity on steering potential. The total number of nucleotides translocated is plotted against the simulation time. Black triangles, white triangles, diamonds, and squares indicate simulations carried out at a 1.2 V electrostatic field magnified 11, 8, 4 and 2 times, respectively; the circles correspond to the simulation carried out at a 1.2 V bias (no steering force applied). (Inset) Tranlocation velocity versus the effective bias. These simulations were carried out using a poly(dA)58 strand in the A5’ orientation (5’ end of DNA on the trans side; see Fig. 2(a)).
In order to assess the conformational strain introduced by our method, we analyzed the number of nucleotides in the vestibule, constriction and stem (see Fig. 2(a)) as a function of time. The results of this analysis are shown in Fig. 4. We found that for N = 1 or 3, the number of nucleotides in the stem remained nearly constant, indicating that virtually no conformational strain was introduced (the rising number of nucleotides in the vestibule results from DNA relaxation; compare the N = 0 case.) The strain remained modest for higher values of N. We thus conclude that N = 3, i.e., an effective bias of 4.8 V, is optimal for G-SMD simulations, as it does not distort the conformation of the DNA in α-hemolysin while still substantially accelerating its movement. In all simulations, the hydrophobic stacking of the DNA bases is broken in the constriction, as it is too narrow for the DNA strand to fit in in the base-stacked conformation. In the stem and in the vestibule the DNA strand is split into groups of 2–4 nucleotides that preserve the base-stacking pattern. The structure of the lipid bilayer and of the protein is unaffected by the steering potential, as it is applied to atoms of DNA only.
Figure 4.
Influence of the driving potential on the conformation of DNA in α-hemolysin. The number of nucleotides in the vestibule (a) and in the stem (b) of the channel are shown as functions of the simulation time. The symbols indicate the scaling of the driving potential: N = 0 (circles), N = 1 (squares), N = 3 (diamonds), N = 7 (white triangles) and N = 10 (black triangles). Each trace for N = 7 or 10 is an average over four trajectories. These simulations correspond to DNA in the A5’ orientation (5’ end of DNA on the trans side; see Fig. 2(a)).
We next investigate whether G-SMD can provide quantitative insights into the mechanics of DNA translocation, despite the dramatic reduction of the translocation timescale. It has been experimentally shown that the global orientation of a DNA strand in the α-hemolysin channel has a deterministic effect on the velocity of DNA translocation: when a single DNA strand of adenine nucleotides (poly(dA) strand) is driven by a transmembrane bias32, the translocation velocity of the DNA strand in the direction of its 3’-end is about 16% higher than that in the direciton of its 5’-end. To test whether G-SMD can capture this rather subtle difference of the translocation velocities, we set up two systems in which a poly(dA)58 strand was threaded through α-hemolysin in two global orientations, (cis) 3’-dA58-5’ (trans) (referred to as A5’) and (cis) 5’-dA58-3’ (trans) (A3’). Refer to Fig. 2 for the cis/trans convention used in this paper. For each orientation of the poly(dA)58 strand, we carried out four G-SMD simulations applying an effective bias equivalent to 9.6 V, and four simulations at 13.2 V. In Fig. 5, we plot the cumulative number of translocated nucleotides for the poly(dA)58 systems at a 13.2 V effective bias, indicating by the color of the lines the orientation of the DNA strand in the pore. In the same figure, we plot a linear regression fit to the cumulative current curves averaged over the four simulations (symbols). The slope of the fit yields the average translocation velocity of 11.2 ± 0.4 nucleotides/ns for A3’ and 8.8 ± 0.4 nucleotides/ns for A5’; the error is the 90% confidence interval estimated from the standard deviation of the current traces of the four runs taken together. The same systems at a 9.6 V effective bias produced translocation velocities of 7.4 ± 0.2 nucleotides/ns for A3’ and 6.4 ± 0.2 nucleotides/ns for A5’, or a ratio of 1.16 ± 0.05, in excellent agreement with the experimental ratio of 1.16 ± 0.05.32 Hence, we conclude that although the absolute values of the translocation velocities are much higher than the experimental ones, their ratio is in agreement with experiment32.
Figure 5.
Effect of global orientation on DNA translocation velocity. The total number of nucleotides translocated is plotted against the simulation time. The chart plots the results of eight simulations, four for each of the two global orientations of a poly(dA)58 strand. Red and blue lines correspond to A3’ (3’ end of DNA on the trans side; see Fig. 2(a)) and A5’ orientations of the strand; squares and circles indicate a linear regression fit to the above sets of four trajectories. The insets illustrate the conformations of a poly(dA)58 strand in the α-hemolysin pore.
It was suggested in Ref. 32 that tilting of the DNA bases towards the 5’-end of the strand in the constriction of α-hemolysin is the molecular origin of the observed directionality of the DNA translocation. However, the conventional MD simulation employed in that study could sample only a rather small part of the translocation trajectory, in which the displacement of DNA originated mostly from the relaxation of the DNA conformation in response to the applied electric field. Using G-SMD we could, for the first time, not only simulate the entire translocation of a DNA strand through α-hemolysin but also carry out our simulations multiple times. Visual analysis of the MD trajectories confirmed the original suggestion of Mathé et al.32 that reorientation of the DNA bases in the constriction of α-hemolysin is responsible for the observed directionality. In the insets to Fig. 5 we display conformations of two DNA strands identical in sequence but different in their global orientations. When a DNA strand enters the pore with its 5’-end first, the base stacking is more likely to be disrupted due to the preferential tilt of the DNA bases towards the 5’-end in the constriction. Animations illustrating translocation of DNA through α-hemolysin in both orientations are available in the supplementary information58.
Another factor that can affect the velocity of DNA translocation is the sequence of the DNA strand. The most probable translocation rates of poly(dA)100, poly(dAdC)50 and poly(dC)100 single-stranded DNA at 25°C were measured at 1.92, 1.36 and 0.76 µsec/base, respectively, at a 120 mV driving bias15. The global orientation of the strands was not resolved in those experiments. To investigate whether G-SMD could capture the effect of the DNA sequence on the translocation velocity, we carried out twelve additional simulations on the system that included a poly(dC)58 strand threaded through α-hemolysin in two global orientations, (cis) 3’-dC58-5’ (trans) (C5’) and (cis) 5’-dC58-3’ (trans) (C3’), as well as a system of poly(dAdC)29 in one orientation, (cis) 5’-(dAdC)29-3’ (trans) (AC3’). These simulations were carried out using a steering potential of intermediate strength (N = 7) corresponding to a 9.6 V effective bias. Four simulations at N = 3 were also conducted. In Fig. 6 we plot the cumulative currents for each of the three sequences in the same global orientation. We found that the poly(dC)58 strand permeates the pore faster than the poly(dAdC)29 strand, which in turn permeates the pore faster than poly(dA)58, which is in qualitative agreement with experiment15. As the global orientation of poly(dC) strands was not resolved in experiment, direct quantitative comparison between simulation and experiment is not possible. Nevertheless, we note that the ratios of the translocation velocities obtained with G-SMD are systematically smaller than the ratios of the most probable translocation times observed in experiment. For example, in our simulations we observed poly(dC)58 to move about 1.65 times faster than poly(dA)58 in the direction of the 3’-end of the strand, whereas the ratio of the most probable translocation times is about 2.5 in experiment15. At this time we do not know the exact reason for this quantitative discrepancy. We note, however, that our G-SMD simulations were designed to minimize the interactions between DNA and the cap of α-hemolysin, which can arrest the translocation process (see supplementary information58.) Also, we observed no anomalously long translocation events for poly(dA) that, in experiment, result in a very broad distribution of the poly(dA) translocation times.
Figure 6.
Effect of sequence on DNA translocation velocity. The total number of nucleotides translocated is plotted against the simulation time. The chart plots the results of 12 simulations, 4 for each of poly(dC)58, poly(dAdC)29 and poly(dA)58, all in the (cis) 5’-3’ (trans) orientation (see Fig. 2(a)). Traces of the four trajectories are shown for each sequence, represented by thin lines, as well as linear regression fits indicated by black diamonds, red squares and blue circles, corresponding to poly(dC)58, poly(dAdC)29 and poly(dA)58, respectively.
The ratio of the translocation velocities at different strengths of the steering potential is shown in Fig. 7. As in the case of poly(dA)58, for poly(dC)58 we observed faster translocation in the direction of the 3’ end of the strand at both N = 3 and N = 7. The ratio of the 3’- to 5’-first translocation velocities for poly(dA)58 is in the 1.1–1.2 range for N = 3, 7 and 10. Overall, the ratio of the translocation velocities was not observed to change dramatically with the scaling of the steering potential for the scaling factors considered. We note, however, that for very large steering potentials, the hydrodynamic drag force on DNA should become comparable to the friction forces between the stem of α-hemolysin and DNA, which obviously will reduce the influence of the sequence on the translocation velocity.
Figure 7.
DNA current ratios. Selected DNA current ratios are shown comparing orientation or sequence at different effective biases, e.g., IC3’/IC5’ is the ratio of the nucleotide current of the (cis) 5’-dC58-3’ (trans) system to that of the (cis) 3’-dC58-5’ (trans) system. Error bars indicate 90% confidence interval estimated from the standard deviations of the current traces.
Some of the permeation traces noticeably deviate from a straight line; an exemplary trajectory is presented in Fig. 8. In some parts of the trajectory, the velocity of DNA translocation changes abruptly. Such “steps” in the DNA translocation, however, do not correspond to the translocation of individual nucleotides, as there are fewer steps in the translocation traces than nucleotides translocated through the pore. To verify that our procedure for computing DNA currents, which involves integration over the entire volume of α-hemolysin, did not conceal some events that could be associated with the translocation of individual nucleotides, we computed the DNA current through the constriction of α-hemolysin only. One such trace in shown Fig. 8 as a dashed line. The steps in the resulting traces are sharper, but the number of steps remains the same. Visual inspection of the translocation trajectories with VMD59 revealed that the steps are associated with broken base stacking of the DNA strand entering the constriction, indicated by the arrows in Fig. 8. Two example base conformations are shown as insets. Thus, our G-SMD simulations suggest that permeation of DNA through the constriction of α-hemolysin takes place in pulses of 2–4 nucleotides that preserve the base stacking pattern of an unconfined strand.
Figure 8.
Steps in DNA current traces. Exemplary traces of DNA current are shown, both through the entire channel (solid line) as well as through the constriction only (dashed line). Rather than a smooth linear trace, simulations of this system displayed a pronounced step-like current. Inspection with the program VMD59 revealed the steps to be associated with breaks in the base stacking structure of the DNA entering the protein constriction, causing temporary halts in the translocation followed by surges of DNA current. Times at which the base stacking was broken in the constriction are marked by the arrows. The insets show the conformations of the nucleotides in the constriction (Thr145 to Val149) for the structure breaks at 0 ns (left) and 1.9 ns (right). This simulation corresponds to DNA in the A5’ orientation (5’ end of DNA on the trans side; see Fig. 2(a)) at a 9.6 V effective bias.
To further explore the capabilities of the G-SMD method, we attempted to simulate permeation of a DNA hairpin and of an α-helical peptide through α-hemolysin. Figs. 9 and 10, as well as the animations available in the supplementary information58, illustrate the resulting MD trajectories.
Figure 9.
G-SMD simulation of DNA hairpin translocation through α-hemolysin. This simulation was performed using the scaling factor N = 15. The DNA hairpin is shown in yellow. The snapshots illustrate the conformation of the hairpin at the beginning of the simulation (a), and after 1.4 (b), 2.0 (c), and 4.2 ns (d).
Figure 10.
G-SMD simulation of peptide translocation through α-hemolysin. For the first 17.7 ns of this simulation the scaling factor N was set to 10. The scaling factor was increased to 20 for the last 3.8 ns. The peptide atoms are shown as van der Waals spheres colored according to the residue type: positively charged (blue), uncharged polar (green), and non-polar (white). The snapshots show the conformation of the peptide at the beginning of the simulation (a), and after 3 (b), 17.7 (c), and 21.5 ns (d).
It has been suggested that the loop of the hairpin is too big to fit through the opening in the α-hemolysin cap9, and that, in order to permeate through the pore, the hairpin has to unzip. To test whether our G-SMD method can capture these key features of the permeation kinetics, we placed a DNA hairpin half-way through the entrance to α-hemolysin’s vestibule from the cis side, such that the hairpin loop protruded outside of the pore, as shown in Fig. 9(a). Using the standard protocol of the G-SMD method, we applied an external potential to steer permeation of the hairpin, using a rather large scaling factor of N = 15 (the translocation times of DNA hairpins in experiment are in the millisecond range). Within the first nanosecond of the simulation, the hairpin was observe to slide into the cap’s mouth; the terminal base pair of the hairpin frayed within the first 0.4 ns of the simulation. The translocation halted, however, when the loop reached the level of Lys8 residues of α-hemolysin. After about 1.2 ns, one end of the DNA was captured by the constriction of the channel. The electrostatic force exerted on the captured nucleotides pulled down the rest of the strand, as shown in Fig. 9(b). At this time, half of the hairpin loop slid down into the vestibule and stem as the hairpin unzipped. The unzipped hairpin moved quickly through the constriction in noticeable steps. At one point more than one DNA strand was observed to pass through the constriction of α-hemolysin, which could be an artifact of using a steering potential that is too high. Future simulations will reveal if such events can indeed take place. After about 2.0 ns, all DNA left the vestibule of the pore, shown in Fig. 9(c). Within the next nanosecond, the hairpin slid down the α-hemolysin stem and adhered to the ring of charged residues near the exit from the stem on the trans side, shown in Fig. 9(d). Although a more careful simulation using a smaller scaling factor needs to be performed before any definitive conclusion about the kinetics of hairpin translocation can be drawn, this preliminary simulation demonstrates the wealth of information that can be obtained with G-SMD.
The last system that we used to explore the capabilities of G-SMD included a 23-residue α-helix that was initially placed near the entrance to the α-hemolysin stem from the trans side, as shown in Fig. 10(a). As the total charge of the peptide is +4e, we used the same potential as that used to induce DNA translocation from the cis side. Under a steering field with N = 10, the peptide was observed to move very quickly into the pore, until its N-terminus reached the pore’s constriction, as shown in Fig. 10(b). The helix maintained its α-helical structure. For the next 14.7 ns, the peptide moved very slowly through the constriction region of the pore. In order to squeeze through the constriction, a few turns of the α-helix unwound. Although after 17.7 ns more than a half of the peptide’s residues passed through the α-hemolysin constriction (Fig. 10(c)), the translocation appeared to be arrested due to the interactions between the charged groups of the peptide and the charged groups of α-hemolysin at the bottom of the vestibule. Increasing the strength of the steering field by a factor of two (N = 20) did not appear to dramatically accelerate the translocation. The artifacts of such a strong steering potential are evident in Fig. 10(d): the charged residues of the peptide aligned along the field lines of the applied potential. Although in this simulation we were not able to observe a complete permeation event, the simulation revealed the transformation of the secondary structure of the peptide in the stem and the constriction regions of α-hemolysin. A longer simulation with a smaller steering field is required to reveal how the peptide leaves the pore through the vestibule of α-hemolysin.
In both simulations, one common artifact of the G-SMD simulations became apparent: under some conditions, the translocation can get arrested in one of the local minima of the steering potential. Adding a stochastic component to the driving potential should enable escape from such local minima, which is the subject of ongoing research.
IV. DISCUSSION AND CONCLUSIONS
We have developed and tested the extension of the Steered Molecular Dynamics method to simulations of electric field-driven permeation of large solutes through membrane channels. In tens of nanoseconds this method can provide a realistic account of permeation events that would require millisecond simulations with standard MD. The decoupling of the electric field driving the solutes through the channel from the electric field acting on the rest of the system allows us to apply high effective biases without affecting the system’s integrity, greatly reducing the permeation timescale. As the SMD forces in our method act on the entire solute via a smooth 3-D potential, the strain introduced into the solute’s conformation is small, comparable to the strain observed during a standard MD simulation.
Using the G-SMD method we investigated the mechanics of DNA permeation through α-hemolysin. Our simulations revealed that the base-stacking of nucleotides in single DNA strands has a significant effect on the DNA translocation velocity in α-hemolysin. Our G-SMD results support the molecular mechanism proposed in Ref. 32 to explain the dependence of the DNA translocation velocity on the global orientation of the DNA strands in the channel, i.e., the preferential tilt of the DNA bases towards the 5’-end of the strand near the channel’s constriction. Due to the dramatic reduction of the translocation timescale and small distortions that G-SMD introduces into the solute’s conformation, we could carry out multiple simulations of the same permeation process and characterize the outcome in statistical terms. The outcome of such simulations can provide the sequence-specific information required for a realistic coarser-scale description of the translocation process40–44,60–63.
Our initial motivation for developing the G-SMD method was to enable characterization of the conformations of large solutes during their transport through α-hemolysin. The results of our simulations demonstrate that the G-SMD method can adequately describe permeation not only of linear DNA strands but also of DNA hairpins and small peptides. The ensemble of conformations resulting from a G-SMD run can be related to the measured ionic current blockages using standard MD34 or other methods64,65. Next, we investigated if, in addition to screening possible conformations of the solute in the pore, the G-SMD method can adequately describe permeation kinetics and relative translocation velocities. For this purpose we carried out G-SMD simulations on DNA strands of different sequences and global orientations. The obtained ratios of the simulated translocation velocities of DNA strands in different global orientations were in excellent quantitative agreement with experiment. The ratio of the translocation velocities of DNA homopolymers of different sequence were found in semi-quantiative agreement with experiment. Such good agreement with experiment is encouraging given that our method reduces the permeation time 1000-fold.
G-SMD has proven to be an effective technique for probing certain processes that cannot be accurately described using SMD. As with SMD, however, the magnitude of the steering forces must be carefully chosen to ensure that the simulated trajectories correspond to experiment. Ideally, the magnitude of the applied forces and the timescale of the simulation should allow relaxation forces to dissipate the strain introduced by the G-SMD protocol.
The G-SMD method extends the applicability of atomic-scale simulations to processes that were previously off-limits. Among the topics to be addressed in the future are mechanisms of protein translocation through membrane channels, selectivity of ion channels and transporters, simulations of nanopore devices for high-throughput sequencing of DNA and proteins, and, eventually, the transport of biomolecules in nanofluidic systems. In addition to studies of membrane transport, the G-SMD method is expected to find application in multiscale modeling, cryo-electron microscopy data fitting, in developing implicit models of biomaterials, and for simulations of nanoscale hydrodynamics. The G-SMD method will be further developed to enhance random forces producing stochastic motion of the solutes in the channel, to employ a self-consistent electrostatic potential (i.e., one calculated during the G-SMD simulation) to steer permeation of solutes, and to provide methods for extracting the Potential of Mean Force from G-SMD trajectories.
Supplementary Material
Acknowledgments
This work was supported by the grants from the National Institutes of Health (PHS 5 P41 RR05969 & R01-HG003713), and the Department of Physics at the University of Illinois. The authors gratefully acknowledge supercomputer time at the Pittsburgh Supercomputer Center and the National Center for Supercomputing Applications provided via Large Resources Allocation Committee grant MCA05S028, as well as time on the Turing cluster at the University of Illinois.
References
- 1.Bhakdi S, Tranum-Jensen J. Microbiol. Rev. 1991;55:733. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc. Natl. Acad. Sci. USA. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akenson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys. J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezrukov SM. J. Membr. Biol. 2000;174:1. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]
- 5.Gu Q, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 6.Braha O, et al. Nature Biotech. 2000;18:1005. doi: 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- 7.Kang XF, Cheley S, Guan XY, Bayley H. J. Am. Chem. Soc. 2006;128:10684. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 8.Movileanu JSL, Schmittschmitt JP, Bayley H. Biophys. J. 2005;89:1030. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vercoutere W, et al. Nature Biotech. 2001;19:248. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Dunning JE, Huang APH, Nyamwanda JA, Branton D. Proc. Natl. Acad. Sci. USA. 2004;101:13472. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Henrickson SE, Weetall HH, Robertson B. Anal. Chem. 2001;73:2268. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 12.Winters-Hilt S, Akeson M. DNA and Cell Biology. 2004;23:675. doi: 10.1089/dna.2004.23.675. [DOI] [PubMed] [Google Scholar]
- 13.Tropini C, Marziali A. Biophys. J. 2007;95:1632. doi: 10.1529/biophysj.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornblower B, et al. Nature Mater. 2007;4:315. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 15.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc. Natl. Acad. Sci. USA. 2000;97:1079. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashkenasy N, Sànchez-Quesada J, Bayley H, Ghadiri MR. Angew. Chem. Int. Ed. Engl. 2005;44:1401. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Astier Y, Braha O, Bayley H. J. Am. Chem. Soc. 2006;128:1705. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 18.Deamer D, Branton D. Acc. Chem. Res. 2002;35:817. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 19.Bayley H. Curr. Op. Chem. Biol. 2006;10:628. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 20.de Groot BL, Grubmüller H. Science. 2001;294:2353. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 21.Tajkhorshid E, et al. Science. 2002;296:525. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 22.Ilan B, Tajkhorshid E, Schulten K, Voth GA. Proteins: Struct., Func., Bioinf. 2004;55:223. doi: 10.1002/prot.20038. [DOI] [PubMed] [Google Scholar]
- 23.Bernèche S, Roux B. Nature. 2001;414:73. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 24.Im W, Roux B. J. Mol. Biol. 2002;319:1177. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 25.Domene C, Bond PJ, Sansom MSP. In: Membrane protein simulations: Ion channels and bacterial outer membrane proteins, in APC. Richards FM, Eisenberg DS, Kuriyan J, editors. volume 66. New York: Elsevier Academic Press; 2003. pp. 159–193. [DOI] [PubMed] [Google Scholar]
- 26.Allen T, Andersen O, Roux B. Proc. Natl. Acad. Sci. USA. 2004;101:117. doi: 10.1073/pnas.2635314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monticelli L, Robertson K, MacCallum J, Tieleman D. FEBS Lett. 2004;564:325. doi: 10.1016/S0014-5793(04)00271-6. [DOI] [PubMed] [Google Scholar]
- 28.Noskov S, Berneche S, Roux B. Nature. 2004;431:830. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 29.Robertson KM, Tieleman DP. FEBS Lett. 2002;528:53. doi: 10.1016/s0014-5793(02)03173-3. [DOI] [PubMed] [Google Scholar]
- 30.Ceccarelli M, Danelon C, Laio A, Parrinello M. Biophys. J. 2004;87:58. doi: 10.1529/biophysj.103.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen MP, Tildesley DJ. Computer Simulation of Liquids. New York: Oxford University Press; 1987. [Google Scholar]
- 32.Mathè J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Proc. Natl. Acad. Sci. USA. 2005;102:12377. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JC, et al. J. Comp. Chem. 2005;26:1781. [Google Scholar]
- 34.Aksimentiev A, Schulten K. Biophys. J. 2005;88:3745. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isralewitz B, Izrailev S, Schulten K. Biophys. J. 1997;73:2972. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grubmüller H, Heymann B, Tavan P. Science. 1996;271:997. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 37.Marrink S-J, Berger O, Tieleman P, Jähnig F. Biophys. J. 1998;74:931. doi: 10.1016/S0006-3495(98)74016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarzynski C. Phys. Rev. Lett. 1997;78:2690. [Google Scholar]
- 39.Park S, Schulten K. J. Chem. Phys. 2004;120:5946. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 40.Muthukumar M, Kong CY. Proc. Natl. Acad. Sci. USA. 2006;103:5273. doi: 10.1073/pnas.0510725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong Y, Shen Y, Warth TE, Ma J. Proc. Natl. Acad. Sci. USA. 2002;99:5999. doi: 10.1073/pnas.092051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Peng E. Appl. Phys. Lett. 2003;82:1308. [Google Scholar]
- 43.Milchev A, Binder K, Bhattacharya A. J. Chem. Phys. 2004;121:6042. doi: 10.1063/1.1785776. [DOI] [PubMed] [Google Scholar]
- 44.Kostev S, Kolomeisky A. J. Chem. Phys. 2006;125:084906. doi: 10.1063/1.2338539. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Schulten K, Tajkhorshid E. Structure. 2005;13:1107. doi: 10.1016/j.str.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Gumbart J, Schulten K. Biophys. J. 2006;90:2356. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aksimentiev A, Heng JB, Timp G, Schulten K. Biophys. J. 2004;87:2086. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Hermans J. Proteins: Struct., Func., Gen. 1996;24:433. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 50.Heng JB, et al. Biophys. J. 2006;90:1098. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polygen Corporation, 200 Fifth Av., Waltham, MA 02254, Quanta. 1988. [Google Scholar]
- 52.Batcho PF, Case DA, Schlick T. J. Chem. Phys. 2001;115:4003. [Google Scholar]
- 53.Cornell WD, et al. J. Am. Chem. Soc. 1995;117:5179. [Google Scholar]
- 54.MacKerell AD, Jr, et al. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 55.Brünger AT. X-PLOR, Version 3.1: A System for X-ray Crystallography and NMR. Yale University: The Howard Hughes Medical Institute and Department of Molecular Biophysics and Biochemistry; 1992. [Google Scholar]
- 56.Martyna GJ, Tobias DJ, Klein ML. J. Chem. Phys. 1994;101:4177. [Google Scholar]
- 57.Crozier PS, Henderson D, Rowley RL, Busath DD. Biophys. J. 2001;81:3077. doi: 10.1016/S0006-3495(01)75946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.See EPAPS Document No.__________ for details of all simulations, information on an arrested translocation, and movies of DNA and peptide permeation. This document may be retrieved via the EPAPS homepage (http://www.aip.org/pubservs/epaps.html) or from ftp.aip.org in the directory /epaps/. See the EPAPS homepage for more information.
- 59.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 60.Lubensky DK, Nelson DR. Biophys. J. 1999;77:1824. doi: 10.1016/S0006-3495(99)77027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slonkina E, Kolomeisky AB. J. Chem. Phys. 2003;118:7112. [Google Scholar]
- 62.Ambjörnsson T, Apell SP, Konkoli Z, Marzio EAD, Kasianowicz JJ. Biophys. J. 2002;117:4063. [Google Scholar]
- 63.Fromenborn O, Klafter J. Biophys. J. 2004;86:3576. doi: 10.1529/biophysj.103.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bransburg-Zabary S, Nachliel E, Gutman M. Biophys. J. 2002;83:3001. doi: 10.1016/S0006-3495(02)75306-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bransburg-Zabary S, Nachliel E, Gutman M. Solid State Ionics. 2004;168:235. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.