Abstract
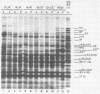
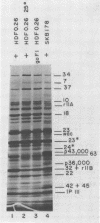
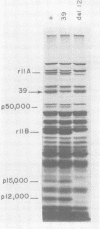
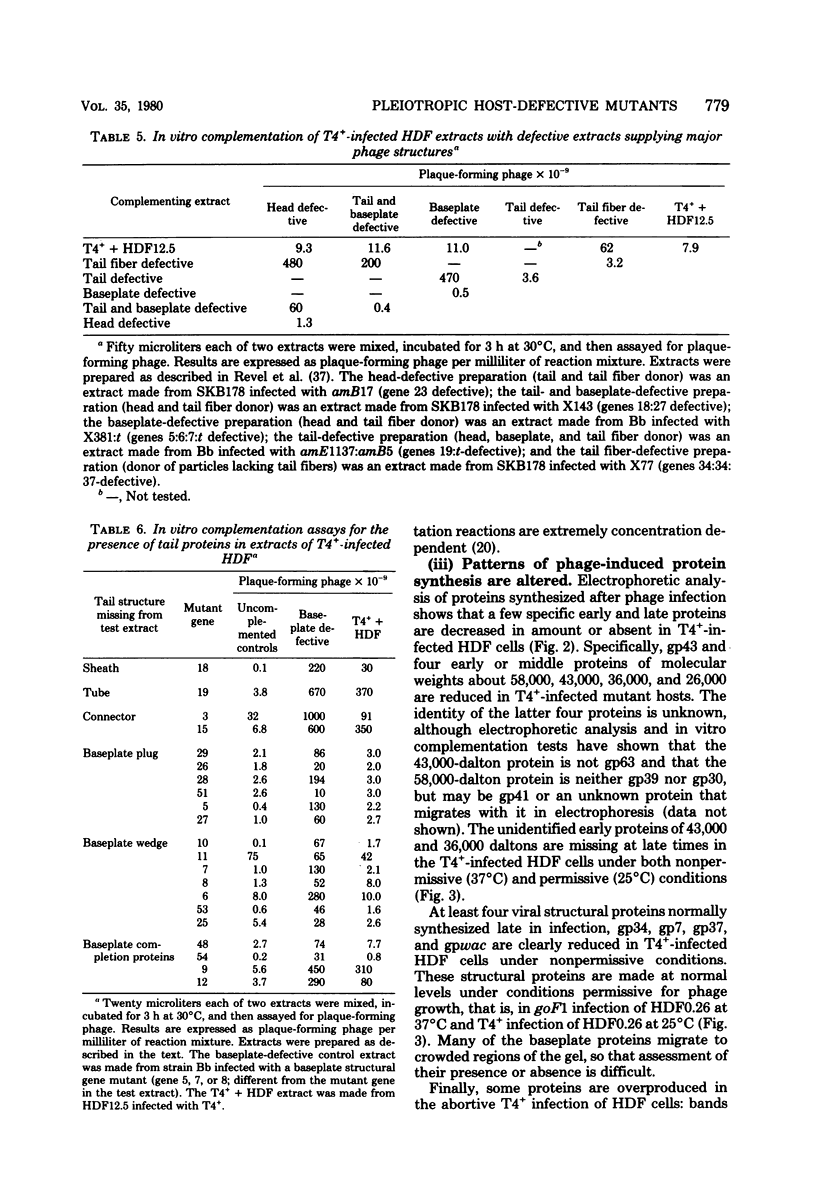
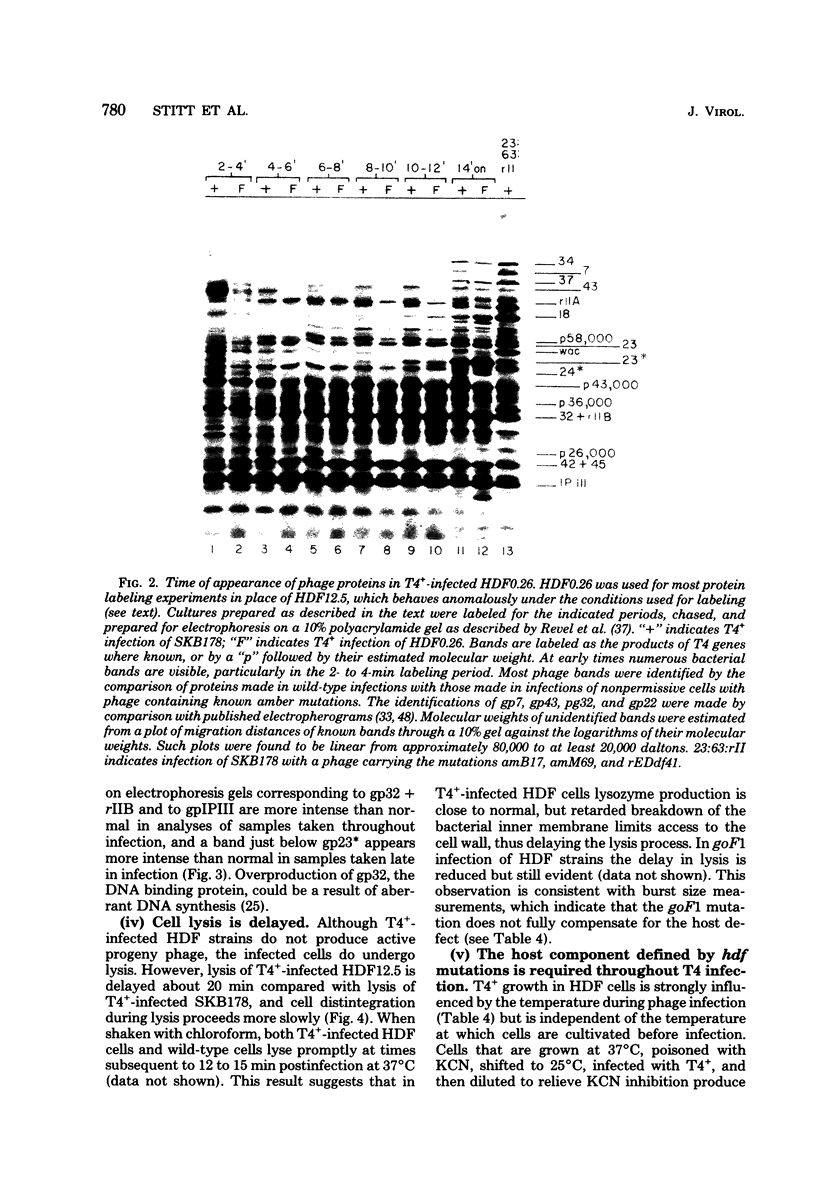
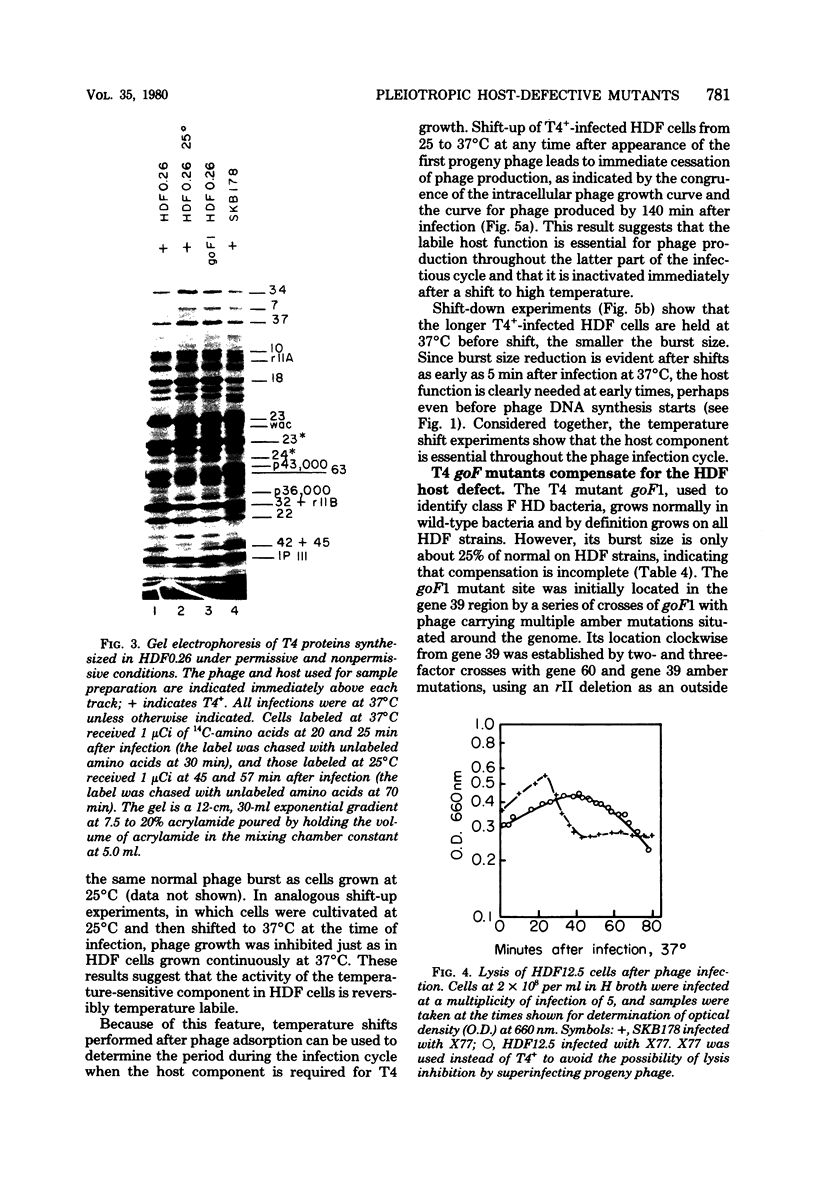
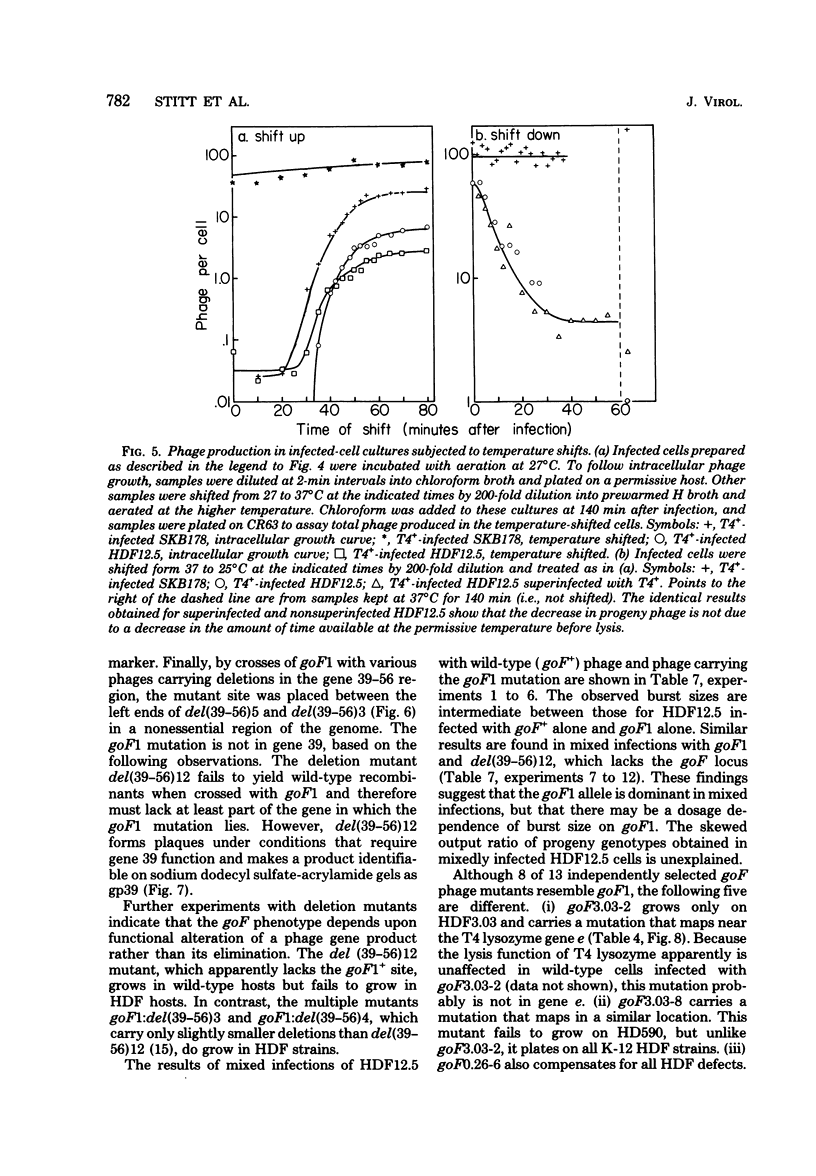
Mutant host-defective Escherichi coli that fail to propagate bacteriophage T4 and have a pleiotropic effect on T4 development have been isolated and characterized. In phage-infected mutant cells, specific early phage proteins are absent or reduced in amount, phage DNA synthesis is depressed by about 50%, specific structural phage proteins, including some tail and collar components, are deficient or missing, and host-cell lysis is delayed and slow. Almost all phage that can overcome the host block carry mutantions that map in functionally undefined 'nonessential' regions of the T4 genome, most near gene 39. The mutant host strains are temperature sensitive for growth and show simultaneous reversion of the ts phenotype and the inability to propagate T4+. The host mutations are cotransduced with ilv (83 min) and may lie in the gene for transcription termination factor rho.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961 Mar;47(3):403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. P1 transduction: formation of heterogenotes upon cotransduction of bacterial genes with a P2 prophage. Virology. 1972 Jan;47(1):76–93. doi: 10.1016/0042-6822(72)90241-3. [DOI] [PubMed] [Google Scholar]
- Brody E., Daegelen P., d'Aubenton-Carafa The role of termination factor rho in the development of bacteriophage T4 [proceedings]. Arch Int Physiol Biochim. 1978 Oct;86(4):897–898. [PubMed] [Google Scholar]
- Bujard H., Mazaitis A. J., Bautz E. K. The size of the rII region of bacteriophage T4. Virology. 1970 Nov;42(3):717–723. doi: 10.1016/0042-6822(70)90317-x. [DOI] [PubMed] [Google Scholar]
- Caruso M., Coppo A., Manzi A., Pulitzer J. F. Host--virus interactions in the control of T4 prereplicative transcription. I. tabC (rho) mutants. J Mol Biol. 1979 Dec 25;135(4):959–977. doi: 10.1016/0022-2836(79)90522-9. [DOI] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Hanawalt P. C., Guerola N. Mutagenesis of the replication point by nitrosoguanidine: map and pattern of replication of the Escherichia coli chromosome. J Mol Biol. 1968 May 14;33(3):705–719. doi: 10.1016/0022-2836(68)90315-x. [DOI] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F., Takahashi H. Abortive bacteriophage T4 head assembly in mutants of Escherichia coli. J Mol Biol. 1973 May 5;76(1):61–87. doi: 10.1016/0022-2836(73)90081-8. [DOI] [PubMed] [Google Scholar]
- Cox G. B., Gibson F., Pittard J. Mutant strains of Escherichia coli K-12 unable to form ubiquinone. J Bacteriol. 1968 May;95(5):1591–1598. doi: 10.1128/jb.95.5.1591-1598.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDGAR R. S., FEYNMAN R. P., KLEIN S., LIELAUSIS I., STEINBERG C. M. Mapping experiments with r mutants of bacteriophage T4D. Genetics. 1962 Feb;47:179–186. doi: 10.1093/genetics/47.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hendrix R. W., Kaiser A. D., Wood W. B. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol. 1972 Sep 13;239(89):38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- Grasso R. J., Buchanan J. M. Synthesis of early RNA in bacteriophage T4-infected Escherichia coli B. Nature. 1969 Nov 29;224(5222):882–885. doi: 10.1038/224882a0. [DOI] [PubMed] [Google Scholar]
- Guerola N., Ingraham J. L., Cerdá-Olmedo E. Induction of closely linked multiple mutations by nitrosoguanidine. Nat New Biol. 1971 Mar 24;230(12):122–125. doi: 10.1038/newbio230122a0. [DOI] [PubMed] [Google Scholar]
- Homyk T., Jr, Weil J. Deletion analysis of two nonessential regions of the T4 genome. Virology. 1974 Oct;61(2):505–523. doi: 10.1016/0042-6822(74)90286-4. [DOI] [PubMed] [Google Scholar]
- Inoko H., Imai M. Isolation and genetic characterization of the nitA mutants of Escherichia coli affecting the termination factor rho. Mol Gen Genet. 1976 Jan 16;143(2):211–221. doi: 10.1007/BF00266924. [DOI] [PubMed] [Google Scholar]
- Inoko H., Shigesada K., Imai M. Isolation and characterization of conditional-lethal rho mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1162–1166. doi: 10.1073/pnas.74.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Imada M., Tsugita A. The amino acid sequence of T4 phage lysozyme. IV. Dilute acid hydrolysis and the order of tryptic peptides. J Biol Chem. 1970 Jul 25;245(14):3479–3484. [PubMed] [Google Scholar]
- Jayaraman R. Transcription of bacteriophage T4 DNA by Escherichia coli RNA polymerase in vitro: identification of some immediate-early and delayed-early genes. J Mol Biol. 1972 Sep 28;70(2):253–263. doi: 10.1016/0022-2836(72)90537-2. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975 Dec 25;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- Koplow J., Goldfine H. Alterations in the outer membrane of the cell envelope of heptose-deficient mutants of Escherichia coli. J Bacteriol. 1974 Feb;117(2):527–543. doi: 10.1128/jb.117.2.527-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K. N., McEntee K., Geballe A. P., Cozzarelli N. R. Lambda transducing phages for the nalA gene of Escherichia coli and conditional lethal nalA mutations. Mol Gen Genet. 1978 Nov 29;167(2):129–137. doi: 10.1007/BF00266906. [DOI] [PubMed] [Google Scholar]
- Krisch H. M., Bolle A., Epstein R. H. Regulation of the synthesis of bacteriophage T4 gene 32 protein. J Mol Biol. 1974 Sep 5;88(1):89–104. doi: 10.1016/0022-2836(74)90296-4. [DOI] [PubMed] [Google Scholar]
- Linder C. H., Sköld O. Control of early gene expression of bacteriophage T4: involvement of the host rho factor and the mot gene of the bacteriophage. J Virol. 1980 Feb;33(2):724–732. doi: 10.1128/jvi.33.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C. H., Sköld O. Evidence for a diffusible T4 bacteriophage protein governing the initiation of delayed early RNA synthesis. J Virol. 1977 Jan;21(1):7–15. doi: 10.1128/jvi.21.1.7-15.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson T., Richardson J., Goodin D. Mutant of bacteriophage T4D affecting expression of many early genes. Nature. 1974 Jul 5;250(461):48–50. doi: 10.1038/250048a0. [DOI] [PubMed] [Google Scholar]
- Mindich L., Sinclair J. F., Cohen J. The morphogenesis of bacteriophage phi6: particles formed by nonsense mutants. Virology. 1976 Nov;75(1):224–231. doi: 10.1016/0042-6822(76)90021-0. [DOI] [PubMed] [Google Scholar]
- Mufti S., Bernstein H. The DNA-delay mutants of bacteriophage T4. J Virol. 1974 Oct;14(4):860–871. doi: 10.1128/jvi.14.4.860-871.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M., Huang W. M. The identification of prereplicative bacteriophage T4 proteins. J Biol Chem. 1973 Aug 10;248(15):5499–5501. [PubMed] [Google Scholar]
- Pulitzer J. F., Coppo A., Caruso M. Host--virus interactions in the control of T4 prereplicative transcription. II. Interaction between tabC (rho) mutants and T4 mot mutants. J Mol Biol. 1979 Dec 25;135(4):979–997. doi: 10.1016/0022-2836(79)90523-0. [DOI] [PubMed] [Google Scholar]
- Pulitzer J. F., Yanagida M. Inactive T4 progeny virus formation in a temperature-sensitive mutant of Escherichia coli K12. Virology. 1971 Sep;45(3):539–554. doi: 10.1016/0042-6822(71)90170-x. [DOI] [PubMed] [Google Scholar]
- Revel H. R., Stitt B. L., Lielausis I., Wood W. B. Role of the host cell in bacteriophage T4 development. I. Characterization of host mutants that block T4 head assembly. J Virol. 1980 Jan;33(1):366–376. doi: 10.1128/jvi.33.1.366-376.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W., Bolle A., Epstein R. Transcription during bacteriophage T4 development: a demonstration that distinct subclasses of the "early" RNA appear at different times and that some are "turned off" at late times. J Mol Biol. 1970 Apr 28;49(2):271–295. doi: 10.1016/0022-2836(70)90246-9. [DOI] [PubMed] [Google Scholar]
- Simon L. D., Gottesman M., Tomczak K., Gottesman S. Hyperdegradation of proteins in Escherichia coli rho mutants. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1623–1627. doi: 10.1073/pnas.76.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L. D., Snover D., Doermann A. H. Bacterial mutation affecting T4 phage DNA synthesis and tail production. Nature. 1974 Dec 6;252(5483):451–455. doi: 10.1038/252451a0. [DOI] [PubMed] [Google Scholar]
- Simon L. D. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. 3. Membrane-associated intracellular bacteriophages. Virology. 1969 Jun;38(2):285–296. doi: 10.1016/0042-6822(69)90370-5. [DOI] [PubMed] [Google Scholar]
- Takahashi H. Genetic and physiological characterization of Escherichia coli K12 mutants (tabC) which induce the abortive infection of bacteriophage T4. Virology. 1978 Jun 15;87(2):256–265. doi: 10.1016/0042-6822(78)90131-9. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Yoshikawa H. Genetic study of a new early gene, comC-alpha, of bacteriophage T4. Virology. 1979 May;95(1):215–217. doi: 10.1016/0042-6822(79)90417-3. [DOI] [PubMed] [Google Scholar]
- Takano T., Kakefuda T. Involvement of a bacterial factor in morphogenesis of bacteriophage capsid. Nat New Biol. 1972 Sep 13;239(89):34–37. doi: 10.1038/newbio239034a0. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Positive control of transcription by a bacteriophage sigma factor. Nature. 1970 Mar 14;225(5237):1009–1012. doi: 10.1038/2251009a0. [DOI] [PubMed] [Google Scholar]
- Vanderslice R. W., Yegian C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974 Jul;60(1):265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- Ward S., Luftig R. B., Wilson J. H., Eddleman H., Lyle H., Wood W. B. Assembly of bacteriophage T4 tail fibers. II. Isolation and characterization of tail fiber precursors. J Mol Biol. 1970 Nov 28;54(1):15–31. doi: 10.1016/0022-2836(70)90443-2. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Function of the bacteriophage T4 transfer RNA's. J Mol Biol. 1973 Mar 15;74(4):753–757. doi: 10.1016/0022-2836(73)90065-x. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Kim J. S., Abelson J. N. Bacteriophage T4 transfer RNA. 3. Clustering of the genes for the T4 transfer RNA's. J Mol Biol. 1972 Nov 28;71(3):547–556. doi: 10.1016/s0022-2836(72)80022-6. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]