Abstract
The Myc pathway, often deregulated in cancer, is critical in determining cell fate by coordinating a gene expression program that links the control of cell proliferation with cell fate decisions. As such, precise control of the Myc pathway activity must be achieved to ensure faithful execution of appropriate cellular response and to prevent progressing toward a malignant state. With recent highlighted roles of microRNAs (miRNAs) as critical components of gene control, we sought to evaluate the extent to which miRNAs may contribute in the execution of Myc function. Combined analysis of mRNA and miRNA expression profiles reveals an integration whereby the Myc-mediated induction of miRNAs leads to the repression of various mRNAs encoding tumor suppressors that block cell proliferation including p21, p27 and Rb. In addition, the pro-apoptotic PTEN tumor suppressor gene is also repressed by a Myc-induced miRNAs suggesting that Myc-induced miRNAs contribute to the precise control of a transcriptional program that coordinates the balance of cell proliferation and cell death.
Introduction
Cancer is a disease of gene mutation, arising as a result of complex alterations in signaling pathways that are essential for making critical cell fate decisions under appropriate circumstances (1). The Myc pathway represents one key aspect of the signaling events that dictate important cellular outcomes in normal cells and its dysregulation is a frequent etiology in cancer development (2). A key role for Myc is to integrate various environmental external signals so as to orchestrate an extensive transcription program that elicits cell proliferation, differentiation, or cell death (3, 4).
The Myc transcription factor recognizes a canonical motif known as the E-box (CACGTG) where it forms a heterodimer with a partner protein Max, to further recruit additional co-factors that are necessary for transcriptional activation of target genes, many of which are critical for S phase entry (5, 6). Equally important is the ability of Myc to repress the expression of various target genes, including those encoding key cell cycle arrest proteins such as CDK inhibitors (p21 and p27), as well as additional tumor suppressor genes that act negatively to regulate the onset of proliferation (7–9). Indeed, ectopic expression of many Myc repressed target genes have been shown to lead to defects in cell cycle progression and proliferation (10–12).
Despite such signficance, much less is known of the mechanisms mediating the repression of gene expression by Myc. It has been proposed that Myc may achieve transcriptional repression in part through recruitment of additional co-factors such as MIZ1 or Sp1 (11, 13, 14), histone modifying factors, or DNA methyltransferases, to possibly explain the conundrum of how a single molecule can act both as an activator and a repressor in the same cellular context (15, 16).
Adding to this complexity is the recent identification of microRNAs (miRNAs) as important contributors to Myc activity. miRNAs are small 19–24 nucleotide long non-coding RNAs which individually target hundreds of genes by virtue of forming a partial complementary binding to the 3’-UTR of its cognate genes (17). miR-17–92 cluster, which encode 6 individual miRNAs, is directly activated by Myc as a way to fine tune the activity of yet another Myc target E2F1 (18). Furthermore, these miRNAs, along with its paralogous cluster miR-106b-25, have been implicated in the control of the transforming growth factor beta (TGF-β) signal pathway. (19, 20) More recent studies have shown that repression of many miRNAs by Myc contribute to conferring cells with proliferative advantage in tumor cells (21, 22). In this study, we set out to examine the role of miRNAs in impacting the dynamics of the Myc transcriptional network of normal proliferating cells through an integrative approach of examining the genome-wide expression profiles of messenger RNA (mRNA) and miRNA. By combined analysis of the two complementary data, we not only identify novel miRNAs that are induced by Myc, but also a regulatory connection that appears to ensure faithful execution of the Myc proliferative program by mediating the repression of key cell cycle arrest genes and apoptotic genes.
Materials and Methods
Cell culture
Human Mammary Epithelial Cells (HMECs) were cultured as previously described (23). Prior to infection with recombinant adenovirus expressing Myc or GFP, cells were grown in 0.25% serum starvation media (without EGF) for 48 hours. WI38 fibroblasts and Hela cells were maintained in DMEM media with 10% Fetal Bovine Serum in 5% CO2. Mouse Embryonic Fibroblast (MEF) harboring c-Myc (Myc-MEFs) were cultured as previously described. (24).
miRNA array hybridization and data analysis
Experiments for generating miRNA expression profiles reflecting MYC activities were carried out as previously described (25). For RNA preparation, mirVana miRNA Isolation Kit (Ambion) was used to extract total RNA retaining smaller RNA population. Concentration and the quality of the resulting RNAs were assessed both through NanoDrop (Thermo Scientific) and Agilent 2100 bioanalyzer. miRNA array hybridization was carried out according to the manufacture’s protocol (Exiqon). High quality RNA derived from lung, breast, brain, heart and prostate tissues (Ambion) were pooled and used as a reference to label green channel across the samples. The resulting GPR files were loess normalized using the Limma package in R (26). The normalized data were labeled 1 or 0 depending on the status of Myc. We correlated the Myc status with each miRNA probe and calculated the p-value of the significance of the observed correlation against a null hypothesis of no correlation.
Myc expression profile which was previously generated on an U133 plus2.0 array (GSE3151) was MAS5 normalized with R. Software to implement Gene Set Enrichment Analysis was downloaded from Broad Institute website. The samples were given phenotypic label depending on the Myc status and C3 gene set was selected for the analysis (27). The detailed description of the parameters used to run the analysis is provided in the Table S6. Gene sets whose nominal p values <0.05 and FDR <0.25 were considered to be significant for further investigation.
PTEN luciferase reporter assay
3’-UTR sequence of the PTEN was retrieved through NCBI nucleotides. The PTEN 3’-UTR were amplified using primers (for sequences, see Table S5) flanking XhoI and NotI restriction sites and cloned into siCHECK-2 Vectors (Promege). Site directed mutagenesis was carried out by using GeneTailor Site-Directed Mutagenesis System (Invitrogen) according to the manufacturer’s protocol, and verified by sequencing. Dual luciferase assay was carried out by co-transfecting HeLa cells with either the wild-type or the mutant luciferase construct, in combination with designated miRNA mimics (Dharmacon), and harvested according to the manufacture’s protocols (Promega). All luciferase assays were carried out in Hela cells 24 hours post-transfection in three independent experiments. Renilla luciferase activity was normalized to firefly luciferase to control for transfection variation.
Transfection
Cells were split 24 hours prior to all transfection assays at a confluence of 50–70%. ASOs targeting miR-18a, miR-20a, miR-23b, miR-130a, miR-193b, and miR-302c were obtained through Integrative DNA technology (IDT). For siRNA targeting Myc and miRNA mimics of miR-23b and miR-193b, and negative control oligos (Cat#: CN-001000-01-05), were purchased from Dharmacon. For transfection of oligos and plasmids, lipofectamine 2000 was used and the instructions were followed according to the protocols provided by the manufacturer. (Invtirogen) ASOs were transfected at a concentration of 400nM and the mimics were co-transfected at 200nM with psiCHECK-PTEN 3’-UTR vector (400ng). For combined transfection of miRNAs, 200nM of indicated ASOs were used. For ectopic PTEN expression, 1 ug of plasmids containing either a full length PTEN (Origene) or miR-23b and miR-193b mutant PTEN 3’-UTR were co-transfected with 200nM of corresponding miRNA mimics. For Myc inhibition, 100nM of siRNA targeting Myc was used prior to measurement of proliferation.
BrdU immunostaining
BrdU immunostaining was assessed on asynchronously growing WI38 fibroblasts. 24 hours after 1.5 × 105 cells were split into a 6-well plate, cells were either mock transfected or transfected with ASOs. To achieve a steady-state inhibition of individual miRNAs and also to avoid contact inhibition upon confluence, the transfection protocols were repeated 48 hours after the initial transfection. The cells were pulsed labeled for 1 hour with 10uM of BrdU 24 hours post second transfection. Immunostaining protocol was carried out as previously reported (28).
Caspase-3 assay
Active Caspase-3 staining assay (BD Pharmingen) was carried out according to manufacturer’s protocol. Apoptosis upon transfection of either PTEN construct with full length 3’-UTR (Origene), or ASOs inhibiting the activity of miR-23b, miR-193b, and miR-302c was also determined. The latter was used as an internal control, alongside mock transfection. Floating cells, as well as attached cells were collected 48 hours after transfection. Cells stained with PE-conjugated anti-Caspase-3 were subject to flow cytometry analysis on Canto II (BD Pharmingen) and subsequent data was analyzed on the flow cytometry analyzing software (Flowjo). Fraction of cells which were labeled positive for PE-staining was considered to be apoptotic.
Results
A comparison of mRNA and miRNA expression profiles induced by Myc
As one approach to explore the molecular architecture of the Myc pathway, we focused on a previously generated gene expression profile that reflects changes in mRNA levels as a consequence of Myc activity (25). This expression signature includes genes whose level of expression increases (red color) as well as genes that are repressed (blue color) by the action of Myc (Fig. 1). Examination of the functional annotation of the group of genes that were induced by Myc demonstrated that many have known roles in metabolism and nucleotide biosynthesis, an aspect which has been previously established to be important for Myc mediated proliferation (Table S1) (5, 6, 29). Although there was no predominant enrichment for specific biological annotation for genes that were repressed by Myc, they included many previously known Myc repressed target genes, such as Cyclin Dependent Kinase inhibitors(CDKIs), cell cycle arrest genes, as well as components of the TGF-β signaling pathway, etc., many of whose activity negatively affect the activity of the Myc pathway, suggestive of an active mechanism by which Myc represses the activity of these functionally related genes (7–9).
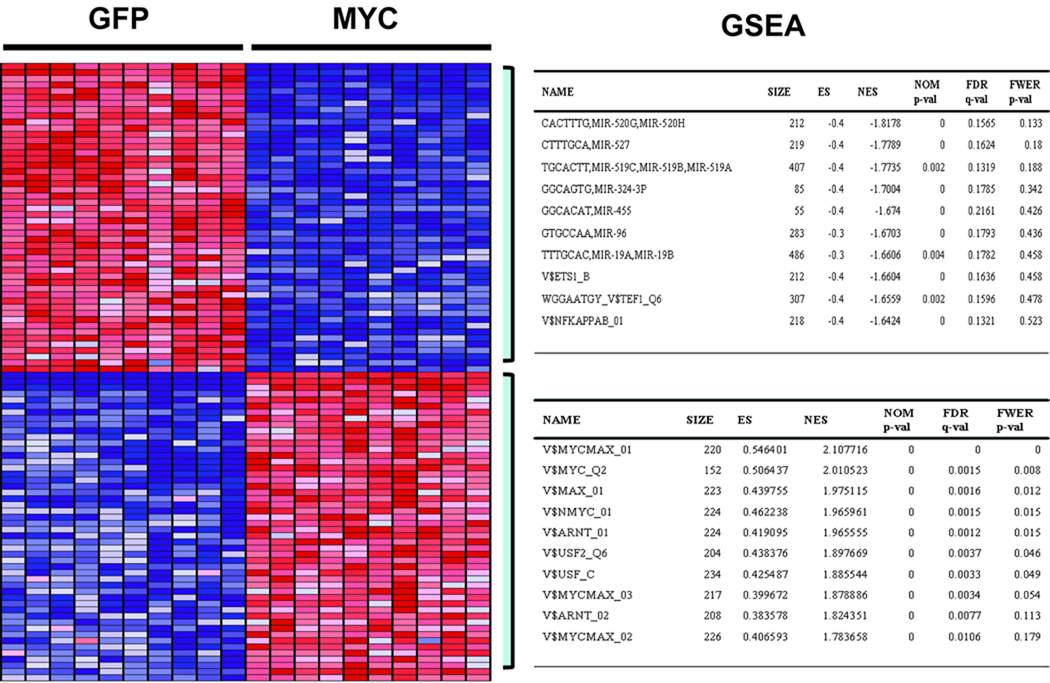
Fig. 1.
Representative heatmap of top 50 genes that are differentially regulated by Myc as identified by the Gene Set Enrichment Analysis (GSEA). The tables on the right panel summarize top 10 results obtained through the GSEA analysis. Each column on the heatmap represents replicate samples of designated biological status (Myc vs. GFP control), with rows representing individual genes. Red indicates activation, while the blue repression. The top table on the right panel summarizes top10 GSEA results for significantly enriched transcription factor (TF) or microRNA target site motifs of Myc repressed genes while the bottom table that of the activated genes.
To focus on the mechanisms that underlie the generation of this Myc profile, including the genes induced or repressed by Myc, we made use of Gene Set Enrichment Analysis (GSEA), a computational tool developed to derive biological significance from phenotypes reflected in transcriptional profiles by utilizing various a priori established gene set (30). We focused on gene sets representing regulatory motifs for transcription factor and miRNA binding that were previously defined by species conservation (27). As expected, there was a strong enrichment for canonical Myc binding sites within the group of genes that were activated by Myc (Fig. 1). In contrast, the group of genes that were repressed by Myc did not exhibit enrichment for Myc binding sites but rather a substantial enrichment for miRNA binding sites in their 3’-UTR sequences. A similar result was seen in a second independent dataset (Table S2, S3). This result suggested a possible contribution of miRNAs in mediating repression of Myc target genes.
A role for Myc-induced miRNAS in cell proliferation
To address a potential role for miRNAs in a Myc regulatory program, we made use of the same experimental approach to generate RNA samples that were used to generate the mRNA expression profiles, but now focusing on low molecular weight RNAs and hybridization to a miRNA microarray platform. We utilized LNA-based miRNA arrays (Exiqon) that have previously been shown to have high specificity and sensitivity (31). As with the mRNA expression profile, the microarray was performed in 10 experimental replicates for each condition (Myc off and Myc on), in order to minimize any variations that may result from technical or experimental noises. Differentially expressed genes were identified by assigning a 1, 0 class to each phenotype and calculating correlation coefficient of individual miRNAs according to Myc status. Only genes with p-value of less than p<0.05 (see Experimental Procedures) were selected for further analysis. As depicted in Fig. 2, a number of miRNAs displayed statistically significant correlation (p<0.05) with the status of Myc expression (for full list, see Table S4). Quantitative real-time PCR analysis on 9 selected miRNAs, preferentially activated by Myc, showed an overall good correlation with the microarray results, indicating that the results reflected by the microarray analysis were reliable (Fig. S1).
Fig. 2.
miRNA expression profiles reflecting the activity of Myc. Heatmap representing miRNA expression profile that differentially correlates with Myc (p < 0.05) with 10 replicate samples of indicated Myc status (Myc on vs. off), while the individual miRNAs are depicted in each rows. Red indicates induction and the green repression. The bar right to the heatmap and the corresponding miRNA list that reflects miRNAs that overlap with the GSEA results of miRNA motifs enriched in the Myc repressed genes (Fig 1. and Table S2, S3).
Previous work has shown that members of the miR-17–92 cluster are a direct target of Myc. (18) Indeed, the analysis depicted in Fig. 2 shows that members of this cluster of miRNAs, specifically miR-18a and miR-20a, were indeed activated by Myc. We also found induction of many miRNAs that were previously shown to be induced by serum (32, 33), consistent with a role for Myc in the response to serum. We also observed overlap in several miRNAs that are induced by N-Myc (let-7f, miR-100, miR-221, miR-92, and members of the miR-17–92 cluster), raising the possibility that some miRNAs may be common target of both N-Myc and c-Myc (34).
A number of the miRNAs that were induced by Myc overlapped with the predictions by GSEA of miRNAs potentially targeting Myc-repressed mRNAs, suggesting that miRNAs induced by Myc may indeed participate in mediating repression of Myc target genes. To further examine the functional implications of Myc induced miRNAs to Myc pathway activity, we chose to focus on miRNAs whose average raw expression value was at least two fold above background (miR-18a, miR-20a, miR-100, miR-183, miR-193b, miR-130a, miR-221, miR-222, miR-23b). Examination of the predicted targets of these miRNAs, also repressed by Myc as reflected in the Myc expression profile, revealed a potential connection between the Myc induced miRNAs with Myc targets which play a role in cell cycle progression (Table 1, Fig. S2). For example, the Myc-induced miR-20a is predicted to target the cyclin kinase inhibitor p21 (CDKN1A), a gene encoding a negative regulator of cell cycle progression. Likewise, Myc induces the expression of miR-221 and miR-222 which are predicted to target p27 and p57 (CDKN1B, CDKN1C), additional cyclin kinase inhibitors that have prominent roles in triggering cell cycle arrest. As miR-221/222 - p27 and p57 connection has been well established, this further supports a widespread mechanism of Myc induced miRNAs in suppressing the expression of key cell cycle arrest genes (33, 35–37). Taken together, our integrated analysis of miRNA and mRNA profile suggests that, in additional to the induction of various protein-encoding genes that mediate cellular proliferation, Myc induces a series of miRNAs that inhibit genes responsible for negative control of cellular proliferation.
Table 1.
Myc induced miRNAs and their predicted targets.
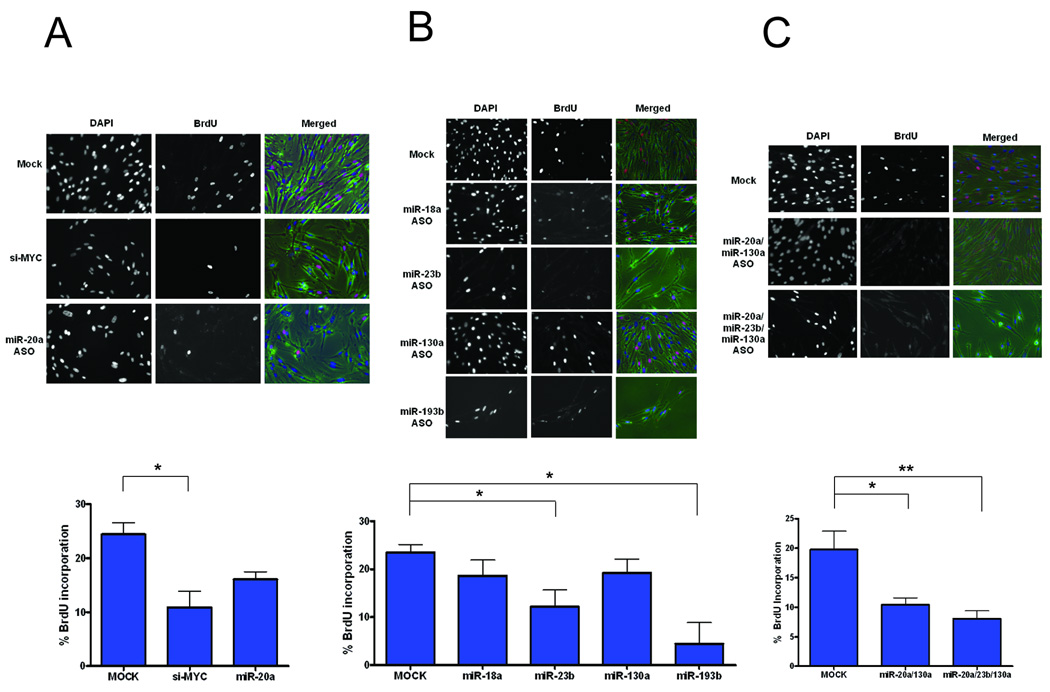
To directly address the role of Myc-induced miRNAs in facilitating the induction of cell proliferation, we made use of asynchronously growing normal WI38 fibroblasts, as these cells have largely intact cell cycle machinery. As a control, we first assessed the extent to which inhibition of Myc would reduce the proliferation capacity of these cells. As shown in Fig. 3A, inhibition of Myc displayed a marked decrease in proliferation as measured by Bromodeoxyuridine (BrdU) incorporation. We then measured the effect of inhibition of various miRNAs that were induced by Myc. Inhibition of miR-20a by anti-sense oligos (ASO hereafter) led to a modest reduction in proliferation in WI38 fibroblasts, as previously shown in other cell types (Fig. 3A) (38). Inhibition of miR-18a had little impact on the proliferation of WI38 fibroblasts and likewise for miR-130a (Fig. 3B). However, inhibition of miR-23b and miR-193b led to a marked effect on proliferation, both as assessed visually through the microscope, as well as through BrdU quantification, with the miR-193b inhibition leading to a most profound effect. Since it is possible that the function of the Myc-induced miRNAs could be at least partially redundant, we also examined whether the combined inhibition of miRNAs with multiple targets would have greater impact. Indeed, when the miRNAs were inhibited in combination, the effect was more pronounced as shown in Fig. 3C. This was not due to increase in the concentration of ASO as transfecting same concentration of single miRNAs did not affect further brdU incorporation (data not shown). Taken together, we conclude that Myc induced miRNAs, either individually or collectively, impact the cell proliferation by targeting various cell cycle arrest genes.
Fig. 3.
Impact of Myc induced miRNAs in proliferation. Ability of the following experimental treatments to impair proliferation in normal WI38 fibroblasts was determined by immunostaining after 1 hour pulse labeling with BrdU. Representative images of BrdU and DAPI labeled WI38 fibroblasts under the fluorescent microscope are shown on top. The bar graph below the images depicts quantification of average ratio of BrdU positive cells normalized to the number of DAPI positive cells counted across multiple fields. Results were carried out in three independent experiments from which mean and the standard errors were calculated. (*p<0.05, **p<0.005, two-tailed t-test).
A) siRNA targeting Myc and anti-sense oligo targeting miR-20a serves as a positive control.
B) Effect of inhibiting miR-18a, miR-23b, miR-130a and miR-193b on BrdU incorporation was assessed by transfecting 400nM of anti-sense inhibitors.
C) Combined inhibition of Myc induced miRNAs (miR-20a/miR-130a and miR-20a/130a/23b) was examined.
A role for Myc-induced miRNAs in coordinating the balance of proliferation and cell death
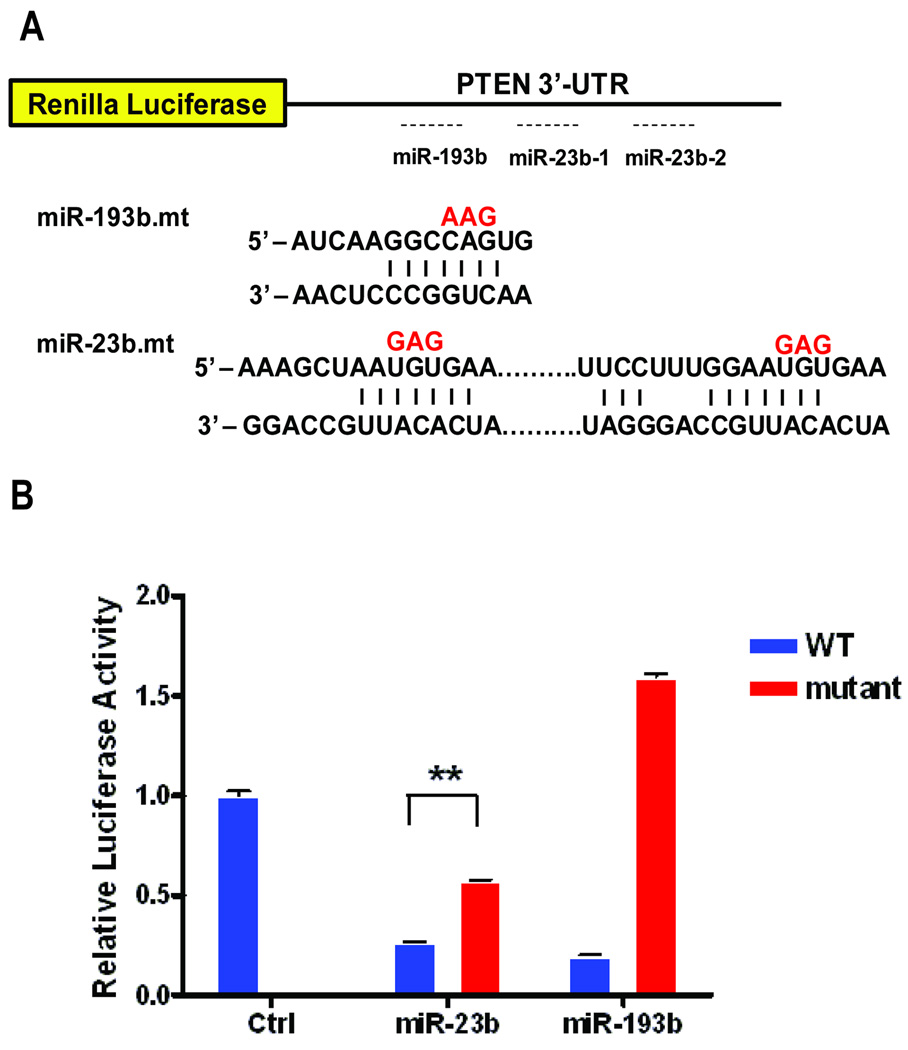
A large body of previous work has shown that activation of Myc in the absence of Phosphoinositide 3-kinases (PI3K)/Akt pathway activity leads to apoptosis, consistent with a role for PI3K/Akt in providing survival signals in the context of normal cellular proliferation (4). The phosphatase and tensin homolog (PTEN) gene, among the most frequently mutated genes in cancer, acts as an important regulator of the survival signals by antagonizing PI3K activity (39). Ectopic expression of PTEN in both normal and cancer cells have been shown to induce cell cycle arrest or apoptosis depending on the cellular context (39). We noted a decrease in the level of PTEN transcripts in the Myc expressing cells and this was further confirmed at the protein level by western blot analysis (Fig. S3). Based on our analysis of Myc-induced miRNAs (Table 1), three miRNAs (miR-20a, miR-23b, miR-193b) were predicted to target PTEN (Fig. 4A). We chose to focus on miR-23b and miR-193b, miRNAs not previously implicated in the control of PTEN regulation, by first determining if these miRNAs did indeed target PTEN. To this end, we cloned the initial 2.2 kb fragment of the PTEN 3’-UTR sequence spanning the miRNA target sites upstream of the Renilla luciferase construct. This construct served as a reporter for miRNA targeting by measuring the extent to which there was a reduction in the luciferse activity following co-introduction of miR-23b and miR-193b. As shown in Fig. 4B, expression of miR-23b and miR-193b mimics each led to a substantial reduction in luciferase activity with the construct containing wild-type PTEN 3’-UTR (blue) relative to the control miRNA, while having little or no impact on the 3’-UTR that have mutation in the designated target sites (red), indicating that these miRNAs do indeed target PTEN.
Fig. 4.
Dual luciferase reporter assay to assess the extent to which miR-23b and miR-193b target PTEN 3’-UTR. A) There are two conserved miR-23b sites and one semi-conserved miR-193b sites in the PTEN 3’-UTR. B) 3’-UTR of either wild-type or base-pair substituted mutant constructs were introduced (as indicated in red in Fig. 4A), in conjunction with corresponding miRNA mimics as indicated to assess the ability of the respective miRNAs to specifically repress PTEN 3’-UTR activity as measured by Renilla luciferase activity. Each assay was normalized to Firefly luciferase activity, and the mean and standard errors were calculated across three independent experiments. (**p<0.005, two-tailed t-test).
To test whether these miRNAs may play a role in apoptosis by regulating PTEN levels in the context of Myc induced apoptosis, we made use of mouse embryonic fibroblasts (MEFs) that stably express the Myc oncogene (Myc-MEF) (24). As shown in Fig. 5A, there was a strong induction of cell death in these Myc-expressing cells upon serum withdrawal, as measured by Caspase-3 assay using flow cytometry. Furthermore, inhibition of PI3K activity with LY290024 in the presence of 10% serum, also led to an induction of apoptosis confirming the role of PI3K signaling in Myc induced apoptosis (Fig. 5B) (40). Finally, consistent with the role of PTEN in antagonizing PI3K activity, ectopic expression of PTEN also led to an induction of apoptosis (Fig. 5C).
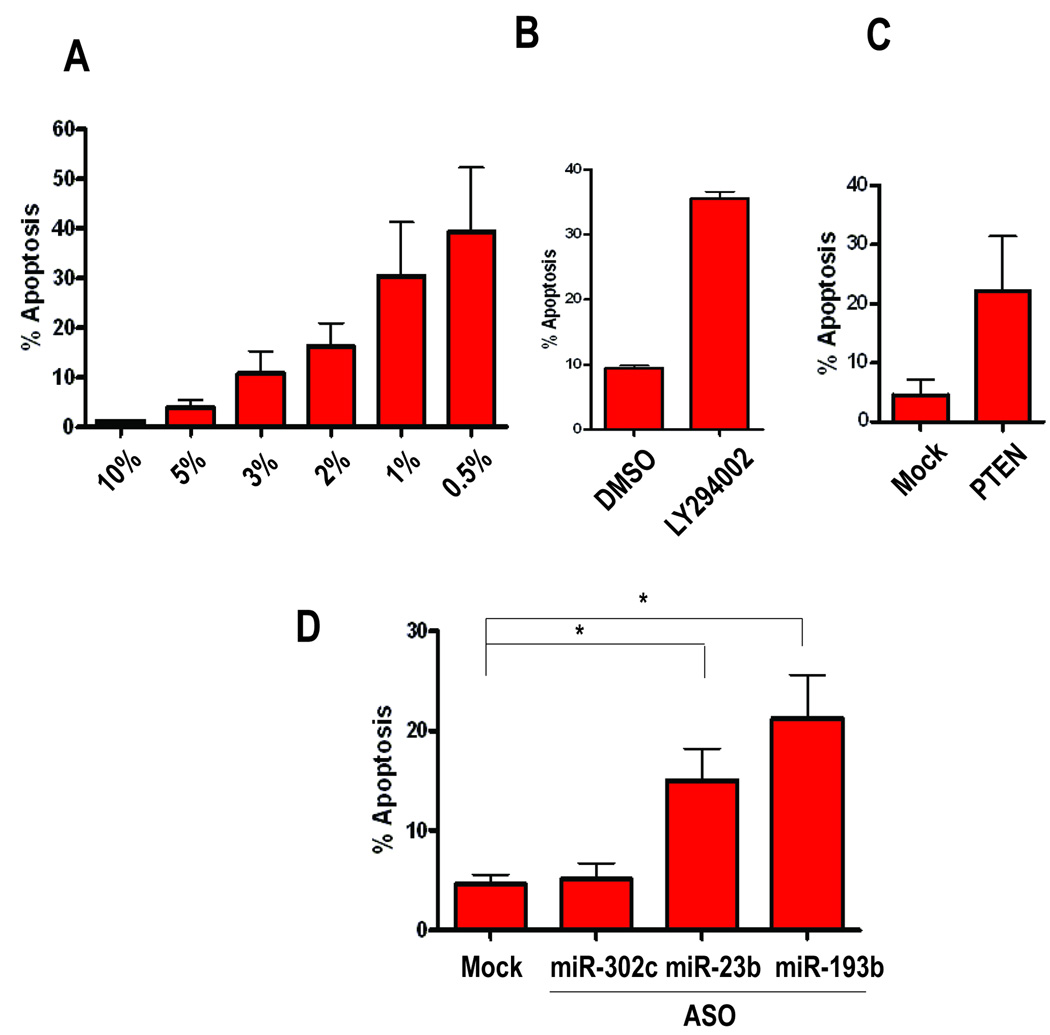
Fig. 5.
Role of Myc induced miRNAs in Myc induced apoptosis. Stable cell line constitutively expressing Myc derived from Mouse Embryonic Fibroblast (Myc-MEFs) was tested for its ability to recapitulate Myc induced apoptosis by A) adding different concentration of indicated serum dose, B) inhibiting PI3K signaling by 20uM of PI3K inhibitor LY294002 in 10% serum, or C) by ectopic introduction of PTEN in 10% serum.
D) Effects of anti-sense mediated inhibition of Myc induced miRNAs on Myc induced apoptosis. Anti-sense oligos targeting indicated miRNAs were transfected in Myc-MEFs asynchronously growing in 10% serum and collected 48 hours post-transfection. miR-302c was transfected as a negative control along with the Mock transfected. % of cells undergoing apoptosis was assessed by measuring cleavage of Caspase-3 activity as assessed by flow cytometry. Results were carried out in three independent experiments from which mean and the standard errors were calculated. (*p<0.05, two-tailed t-test).
To directly assess the role of the Myc-induced miRNAs predicted to target PTEN in controlling the induction of apoptosis, we blocked the activity of miR-23b and miR-193b with the use of ASOs in the Myc-MEFs. Consistent with the role of PTEN in apoptosis (Fig. 5D), the inhibition of miR-193b, and to a lesser degree miR-23b, led to an increase in the fraction of the apoptotic cells. The effect was specific because transfection of the same concentration of miR-302c (Fig. 5D) or miR-18a (data not shown), miRNAs not predicted to target PTEN, had no effect on the level of apoptosis. Western blot analysis indicated that indeed, PTEN levels were elevated upon inhibition of miR-23b and miR-193b, suggesting that the phenotype observed is specific to PTEN activity (Fig. S4).
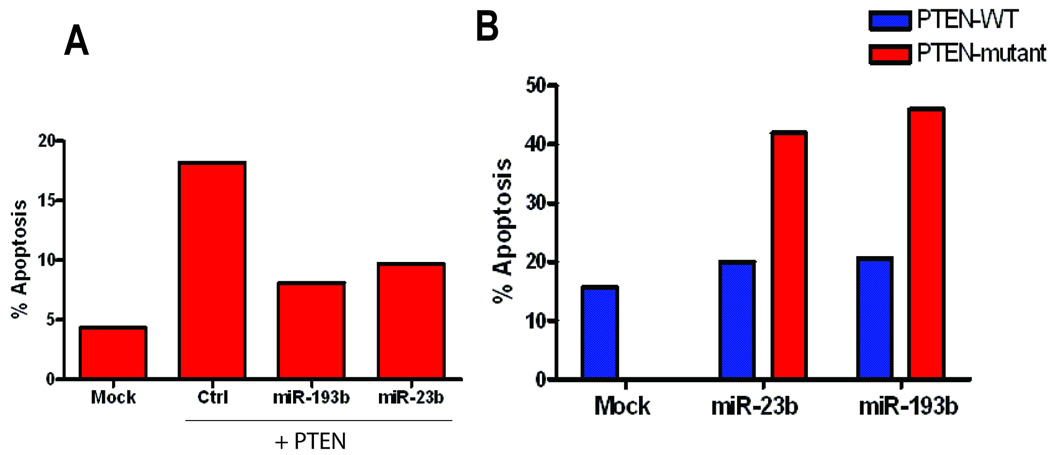
To further assess whether PTEN-induced apoptosis in the context of Myc activation can be rescued by these PTEN targeting miRNAs, we co-transfected a PTEN construct with full length wild type 3’-UTR, in combination with either a control or miRNA mimics of miR-23b and miR-193b. Introduction of miR-23b and miR-193b mimics with PTEN reduced the number of cells undergoing apoptosis in comparison to a control miRNA mimic (Fig. 6A). To further test the specificity of the ability of miR-23b and miR-193b to rescue apoptosis induced by relieving PTEN expression, we generated mutant constructs that contain three base-pair substitutions in the corresponding miRNA binding sites in their 3’-UTR. Indeed, when we co-introduced miR-23b or miR-193b with the either a wild-type PTEN or a mutant PTEN construct, the mutant constructs escaped the rescue and led to an increase in apoptosis relative to the wild-type construct (Fig. 6B). Based on these results, we conclude that Myc induces a group of miRNAs that contribute to a proliferation program by not only inhibiting the expression of anti-proliferative genes but also by blocking the activity of PTEN, thus allowing activation of PI3K and the provision of cell survival signals.
Fig. 6.
Suppression of PTEN induced apoptosis by miR-23b and miR-193b. A) Indicated miRNA mimics, along with PTEN expression vector was co-transfected to assess the extent to which either miR-23b or miR-193b can rescue PTEN induced apoptosis. Subsequent % of cells undergoing apoptosis was assessed by flow-cytometry. B) PTEN constructs containing either an intact 3’-UTR or three base-pair substitutions for miR-23b binding sites or miR-193b binding site were co-transfected along with corresponding miRNA mimics.
Discussion
The critical role for Myc activity in cell proliferation is now well established (2). The expression of the Myc gene, and the accumulation of the Myc protein, is tightly regulated by the stimulation of cell growth and leads to the activation of a large array of genes that provide the essential activities for a normal proliferative process (25, 29, 41, 42). Further, aberrant expression of Myc, without the full complement of proliferative signaling, can also trigger a cell death program providing an important nexus in the determination of cell fate (4, 40). Given this critical role for Myc, many studies have focused on both the mechanisms controlling Myc expression as well as the mechanisms by which Myc activity establishes and controls these proliferation and cell fate decisions (4).
High-throughput expression profiling to assess the global mRNA expression in various contexts of the Myc signaling, coupled with genome-wide chromatin interaction assays, has established a role for Myc in the activation of a vast array of protein-coding genes that enable a proliferation program (7–9). It is also clear that other genes are repressed as a function of Myc activity, including genes that negatively regulate cell proliferation (7–9). Adding to this complexity is the realization that a variety of non-coding miRNAs are also regulated by Myc and have been shown to target a number of key cellular regulatory genes (18, 21, 22, 43). The study we report here extends these observations and importantly, now integrates the analysis of Myc in the control of miRNA expression within the overall context of a mRNA regulatory pathway important for the establishment of a proliferative state in a normal cell.
The significance of Myc-mediated repression has long been recognized, both in the context of normal cellular physiology, as well as in transformation, as many of the repressed target genes represent those that play negative roles in the Myc pathway activity (7–9). Despite extensive investigation, there appears to be not one single mechanism to explain how Myc achieves this function. Rather, various epigenetic, as well as indirect protein-protein interaction mechanisms have been reported to contribute to this process (10, 13–16, 44). For example, p27 has been established as a repressed target of Myc important for antagonizing cell cycle progression, and various models have been proposed as to how this may be achieved (12). Based on our data, induction of miR-221 and miR-222 by Myc may also be involved in ensuring repression of these genes at a post-transcriptional level(33, 35, 37). Likewise, repression of p21 by Myc has long been a topic of interest and our data, as verified by others, suggests another venue by which Myc keeps the level of this protein low(11, 14). Indeed, inhibiting some of these Myc induced miRNAs in an actively growing culture of normal human fibroblast cells, functionally mimics that of ectopically expressing these genes, providing further validations to the notion that Myc utilizes miRNAs to orchestrate transcriptional program to allow into proliferation (Fig. 3).
Our focus in this work has been on the role of Myc-mediated gene regulation in the context of normal cellular proliferation. In contrast, many studies have also focused on the significant role of deregulated Myc activity in cancer. Likewise, many recent studies have detailed the alterations in miRNA expression in cancer cells and tumors (45), particularly the generalized reduction in miRNA levels in many oncogenic states (46). Indeed, a recent report has linked the repression of miR-23a/b with the action of Myc, contributing to enhanced glutamine metabolism (21). The observation that Myc repressed miR-23a/b is in contrast to our observation that Myc induces this miRNA in normal epithelial cell culture. While the mechanistic basis for this distinction is unclear, we surmise that additional regulatory activities that ultimately determine the consequence of Myc action, whether activation or repression, define distinctions between the normal cell state and an oncogenic state.
Perhaps of most interest is the link established by this study in balancing cell proliferation with the control of cell death through the action of several Myc-induced miRNAs that target the tumor suppressor PTEN. Many studies have now delineated a role for the PI3K/Akt pathway in providing cell survival signals, blocking the induction of p53-mediated apoptosis as part of a normal proliferative process (4, 40). Recent work has also identified a role for miRNAs in the miR-17–92 cluster in targeting PTEN (20). Given the role of PTEN in the negative control of PI3K activity, the ability of Myc to induce miRNAs that target PTEN, as documented here, provides a mechanism for synergy in creating the proliferative state (39). Based on these combined results, we propose a novel mechanism by which Myc regulates both the expression of anti-proliferative genes such as the CDK inhibitors and Rb, as well as the level of PTEN to evade apoptosis, through the action of Myc-induced miRNAs.
Supplementary Material
Acknowledgements
We thank Jen-Tsan Ashley Chi, Igor Shats and Jeffery T. Chang, for helpful discussions and critical reading of the manuscript. We also would like to thank Bryan R. Cullen and Pavlina S. Konstantinova for helping with vector constructions and helpful advice. This research was funded (J.R.N and J.W.K) by a grant from the National Institute of Health/National Cancer Institute (5 U54 CA112952) J.W.K was supported by a grant from the Department of Defense (W81XWH-09-1-0102). We are grateful to Kaye Culler for her assistance in the preparation of the manuscript.
Reference
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Murphy DJ, Junttila MR, Pouyet L, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 5.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 6.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 7.Claassen GF, Hann SR. Myc-mediated transformation: the repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- 8.Gartel AL, Shchors K. Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Exp Cell Res. 2003;283:17–21. doi: 10.1016/s0014-4827(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 9.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 10.Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Shen J, Wu M, et al. Repression of transcription of the p27(Kip1) cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene. 2001;20:1688–1702. doi: 10.1038/sj.onc.1204245. [DOI] [PubMed] [Google Scholar]
- 13.Gartel AL, Ye X, Goufman E, et al. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci U S A. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Cetinkaya C, Munoz-Alonso MJ, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 15.Brenner C, Deplus R, Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 19.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 20.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009 doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stampfer M, Hallowes RC, Hackett AJ. Growth of normal human mammary cells in culture. In Vitro. 1980;16:415–425. doi: 10.1007/BF02618365. [DOI] [PubMed] [Google Scholar]
- 24.Mori S, Chang JT, Andrechek ER, et al. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene. 2009 doi: 10.1038/onc.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 27.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DS, Leone G, DeGregori J, et al. Induction of DNA replication in adult rat neurons by deregulation of the retinoblastoma/E2F G1 cell cycle pathway. Cell Growth Differ. 2000;11:625–633. [PubMed] [Google Scholar]
- 29.Liu YC, Li F, Handler J, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castoldi M, Schmidt S, Benes V, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) Rna. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu J, Iyer VR. PI3K signaling and miRNA expression during the response of quiescent human fibroblasts to distinct proliferative stimuli. Genome Biol. 2006;7:R42. doi: 10.1186/gb-2006-7-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina R, Zaidi SK, Liu CG, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulte JH, Horn S, Otto T, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 35.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 36.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 37.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 40.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, et al. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 41.Coller HA, Grandori C, Tamayo P, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 43.Chang TC, Zeitels LR, Hwang HW, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satou A, Taira T, Iguchi-Ariga SM, Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- 45.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 46.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 47.Rogler CE, Levoci L, Ader T, et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009 doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.