Abstract
The biologic underpinnings of posttraumatic stress disorder (PTSD) have not been fully elucidated. Previous work suggests that alterations in the immune system are characteristic of the disorder. Identifying the biologic mechanisms by which such alterations occur could provide fundamental insights into the etiology and treatment of PTSD. Here we identify specific epigenetic profiles underlying immune system changes associated with PTSD. Using blood samples (n = 100) obtained from an ongoing, prospective epidemiologic study in Detroit, the Detroit Neighborhood Health Study, we applied methylation microarrays to assay CpG sites from more than 14,000 genes among 23 PTSD-affected and 77 PTSD-unaffected individuals. We show that immune system functions are significantly overrepresented among the annotations associated with genes uniquely unmethylated among those with PTSD. We further demonstrate that genes whose methylation levels are significantly and negatively correlated with traumatic burden show a similar strong signal of immune function among the PTSD affected. The observed epigenetic variability in immune function by PTSD is corroborated using an independent biologic marker of immune response to infection, CMV—a typically latent herpesvirus whose activity was significantly higher among those with PTSD. This report of peripheral epigenomic and CMV profiles associated with mental illness suggests a biologic model of PTSD etiology in which an externally experienced traumatic event induces downstream alterations in immune function by reducing methylation levels of immune-related genes.
Keywords: epidemiology, methylation, cytomegalovirus, cumulative trauma, psychiatry
Posttraumatic stress disorder (PTSD) is an atypical psychological and physiological response that can occur among persons exposed to a potentially traumatic event (PTE) involving life threat, serious injury, or death. To obtain a diagnosis of PTSD, reactions to the traumatic stressor must include fear, helplessness, or horror. Additionally, persons must experience symptoms from three distinct yet cooccurring domains, including intrusive recollections or reexperiencing of the PTE, accompanied by intense psychological distress or physiologic reactivity; persistent avoidance of feeling, thoughts, or activities that are associated with the trauma, which may involve amnesia or numbness to external stimuli; and increased arousal that may involve insomnia, hypervigilance, or an exaggerated startle response. These cooccurring symptoms must be present for a minimum of 1 month, and, to warrant a PTSD diagnosis, must be severe enough to impair an individual's social, occupational, or interpersonal functioning (1).
Exposure to traumatic stress is a prerequisite for a PTSD diagnosis and distinguishes it from other psychopathologies. Indeed, PTSD has been described as a specific phenotype that develops as a result of a failure to contain the normal stress response (2), resulting in dysregulation of the hypothalamic–pituitary–adrenal (HPA) axis, one of the body's major stress response systems that interacts reciprocally with the immune system to maintain homeostasis (3). PTSD-affected and -unaffected individuals have been shown to have distinct expression patterns in genes involved in immune activation (4, 5) and in genes that encode neural and endocrine proteins (4, 6).
Recent work suggests that epigenetic DNA methylation changes may accompany lifetime experiences and alter gene expression profiles (7). This work has particular promise for the study of PTSD given the direct link between an experience (i.e., a PTE) and a physiologic manifestation of illness. However, to the best of our knowledge, there have been no studies to date that have documented differences in epigenetic methylation patterns among persons with vs. without PTSD, although murine (8, 9) and human (10) studies of genes involved in the HPA axis suggest that epigenetic signatures relevant to the disorder may well exist. In particular, given the body of work suggesting an association between PTSD and alterations in the immune system (3), it is plausible that PTSD may be associated with epigenetic changes in immune system–related gene clusters. If this were the case it would provide clues to a potential mechanism through which an externally experienced traumatic event changes gene expression, thus altering immune function and resulting in other possible physiologic alterations.
Here we report results from a large-scale investigation of epigenetic methylation and immune function profiles among PTSD-affected and -unaffected individuals, drawing on samples obtained from the Detroit Neighborhood Health Study (DNHS). The DNHS is an ongoing, longitudinal epidemiologic study investigating correlates of PTSD and other mental disorders in the city of Detroit (additional information on the study is available in SI Appendix). In wave 1 of this study, prevalences of major depressive disorder (MDD) and generalized anxiety disorder (GAD) were in line with national levels (SI Appendix); however, lifetime and 12-month prevalences of PTSD were more than twice that previously reported in other epidemiologic studies (14.4% and 10.0% vs. 6.8% and 3.5%, respectively) (11) (SI Appendix). This finding may be due in part to an increased exposure to assaultive violence, which has been associated with a higher conditional risk of PTSD compared with other traumatic event types (12): exposure to assaultive violence in the DNHS survey sample was 50.8%, a level that far exceeds that reported for suburban communities surrounding the city of Detroit (13) but is consistent with levels reported for other major US urban areas (12). These findings suggest that PTE exposure can vary across disparate social environments (12), with resulting negative psychological and physiologic consequences. The relatively high burden of PTSD-related psychopathology in the DNHS study population thus offers a unique opportunity to assess the epigenetic correlates of this disorder, which may, in the longer term, assist efforts to identify effective interventions.
Results
We used DNA derived from whole blood from 100 individuals, 23 of whom met the criteria for lifetime PTSD, who participated in wave 1 of the DNHS. Using the humanmethylation27 (HM27) DNA Analysis BeadChip by Illumina, we assessed methylation profiles at more than 27,000 CpG sites covering more than 14,000 genes (additional details regarding experimental protocol and PTSD determination are available in SI Appendix). PTSD-affected and -unaffected individuals did not differ significantly with respect to age, sex, race, or peripheral blood mononuclear cell count (Table S1), and all 100 individuals had been exposed to at least one PTE.
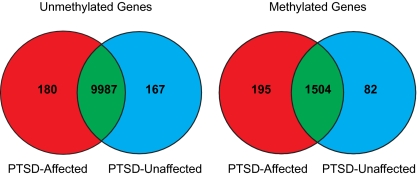
We focused our initial efforts on identifying genes that were uniquely methylated or unmethylated among PTSD-affected vs. -unaffected individuals. DNA methylation beta values are continuous variables between 0 (completely unmethylated) and 1 (completely methylated). In this analysis, we classified probes with beta values <0.2 as unmethylated, and probes with a value of >0.8 as methylated. Although there is currently no consensus regarding how best to model methylation data (14), these values are conservative compared with other cutoffs that have previously been used to determine methylated and unmethylated sites on the HM27 BeadChip (15) and compared with the analogous percentage of methylation reference values often adopted in the cancer literature to represent unmethylated and methylated genes, respectively (e.g., ref. 16). We averaged the methylation levels for each gene in each of the two groups and then determined the number of shared and uniquely methylated/unmethylated genes in each comparison (Fig. 1). The number of uniquely unmethylated genes did not differ significantly between PTSD-affected and -unaffected individuals (χ2 = 0.49, 1 df, P = 0.485); however, there was a significant difference between the two groups with respect to the number of uniquely methylated genes (χ2 = 46.10, 1 df, P < 0.0001). Gene expression analysis of the genes commonly methylated and unmethylated in both groups suggests that, in general, a methylated state corresponds to lower gene expression levels and, conversely, an unmethylated state corresponds to higher gene expression levels (Fig. S1). Pyrosequencing and targeted DNA sequencing of select loci (n = 5) confirmed the direction of initial methylation results from the HM27 BeadChip and the existence of invariant sequence at the probe target regions (SI Appendix).
Fig. 1.
Number of methylated and umethylated genes according to PTSD status. Red indicates the genes uniquely methylated or unmethylated in the PTSD-affected group, blue indicates the genes uniquely methylated or unmethylated in the PTSD-unaffected group, and green indicates the genes commonly methylated or unmethylated in both groups. Methylated genes were defined as those genes with average β values of >0.8, and unmethylated genes were defined as those genes with average β values of <0.2.
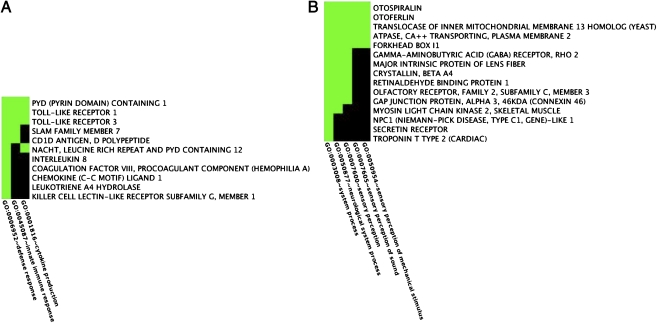
To gain insight into the biologic significance of these uniquely methylated and unmethylated genes, we undertook functional annotation clustering (FAC) analyses. Table 1 presents the results from the top three FACs determined from uniquely methylated and unmethylated genes in each of the two groups (the full set of results is available in Table S2, Table S3, Table S4, and Table S5). Fig. 2 A and B illustrate two sample FACs, and the genes they contain, determined from the PTSD-affected group. Clusters were identified by selecting the overrepresented annotation that conveyed the broadest biologic meaning within each FAC. Consistent with previous findings from gene expression (4, 5) and psychoeneuroimmunologic studies (3), each of the top three FACs determined from uniquely unmethylated genes among PTSD-affected individuals shows a strong signature of immune system involvement. This signature includes genes from the innate immune system (e.g., TLR1 and TLR3), as well as from genes that regulate innate and adaptive immune system processes (e.g., IL8, LTA, and KLRG-1). In contrast, pathways and processes relevant to organismal development in general—and neurogenesis in particular—figure prominently among the genes uniquely unmethylated in the PTSD-unaffected group (e.g., CNTN2 and TUBB2B; Fig. S2). Notably, similar clusters were obtained using an alternative approach based on genes differentially methylated between the two groups at P < 0.01, with annotations in the top five FACs that include signal, cell proliferation, developmental process, neurologic system process, and inflammatory response (complete results of all FAC analyses are available in Table S2, Table S3, Table S4, Table S5, Table S6, Table S7, Table S8, Table S9, and Table S10 and in Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, Fig. S7, Fig. S8, Fig. S9, Fig. S10, Fig. S11, Fig. S12, Fig. S13, Fig. S14, Fig. S15, Fig. S16, Fig. S17, Fig. S18, Fig. S19, Fig. S20, Fig. S21, Fig. S22, and Fig. S23).
Table 1.
Functional annotation cluster analyses of uniquely methylated and unmethylated genes
| Uniquely unmethylated |
Uniquely methylated |
|||||
| Group | Cluster | No. of genes | Enrichment score | Cluster | No. of genes | Enrichment score |
| PTSD affected | Inflammatory response | 26 | 2.38 | Signal | 55 | 2.16 |
| Immune response | 38 | 1.5 | Sensory perception of sound | 15 | 1.07 | |
| Innate immune response | 11 | 1.48 | Response to xenobiotic stimulus | 5 | 1.02 | |
| PTSD unaffected | Developmental process | 50 | 2.67 | Signal | 32 | 1.77 |
| Generation of neurons | 37 | 2.2 | Lipase activity | 6 | 1.22 | |
| Intracellular organelle | 102 | 1.97 | Calcium ion binding | 10 | 1.17 | |
All genes are identified in terms of DAVID IDs; genes can appear in more than one cluster.
Fig. 2.
Sample FAC heat maps in the PTSD-affected group. Shown are the genes and associated annotations for FAC cluster 3 (A) and 2 (B) determined from the uniquely unmethylated and methylated gene sets, respectively.
PTSD often cooccurs with other mental illnesses, in particular depression and other anxiety disorders (17). To investigate the effect that comorbidity may have on our results, we assessed whether genes were significantly differentially methylated among those affected only by PTSD (n = 8), those affected by PTSD and one or more of MDD and GAD (n = 15), and those unaffected by any of these three psychopathologies (n = 53). ANOVA tests confirmed that there were significant differences in methylation levels across the three groups in 345 genes (P < 0.01); however, only three genes showed methylation levels that were significantly different at this alpha level between those affected only by PTSD and those affected by PTSD and another disorder (Tukey honestly significant difference), suggesting that comorbidity did not unduly influence our results.
PTSD is diagnosed in reference to a specific traumatic event rather than multiple potential traumas (1). Epidemiologic studies, however, have demonstrated that, among the PTE exposed, a majority of individuals have been exposed to more than one traumatic event (12, 13) and that multiple such exposures can correlate with symptom severity (18). To investigate whether cumulative trauma is associated with distinctive methylation profiles among PTSD-affected vs. -unaffected individuals, we assessed the correlation between number of PTEs and methylation levels for each CpG site represented on the HM27 microarray. Compared with PTSD-unaffected individuals, PTSD-affected individuals showed nearly six times as many genes (176 vs. 30) with significant (P < 0.01) negative correlations to number of PTEs experienced and nearly seven times as many genes (170 vs. 25) with significant positive correlations to number of PTEs experienced (range of Spearman correlation coefficients, ρ: −0.698 to −0.526 and 0.527 to 0.736 among the PTSD affected; and −0.460 to −0.295 and 0.292 to 0.389 among the PTSD unaffected, respectively). Results from FAC analyses conducted on these four gene sets are presented in Table 2. Here again we see a distinct signature of immune-related methylation profiles among the PTSD-affected group only. More specifically, we see methylation profiles that are suggestive of immune activation among persons with more PTE exposure in the genes that are significantly negatively correlated with increasing number of PTEs—a pattern reflective of that observed for the uniquely unmethylated genes in this same group (Table 1).
Table 2.
Functional annotation cluster analyses of genes significantly correlated with number of PTEs
| Group | Correlation | Cluster | No. of genes | Enrichment score |
| PTSD affected | Negative | Immune response | 38 | 2.14 |
| Defense response | 56 | 2.11 | ||
| Nucleotide receptor activity | 11 | 1.82 | ||
| Positive | I-κB kinase/NF-κB cascade | 10 | 1.38 | |
| Cell proliferation | 13 | 1.34 | ||
| Positive regulation of signal transduction | 21 | 1.31 | ||
| PTSD unaffected | Negative | Transporter activity | 9 | 1.13 |
| Membrane | 17 | 1.05 | ||
| Transmembrane transporter activity | 10 | 0.99 | ||
| Positive | Calcium binding | 6 | 1 | |
| Secreted | 5 | 0.49 | ||
| Nonmembrane bound organelle | 11 | 0.44 |
P < 0.01; all genes are reported in terms of DAVID IDs; genes can appear in more than one cluster.
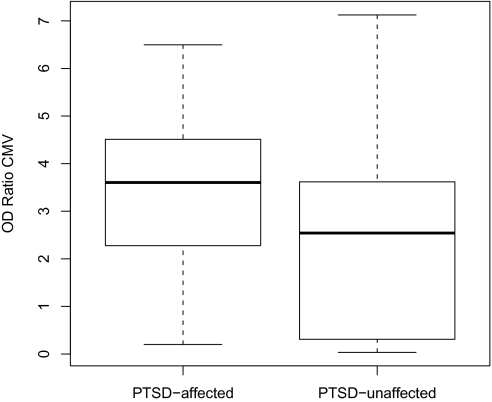
To corroborate these immune-related findings with an independent, physiologic marker of immune function, we assessed antibody levels to CMV. CMV is a persistent herpesvirus highly prevalent in most human populations (19). CMV infection drives many of the alterations associated with an aging immune system (20), and its value as a sentinel biomarker indicative of a compromised immune system is increasingly being recognized (21). If PTSD, as hypothesized here, is the physiologic manifestation of a traumatic insult that results in epigenetic changes and subsequent gene expression patterns that alter immune function, it would be plausible to expect alterations in immune response to CMV among persons affected vs. unaffected by PTSD. In the present study, the titers of CMV-specific antibody, as reflected in optical density ratios, were significantly different between PTSD-affected and -unaffected individuals (mean ratios 3.37 and 2.32, respectively, Welch's t statistic = 2.48, 37 df, P = 0.016; Fig. 3). These results suggest that PTSD is associated with compromised immune reactivity to CMV.
Fig. 3.
Optical density ratios of CMV antibody levels among PTSD-affected and -unaffected individuals. Levels were significantly higher in the PTSD-affected group (Welch's t statistic = 2.48, 37 df, P = 0.016).
Discussion
Drawing on a unique, ongoing epidemiologic study, we have documented a distinct signature reflecting immune activation among PTSD-affected individuals, using both epigenetic markers and a marker indicative of a compromised immune system—the antibody response to CMV. Although preliminary, the findings here are consistent with a model whereby exposure to a PTE induces changes in methylation profiles among some persons and that these changes result in changes in gene expression associated with altered immune function, demonstrated here through differences in immune response to CMV.
Epigenomic Phenotype of PTSD-Affected vs. -Unaffected Individuals.
Among the many analyses performed in this work, the immune-related functions identified in the PTSD-affected group were consistently identified only among gene sets with relatively lower levels of methylation (Tables 1 and 2). Demethylation has previously been shown to correlate with increased expression in several immune system–related genes (reviewed in ref. 22), including some identified here [e.g., IL8 (23)]. In contrast, methylation profiles among the PTSD-unaffected are distinguished by neurogenesis-related functional annotations. Neural progenitor cells have previously been identified in the adult human hippocampus (24); however, stress can inhibit cell proliferation and neurogenesis in this brain region (reviewed in ref. 25), and recent work suggests that adult neurogenesis may be regulated by components of the immune system (reviewed in ref. 26). Thus, immune dysfunction among persons with PTSD may be influenced by epigenetic profiles that are suggestive of immune activation or enhancement and also by an absence of epigenetic profiles that would be consistent with the development of normal neural-immune interactions (27).
Among the genes uniquely methylated in the PTSD-affected group, it is striking that the second most enriched cluster—sensory perception of sound—directly reflects one of the three major symptom clusters that define the disorder (Fig. 3B). Genes in this FAC that may be particularly salient to this symptom domain include otospiralin (OTOS), which shows decreased expression in guinea pigs after acoustic stress (28) and otoferlin (OTOF), mutations in which have been linked to nonsyndromic hearing loss in humans (29). Exaggerated acoustic startle responses, often measured via heart rate or skin conductance after exposure to a sudden, loud tone, have been well documented among the PTSD affected (30) and are indicative of a hyperarousal state that characterizes this symptom domain. Notably, prospective studies have demonstrated that an elevated startle response is a consequence of having PTSD, because the response was not present immediately after exposure to trauma but developed with time among trauma survivors who developed the disorder (30, 31). Although more recent work suggests that such reactions may be influenced by a preexisting hypersensitivity to contextual threat (32), these findings are generally consistent with our hypothesis that PTE exposure may induce epigenetic changes that produce physiologic alterations among PTSD-affected individuals.
Assessment of Comorbidity.
Our finding that methylation levels did not differ significantly between individuals affected by PTSD alone vs. those affected by PTSD and other mood–anxiety disorders lends biologic support to previous epidemiologic findings. Population-based studies have shown that after traumatic event exposure, although there is a higher prevalence of other psychopathologies, PTSD is the sentinel diagnosis, and other diagnoses seldom appear without PTSD (33). In biologic terms, that so few genes show significantly different methylation levels in post hoc comparisons between individuals affected by PTSD alone vs. PTSD and other comorbid disorders suggests that the hypothesized underlying biologic cascade leading to PTSD (i.e., PTE exposure→epigenetic changes→gene expression changes→PTSD) operates among all persons with the disorder, regardless of whether they have other axis I psychological disorders. Future work should verify these results in other, independent cohorts using additional measures of comorbidity.
Maintenance of DNA Methylation and Imprinted Genes.
DNA methylation patterns are maintained and created by DNA methyltransferases, a family of enzymes that, in mammals, catalyze the transfer of a methyl group to cytosine residues predominantly, but not exclusively (34), at CpG dinucleotide sites (35). This enzymatic machinery both sets up methylation patterns early in development and maintains them through subsequent somatic cell division during an individual's lifetime. In this context, it is noteworthy that a gene encoding one of these enzymes (DNMT3B) is both uniquely unmethylated and shows significantly less methylation (P < 0.017) in the PTSD-affected group; and that another, closely related gene (DNMT3L) is significantly more methylated (P = 0.037) among individuals with vs. without PTSD. Although the bulk of these genes’ activity is typically thought to occur early in development (36), recent work has suggested that their enzymatic products may also play a role in DNA methylation maintenance during somatic cell division (35). This suggests that the underlying machinery responsible for creating and maintaining methylation patterns may function differently among those with vs. without PTSD, producing downstream phenotypes with both pathophysiologic and psychopathological dimensions.
Similarly, imprinted genes—genes silenced through epigenetic mechanisms early in development in a parent-of-origin fashion—often function as disease susceptibility genes through deletion and/or inactivation of loci that normally follow a monoallelic expression pattern. Perhaps not surprisingly, a handful of genes known to be involved in well-known diseases of imprinting, specifically Praeder-Willi syndrome (PWS) and Angelmann syndrome (AS), appear on the uniquely methylated and unmethylated gene lists; of interest here, however, is the way in which the methylation pattern seems to “track” both the PTSD phenotype and the phenotype associated with each of these disorders. Two genes, NDN and MAGEL2, localize to the PWS deletion region and are normally expressed from the paternal allele (37, 38). NDN appears on the uniquely unmethylated gene list for those without PTSD, suggesting the likelihood of proper expression; MAGEL2, on the other hand, appears on the uniquely methylated gene list for those with PTSD, suggesting reduced expression in a manner akin to what occurs in PWS. ATP10A, a gene that maps to the genomic region most commonly deleted in AS and that is typically expressed from the maternal allele (39), also appears on the uniquely methylated PTSD list, suggesting again reduced expression in a manner concurrent with AS. These examples suggest that methylation profiles of imprinted genes may contribute to PTSD-associated phenotypes, in a manner that remains to be more fully elucidated.
Study Limitations.
Our study includes a number of limitations that may limit the generalizability of its results. The cross-sectional analyses reported here leave us unable to infer at present whether the observed distinctive methylation profiles and CMV antibody levels are a consequence of PTSD or whether they are indicative of biologic vulnerabilities that existed among the PTSD affected before the onset of their disorder. In addition, given the limited number of PTSD-affected individuals included in this study, we were unable to address in this work whether distinct methylation profiles may exist within the PTSD-affected group that could reflect distinct subtypes and/or phenotypic heterogeneity within this clinical syndrome, as has previously been suggested (2, 40). Finally, our epigenetic data were collected from DNA derived from whole blood, which includes a heterogeneous mixture of cell types; as such, we were unable to assess blood cell–specific differences in methylation status, which would have to be accounted for when replicating the present findings in other, longitudinal cohorts.
Concluding Remarks.
PTSD is an uncommon response to stress. Although nearly 90% of individuals are exposed to a traumatic event during their lifetime (13), only a minority go on to develop the disorder. Nevertheless, this illness is one of the more common and disabling psychopathologies in the United States: with a lifetime prevalence of 6.8% and a 12-month prevalence of 3.5% (11), impairment of proper role functioning due to PTSD is significantly worse than in a number of commonly occurring chronic medical disorders (41). In this study, we found that Detroit residents show lifetime and 12-month PTSD prevalences more than twice these national averages, suggesting that such PTSD-associated impairment may be borne disproportionately among individuals residing in urban social environments. Identification of the biologic underpinnings of PTSD will be crucial for facilitating the development of appropriate psychological and/or pharmacologic interventions, particularly in the wake of an increasing number of military veterans returning home after recent wars worldwide.
Materials and Methods
Participants.
Our analysis made use of a subset of participants from the DNHS (for more details regarding the DNHS, readers are referred to SI Appendix). The sample for this study consisted of 100 DNHS participants. Forty were male and 60 female; 14% were white, 79% African American, and 7% other race; 14% had less than a high school education, whereas 86% were at least a high school graduate; and the average age was 45.8 years (Table S1). The DNHS was approved by the institutional review board at the University of Michigan.
Assessment of PTSD.
Individual assessment of PTSD symptoms were conducted using the PTSD checklist (PCL-C) (42), a 17-item self-report measure of Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) symptoms (13, 43), and additional questions about duration, timing, and impairment or disability due to the symptoms.
Participants were initially asked to identify PTEs that they experienced in the past from a list of 19 events. PTSD symptoms were then assessed by referencing two traumatic events that the respondent may have experienced: one that the participant regarded as the worst and one randomly selected event from the remaining PTEs a respondent may have experienced. Respondents were considered affected by lifetime PTSD if all six DSM-IV criteria were met in reference to either the worst or the random event. All 100 individuals included in this study were exposed to at least one PTE; among these, 23 were PTSD affected and 77 were unaffected. Additional psychopathologies were assessed as described in SI Appendix.
Microarray Analyses.
Bisulfite conversion of whole blood–derived DNA samples was performed using the EZ-96 DNA methylation kit from Zymo Research. One microgram of each sample (including controls) was subjected to bisulfite conversion according to the manufacturer's recommended protocol. Experimental controls included replicates for two samples to assess variation throughout the experimental process (i.e., from initial bisulfite conversion through microarray analysis), as well as one sample of completely unmethylated and completely methylated human DNA, commercially available through Zymo Research, in each of the two 96-well plates used in the bisulfite conversion step. All control replicates were placed on separate microarray chips, and the remaining samples were assigned to microarray chips at random, without regard to PTSD status. Bisulfite-converted DNA samples were subjected to methylation profiling via the HM27 DNA Analysis BeadChip by Illumina according to the manufacturer's recommended protocol. Using this platform, methylation levels were determined for 27,578 CpG dinucleotides spanning 14,495 genes in each of the 100 test samples. The resulting data were background normalized and exported for additional analysis using the R package v2.9.0 (44) and SAS software v9.2 (45). Correlation coefficients of the two replicated samples were 0.81 and 0.89, respectively. Average beta values for the methylated controls was 0.93 and the correlation 0.96. Average beta values and correlation of unmethylated controls were 0.17 and 0.98, respectively.
Initial analyses focused on identifying genes corresponding to probes (CpG sites) that were uniquely methylated or unmethylated among PTSD-affected vs. -unaffected individuals. We calculated the average beta values of the samples in the two groups (PTSD affected and unaffected) and classified probes with average beta values <0.2 as unmethylated and probes with an average beta value of >0.8 as methylated. Each probe was identified to be uniquely methylated or uniquely unmethylated if it was methylated—or unmethylated—in just one group. Because it was possible that a gene may be associated with more than one probe on the HM27 BeadChip, we created two subsets of data consisting only of (i) methylated probes in either the PTSD-affected or -unaffected group, and (ii) unmethylated probes in either the PTSD-affected or -unaffected group. Then, for each subset, we removed duplicate records based on the gene symbol and methylation status in each of the two groups. In this way, genes with two or more probes that showed similar results (both probes uniquely methylated in the PTSD-affected group) were not counted twice. Conversely, genes with two or more probes showing different results (e.g.. one probe is uniquely methylated in the PTSD-affected group, whereas the other probe is uniquely methylated in the PTSD-unaffected group) were counted twice (or more); each result is thus accounted for. The proportion of uniquely methylated and unmethylated genes among PTSD-affected vs. -unaffected individuals was compared using McNemar's χ2 test.
Differential methylation analysis between the PTSD-affected and -unaffected groups was conducted for each probe using the Wilcoxon test. We identified a probe to be significantly differentially methylated if P < 0.01. This analysis was extended to assess differential methylation among participants with no psychopathology, those who were PTSD affected only, and those who were PTSD affected and affected by one or more of MDD and GAD; for this analysis, ANOVA was initially performed followed by the Tukey honestly significant difference test, a post hoc pairwise comparison test.
To assess the correlation between the number of PTEs experienced and methylation levels of each probe among PTSD-affected vs.-unaffected individuals, we stratified by PTSD status and analyzed the correlation between the number of lifetime PTEs and methylation levels of each probe, separately for each group (PTSD affected and PTSD unaffected). Spearman's rho was calculated for each probe, and results were accepted as significant if P < 0.01.
HM27 probes analyzed as described above were assigned to genes on the basis of annotation files available from Illumina on May 20, 2009. Genes identified through the above analyses were assessed for functional significance using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (46, 47). Results were obtained with the FAC tool, with options set to their default values, using annotations available in DAVID as of June–August 2009.
Experimental Procedures: CMV Analyses.
CMV IgG antibody levels were tested using a standard commercial ELISA kit for detecting type-specific IgG antibody in serum (Inverness Medical Innovations; 425200CE). Serum samples were eluted in buffer before being pipetted in duplicate into antigen-coated microtiter wells. Through a series of incubation and wash steps, the CMV antibodies of the sample were anchored to the microtiter plate and linked to a secondary reactive antibody. Absorbance was quantified by a microplate reader set at 450 nm. Concentrations were calculated by optical density (positive ≥1.10, equivocal 0.91–1.09, negative ≤0.9). Antibody levels are expressed in terms of mean optical density ratio. The Inverness Medical Innovations kit exhibits high specificity of 93.9% and a sensitivity of 96.4% when compared with other CMV IgG ELISA procedures. A t test was conducted to compare CMV optical density (OD) ratios between the two groups.
Gene Expression Analysis.
Publicly accessible data on gene expression were obtained from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database. Whole blood–derived gene expression levels from 40 samples were obtained for analysis from the following datasets: GDS2952, GDS2519 (48), GDS1331 (49), and GDS596 (50). All samples were derived from healthy control individuals in these datasets. Commonly methylated and unmethylated probes were obtained from our data as presented in green in Fig. 1. RefSeqs of the GEO data were obtained from Affymetrix NetAffx Analysis Center batch query as of July 27, 2009 and then matched with the RefSeqs (accession) in the Illumina data for additional analysis. Raw gene expression values of commonly methylated and unmethylated probes were log base 2 transformed and normalized by median centering. The values were then averaged for each probe, and a t test was performed to compare the mean expression levels between the two groups.
Supplementary Material
Acknowledgments
We thank Rebecca M. Coulborn for overseeing DNHS specimen collection, Janie Slayden for coordinating the overall DNHS project, and Amy Weckle and Richelo Soliven for handling the DNHS specimen processing and laboratory technical assistance; the many Detroit residents who chose to participate in the DNHS; and Jorge Delva, Larry Gant, Bob Marans, and Trivellore Raghunathan for contributing to the conceptual development of the DNHS. We also thank Dr. Sue Land and staff (Wayne State University, Applied Genomic Technology Center) for running microarrays. This study was supported by National Institutes of Health Grants DA022720, DA022720-S1, MH078152, and MH082729 (to S.G.) and MH070627 and MH078928 (to K.K.). Additional support was provided by the Robert Wood Johnson Health and Society Scholars Small Grant Program and the University of Michigan Office of the Vice President for Research Faculty Grants and Awards Program (M.U.); and by the Wayne State University Research Excellence Fund (D.W.). Funding for CMV testing was generously provided by Grant AG013283 from the University of Michigan Nathan Shock Center (to A.A.). G.P. was supported by Deutsche Forschungsgemeinschaft Grant DFG PA361-11/1 and European Commission Grant EU-LSHG-CT-2007-036894 (“LifeSpan”).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21282).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910794107/-/DCSupplemental.
References
- 1.American Psychiatric A. Diagnostic and Statistical Manual of Mental Disorders. 4th Ed. Washington, DC: American Psychiatric Association; 1994. pp. 424–429. [Google Scholar]
- 2.Yehuda R, LeDoux J. Response variation following trauma: A translational neuroscience approach to understanding PTSD. Neuron. 2007;56:19–32. doi: 10.1016/j.neuron.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Wong CM. Post-traumatic stress disorder: Advances in psychoneuroimmunology. Psychiatr Clin North Am. 2002;25:369–383. doi: 10.1016/s0193-953x(01)00006-5. [DOI] [PubMed] [Google Scholar]
- 4.Segman RH, et al. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry. 2005;10:500–513. doi: 10.1038/sj.mp.4001636. 425. [DOI] [PubMed] [Google Scholar]
- 5.Zieker J, et al. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry. 2007;12:116–118. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
- 6.Yehuda R, et al. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol Psychiatry. 2009;66:708–711. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 9.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: Altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessler RC, Wang PS. The descriptive epidemiology of commonly occurring mental disorders in the United States. Annu Rev Public Health. 2008;29:115–129. doi: 10.1146/annurev.publhealth.29.020907.090847. [DOI] [PubMed] [Google Scholar]
- 12.Breslau N, Wilcox HC, Storr CL, Lucia VC, Anthony JC. Trauma exposure and posttraumatic stress disorder: A study of youths in urban America. J Urban Health. 2004;81:530–544. doi: 10.1093/jurban/jth138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breslau N, et al. Trauma and posttraumatic stress disorder in the community: The 1996 Detroit Area Survey of Trauma. Arch Gen Psychiatry. 1998;55:626–632. doi: 10.1001/archpsyc.55.7.626. [DOI] [PubMed] [Google Scholar]
- 14.Foley DL, et al. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunner AL, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pike BL, et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breslau N, Davis GC, Peterson EL, Schultz L. Psychiatric sequelae of posttraumatic stress disorder in women. Arch Gen Psychiatry. 1997;54:81–87. doi: 10.1001/archpsyc.1997.01830130087016. [DOI] [PubMed] [Google Scholar]
- 18.Suliman S, et al. Cumulative effect of multiple trauma on symptoms of posttraumatic stress disorder, anxiety, and depression in adolescents. Compr Psychiatry. 2009;50:121–127. doi: 10.1016/j.comppsych.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Staras SA, et al. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006;43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 20.Pawelec G, Derhovanessian E, Larbi A, Strindhall J, Wikby A. Cytomegalovirus and human immunosenescence. Rev Med Virol. 2009;19:47–56. doi: 10.1002/rmv.598. [DOI] [PubMed] [Google Scholar]
- 21.Derhovanessian E, Larbi A, Pawelec G. Biomarkers of human immunosenescence: Impact of Cytomegalovirus infection. Curr Opin Immunol. 2009;21:440–445. doi: 10.1016/j.coi.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 22.van Panhuys N, Le Gros G, McConnell MJ. Epigenetic regulation of Th2 cytokine expression in atopic diseases. Tissue Antigens. 2008;72:91–97. doi: 10.1111/j.1399-0039.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira NF, et al. DNA methylation status of the IL8 gene promoter in oral cells of smokers and non-smokers with chronic periodontitis. J Clin Periodontol. 2009;36:719–725. doi: 10.1111/j.1600-051X.2009.01446.x. [DOI] [PubMed] [Google Scholar]
- 24.Manganas LN, et al. Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain. Science. 2007;318:980–985. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirescu C, Gould E. Stress and adult neurogenesis. Hippocampus. 2006;16:233–238. doi: 10.1002/hipo.20155. [DOI] [PubMed] [Google Scholar]
- 26.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Wrona D. Neural-immune interactions: An integrative view of the bidirectional relationship between the brain and immune systems. J Neuroimmunol. 2006;172:38–58. doi: 10.1016/j.jneuroim.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Caravelli A, et al. Down-regulation of otospiralin mRNA in response to acoustic stress in guinea pig. Hear Res. 2004;198:36–40. doi: 10.1016/j.heares.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Varga R, et al. OTOF mutations revealed by genetic analysis of hearing loss families including a potential temperature sensitive auditory neuropathy allele. J Med Genet. 2006;43:576–581. doi: 10.1136/jmg.2005.038612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalev AY, et al. Auditory startle response in trauma survivors with posttraumatic stress disorder: A prospective study. Am J Psychiatry. 2000;157:255–261. doi: 10.1176/appi.ajp.157.2.255. [DOI] [PubMed] [Google Scholar]
- 31.Griffin MG. A prospective assessment of auditory startle alterations in rape and physical assault survivors. J Trauma Stress. 2008;21:91–99. doi: 10.1002/jts.20300. [DOI] [PubMed] [Google Scholar]
- 32.Pole N, et al. Prospective prediction of posttraumatic stress disorder symptoms using fear potentiated auditory startle responses. Biol Psychiatry. 2009;65:235–240. doi: 10.1016/j.biopsych.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breslau N, Davis GC, Peterson EL, Schultz LR. A second look at comorbidity in victims of trauma: The posttraumatic stress disorder-major depression connection. Biol Psychiatry. 2000;48:902–909. doi: 10.1016/s0006-3223(00)00933-1. [DOI] [PubMed] [Google Scholar]
- 34.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 37.MacDonald HR, Wevrick R. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet. 1997;6:1873–1878. doi: 10.1093/hmg/6.11.1873. [DOI] [PubMed] [Google Scholar]
- 38.Boccaccio I, et al. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- 39.Meguro M, et al. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- 40.Nandi A, Beard JR, Galea S. Epidemiologic heterogeneity of common mood and anxiety disorders over the lifecourse in the general population: a systematic review. BMC Psychiatry. 2009;9:31. doi: 10.1186/1471-244X-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Druss BG, et al. Impairment in role functioning in mental and chronic medical disorders in the United States: Results from the National Comorbidity Survey Replication. Mol Psychiatry. 2009;14:728–737. doi: 10.1038/mp.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weathers F, Ford J. Psychometric review of PTSD checklist (PCL-C, PCL-S, PCL-M, PCL-R) In: Stamm BH, editor. Measurement of Stress, Trauma, and Adaptation. Lutherville, MD: Sidran Press; 1996. [Google Scholar]
- 43.Shalev AY, et al. Prospective study of posttraumatic stress disorder and depression following trauma. Am J Psychiatry. 1998;155:630–637. doi: 10.1176/ajp.155.5.630. [DOI] [PubMed] [Google Scholar]
- 44.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 45.Institute S. SAS Software v9.2. Cary, NC: SAS Institute; 2009. [Google Scholar]
- 46.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 47.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Scherzer CR, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc Natl Acad Sci USA. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borovecki F, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.