Abstract
Optimal infectivity of HIV-1 virions requires synthesis of the HIV-1 regulatory protein Nef in some producer cells but not others. A survey of 18 lymphoid cell lines found that Nef was dispensable in three, each of which harbored gammaretroviruses. Nef-dependent cell lines were rendered Nef-independent by a cell-free supernatant from the independent lines or by transfection of cloned murine leukemia virus (MLV). Analysis of MLV deletion mutations identified glycosylated gag (glycogag) as the factor that rescues Nef-defective HIV-1 virions. Glycogag was also demonstrated to be required for the infectivity of MLV virions produced in lymphoid cells. Direct comparison of Nef and glycogag revealed identical dependence for activity on Env-pseudotype and producer cell type. The two proteins colocalize within cells, and both increase the yield of viral cDNA in target cells. The functional similarity of Nef and glycogag is a compelling example of convergent evolution in which two structurally unrelated proteins provide a function necessary for virion infectivity in lymphoid cells.
Nef is a myristoylated protein encoded by HIV-1, HIV-2, and SIV, crucial for virus replication in vivo and rapid AIDS progression (1–3). It performs a remarkable array of activities by exploiting many of its surfaces to interact with several cellular molecules. By interacting with proteins implicated in intracellular trafficking, it modulates cell surface expression of numerous molecules, including the receptor CD4 (4, 5) and MHC-I (6). Alleles derived from most SIV isolates also down-regulate the TCR/CD3 complex (7–9). In addition, Nef alters the activation threshold of lymphocytes (10–12) by interacting with protein kinases (13–16) and modulates apoptotic signals (10, 17).
Nef also has a positive effect on the infectivity of virions (18, 19), a function which remains mechanistically unexplained. Although the ability to down-regulate CD4 can contribute to the effect of Nef on infectivity (20), this activity is visible by using CD4-negative producer cells and was shown to be independent from other Nef effects (18, 21–23), it requires its expression in virus-producing cells and is manifested at an early step of the infection process of target cells (18, 22, 24–26). Nef might play a crucial role during penetration of retroviral cores into the cytoplasm (27). Accordingly, HIV-1 pseudotyped by vesicular stomatitis virus G (VSV-G), which relies on endosomal uptake and, therefore, might enhance cytoplasmic delivery, does not require Nef (28, 29). Although found in virus particles, recent data indicate that Nef itself might not function as a virion protein (30, 31), suggesting that it could induce a yet unknown modification of the particle. The effect of Nef on infectivity was shown to depend on dynamin 2 and clathrin activities in producer cells (32) and on a di-leucine motif critical for the interaction with the clathrin adaptor complexes AP2 (33, 34). The biogenesis and/or trafficking of intracellular vesicles in virus producing cells might therefore play a crucial role in modulating virion infectivity.
Most gammaretroviruses encode an accessory protein (gPr80 or glycogag) from unspliced RNA via an alternative CUG initiation codon upstream and in-frame with the standard Gag polyprotein (35–39). As a result, a leader sequence [88 amino acids in MLV provirus genome (MoMLV)] is added to the N terminus of conventional Gag. The protein encoded has a type II transmembrane topology, with the conventional Gag residues being extracellular or located in the lumen of the ER, and glycosylated. Mature gPr80 is proteolytically cleaved, and only half of the conventional Gag sequence remains attached to the integral transmembrane protein (40). Although not strictly required for virus replication in vitro, gPr80 is crucial in vivo for sustained virus replication and disease progression (41–47). The mechanisms engaged by gPr80 to promote virus replication remain largely unknown. However, a recent report has revealed that gPr80 affects release of both MLV and HIV-1 by facilitating budding from lipid rafts (48).
In this study, a screen of 18 producer cell lines identified three lymphoid cell lines capable of generating HIV-1 that does not require Nef for maximal infectivity. Glycosylated gag expressed from gammaretrovirus genomes harbored by these cell lines was found to functionally replace the infectivity function of Nef, unveiling a role for glycogag as an infectivity factor.
Results
Nef Is Not Required for Optimal Infectivity of HIV-1 Derived from Three Human Lymphoid Cell Lines.
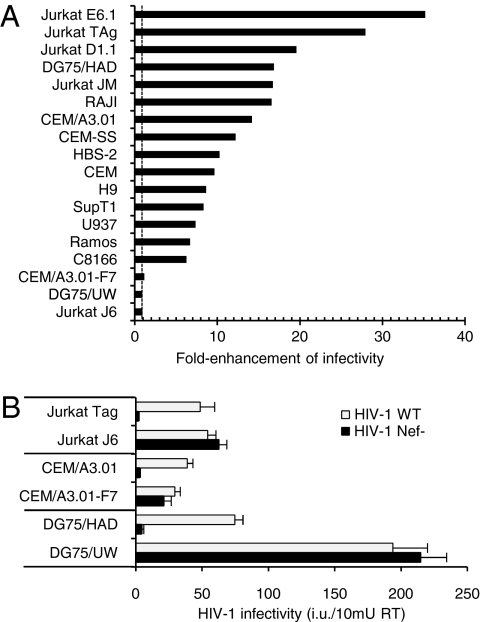
The ability of Nef to enhance HIV-1 infectivity was studied by using a panel of 18 human cell lines of hematopoietic origin, including sublines derived from common progenitors (Table S1). Nef-positive (HIV-1wt) and Nef-defective (HIV-1Nef-) virions limited to a single cycle of replication were produced by transient transfection and inoculated onto TZM-bl reporter cells (Fig. 1A). Nef did not enhance viruses derived from Jurkat J6 (JJ6), CEM/A3.01-F7, and DG75-UW cells (here defined as “Nef-independent”), but increased (6- to 40-fold) the infectivity of HIV-1 derived from the remaining 15 cell lines (defined as “Nef-dependent”), which include other Jurkat, CEM, and DG75 sublines. The absence of Nef activity on virus harbored by JJ6 is in striking contrast with the 20- to 40-fold enhancement observed on HIV-1 derived from the parental cell line (JM) or from other Jurkat sublines (JE6.1, JD1.1, JTAg). Common genomic background between JTAg and JJ6 was confirmed by sequencing the region spanning the V-D-J junction of the T cell receptor gene (SI Text), revealing that the Nef phenotype can be profoundly different, despite identical genetic background of the producer cells.
Fig. 1.
HIV-1 infectivity is Nef-independent when virions are produced from 3 of 18 lymphoid cell lines. (A) Fold enhancement of HIV-1NL4-3 infectivity when virions are produced by using different cell lines. The dotted line denotes no enhancement. Data representative of two independent experiments. (B) The Nef activity on HIV-1 infectivity is redundant in “Nef-independent” producer cell lines. Infectivity of HIV-1 produced in the indicated cells lines was determined on TZM-bl indicator cells. Bars represent the means plus SD from triplicate determinations. Results are representative of two independent experiments.
Measurement of the absolute infectivity of viruses released by sister sublines showed that HIV-1 generated by Nef-independent cell lines is equal to, or more infectious than, HIV-1WT released by Nef-dependent cell lines (Fig. 1B). This evidence indicates that JJ6, CEM/A3.01-F7, and DG75-UW cells have either gained Nef-like infectivity function or lost an inhibiting factor that can be counteracted by Nef.
A Gammaretrovirus in Producer Cells Substitutes for Nef.
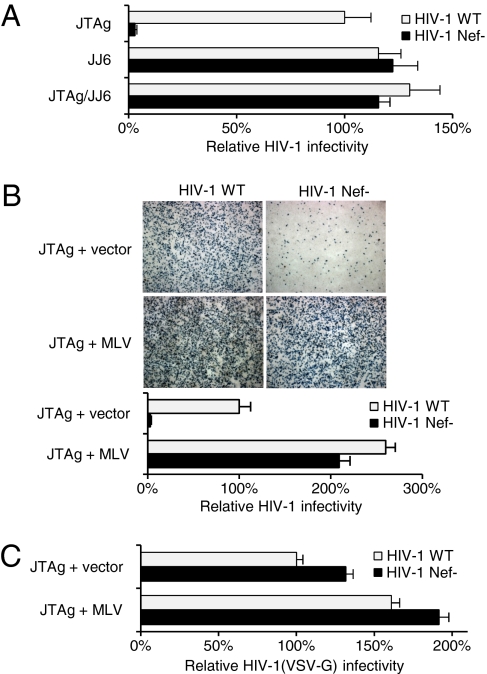
JJ6 and CEM/A3.01-F7 cells were recently found to release xenotropic MLV and gibbon ape leukemia virus (49). A xenotropic MLV had also been detected in DG75-UW cells (50). Using a PCR-based reverse transcriptase assay (51), no RT activity was detected in culture supernatant of other cells used in this study, indicating the presence of gammaretroviruses as a distinctive property of Nef-independent producer cell lines. If the absence of the Nef effect is caused by another virus in the producer cells, the Nef-independent phenotype should be transferable to Nef-dependent cell lines. JTAg cells were therefore exposed to cell-free JJ6 growth medium and cultured until levels of RT activity released indicated that the JJ6 MLV-X had propagated. The newly infected cell line (JTAg/JJ6) produced HIV-1 no longer responsive to Nef and as infectious as HIV-1 derived from JJ6 cells (Fig. 2A). The absence of the Nef phenotype can therefore be transferred from one cell line to another.
Fig. 2.
MLV rescues the infectivity of HIV-1Nef-. (A) Cell-free supernatant from JJ6 renders HIV-1 production from JTAg (JTAg/JJ6) Nef-independent. (B) MLV provirus in producer cells (JTAg+MLV) abolishes the Nef requirement for optimal HIV-1 infectivity. (B Upper) TZM-bl cells infected with RT-normalized HIV-1wt and HIV-1Nef- and stained with X-Gal. (B Lower) Quantification of HIV-1 infectivity. (C) MLV is only minimally active on the infectivity of HIV-1(VSV-G) pseudotypes. Infectivity is expressed as a percentage of the HIV-1wt control and is the mean plus SD from triplicate determinations. Results are representative of two independent experiments.
To demonstrate that the presence of a gammaretrovirus genome in producer cells abrogates the Nef requirement, HIV-1 was produced from JTAg cells cotransfected with a plasmid containing the MoMLV (41). The MLV provirus in producer cells enhanced the infectivity of the HIV-1nef- 60-fold and that of HIV-1wt only 2.5-fold (Fig. 2B). Infectivity of virus particles pseudotyped with VSV-G, which abrogates the Nef dependence, were only minimally responsive to the effect of MLV (Fig. 2C), indicating that the gammaretrovirus genome is active on HIV-1 particles that require Nef.
5′ Region of MLV Genome Is Essential for the Effect on HIV-1 Infectivity.
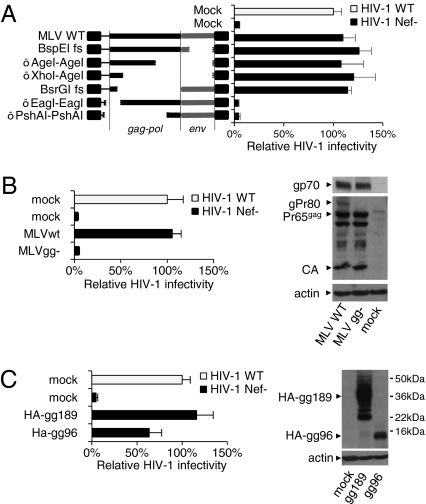
To map the MLV determinant that has Nef-like activity, frameshifts and deletions were introduced by using restriction sites in the MLV provirus to disrupt the coding capacity of MLV ORFs. The mutated genomes were tested for their ability to rescue the defective infectivity of HIV-1Nef- (Fig. 3A). The activity was not affected by mutations disrupting env or 92% of gag. In contrast, activity was abolished by deletions close to the 5′-end of the genome, indicating that the region crucial for the effect on HIV-1 infectivity requires no more than 100 codons of the ordinary Gag protein and might include sequences upstream of gag.
Fig. 3.
MLV Glycogag rescues the infectivity of HIV-1Nef-. (A) Mapping the determinant in MLV that substitutes for Nef. Deletions (Δ) and frameshifts (fs) in the MLV provirus (Left) and the effect on HIV-1 infectivity (Right). (B Left) A mutation that abolishes translation of gPr80 (MLVgg-) impairs the ability of MLV to rescue HIV-1Nef-. (B Right) Western blot of JTAg producer cells. (C) The gag sequence of gPr80 is not required for the effect on HIV-1 infectivity. Infectivity is expressed as a percentage of the HIV-1wt. Bars are means plus SD from triplicate determinations. Results are representative of two independent experiments.
Most gammaretroviruses encode a glycosylated transmembrane Gag molecule (glycogag or gPr80) translated from a CUG initiation codon upstream and in-frame with gag (35–39). To test the possibility that glycogag is capable of rescuing the infectivity defect of HIV-1nef-, a mutant genome unable to encode gPr80 (MLVgg-) was obtained by changing the “CUG” start codon to “CA”; this mutation introduces both a nucleotide change and a frameshift. MLVgg- in producer cells was unable to rescue the infectivity of HIV-1nef- (Fig. 3B), proving that gPr80 is required for the effect on HIV-1.
A vector encoding a minimal active glycogag molecule (based on Fig. 3A) truncated at residue 189 and fused to the HA peptide at its N terminus (HA-gg189; Fig. 3C) had indistinguishable activity from full-length MLV (Fig. 3C), demonstrating that other MLV gene products are dispensable for this function. A further C-terminally truncated molecule (HA-gg96), retaining only 10 aa of the extracellular tail, maintained significant ability to rescue HIV-1Nef- (Fig. 3C), indicating that the extracellular domain is not strictly required for the activity. Of note, although glycogag expression had a robust effect on HIV-1 infectivity, it did not affect the efficiency of virus particle release.
Glycogag Is an Infectivity Factor for MLV.
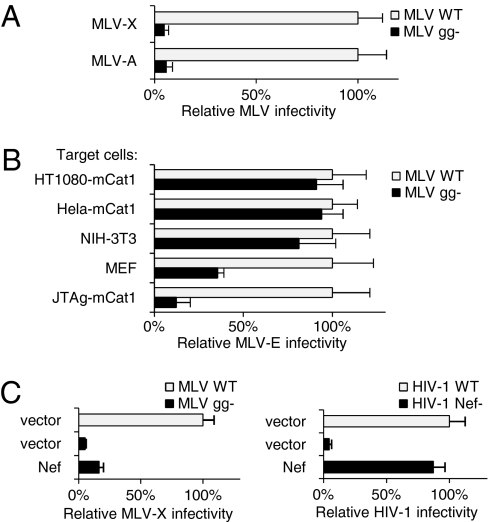
To investigate the function of gPr80 in the context of MLV infectivity, MLVwt and MLVgg- restricted to a single cycle of replication were produced by using JTAg cells. To allow efficient infection of human cells, virions were initially pseudotyped with either xenotropic or amphotropic Envs and inoculated onto HT1080 cells followed by immunofluorescence staining of CAP30 (Fig. 4A and Fig. S1). Loss of glycogag expression did not adversely affect particle production but caused a 15-fold decrease of infectivity, indicating a crucial role of gPr80 for amphotropic and xenotropic infection pathways.
Fig. 4.
gPr80 is an MLV infectivity factor. (A) gPr80 promotes the infectivity of MLV-X and MLV-A produced from JTAg cells. (B) The requirement of gPr80 for infectivity of MLV-E depends on the target cell type. (C) Nef expressed in trans does not rescue the infectivity of MLVgg- (Left) but rescues HIV-1Nef- (Right). Infectivity is expressed as percentage of WT. Bars are the mean plus SD from triplicate determinations. Results are representative of four (A) and three (B and C) independent experiments.
The activity of gPr80 was then tested on MLV pseudotyped with the ecotropic Env (MLV-E). Glycogag was not required for infectivity of MLV-E inoculated on NIH 3T3, HT1080, or HeLa cells expressing the receptor for ecotropic MLV (mCAT1), but a 3-fold and 8-fold reduction was observed when gPr80-defective virus was inoculated on immortalized mouse embryonic fibroblasts (MEF) and JTAg cells expressing the ecotropic receptor (Fig. 4B). The extent of the glycogag requirement for MLV-E infectivity therefore depends on the target cell type.
Because glycogag can substitute Nef for HIV-1, the ability of Nef to replace glycogag for MLV was tested. Nef expressed in trans in producer cells failed to rescue the infectivity of MLV lacking glycogag (Fig. 4C Left). The inability to rescue MLV infectivity was not due to a suboptimal Nef activity, because its expression could fully rescue the infectivity of HIV-1Nef- (Fig. 4C Right) and therefore might indicate mechanistic differences between gPr80 and Nef.
Activities of gPr80 and Nef on Infectivity Have Similar Requirements and Properties.
Having established that gPr80 has a Nef-like activity on HIV-1, the similarity between the two proteins was further investigated.
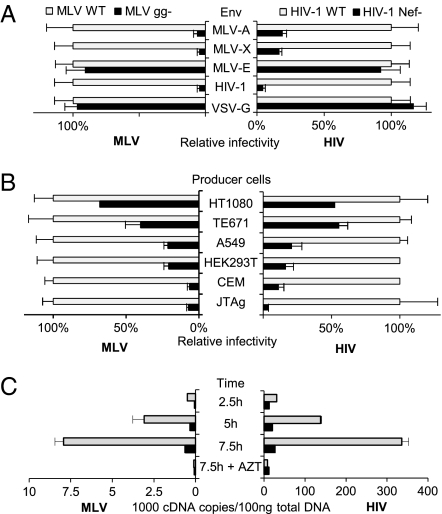
The nature of the envelope glycoprotein can determine the requirements of gPr80 (Fig. 4 A and B) and Nef (28, 29). Their activity on MLV and HIV-1 was therefore tested side-by-side on differently pseudotyped particles. Glycogag was strongly required for infectivity of MLV/HIV-1 pseudotypes inoculated onto NP2-CD4/CXCR4 cells but was dispensable for the infectivity of MLV pseudotyped with VSV-G (MLV/VSV; Fig. 5A Left), recapitulating the Nef requirement on HIV-1. On the other hand, HIV-1 particles pseudotyped with MLV-A or MLV-X Env glycoproteins were responsive to Nef (Fig. 5A Right), which, however, was no longer required for HIV-1 pseudotyped with MLV-E inoculated onto HT1080-mCat1. This evidence shows that Env glycoproteins, which render MLV dependent on gPr80, make HIV-1 responsive to Nef. The requirement of both proteins is therefore similarly determined by Env.
Fig. 5.
The infectivity factors gPr80 and Nef exhibit similar phenotypes. The effect of Env pseudotype (A) and producer cell line (B) on the activity of gPr80 (Left) or Nef (Right). (C) The effect of gPr80 or Nef on steady-state levels of MLV and HIV-1 full-length viral cDNA at the indicated times after infection. Bars represent the mean plus SD from triplicate determinations. Infectivity is expressed as percentage of the WT. Results are representative of three (A) and two (B) independent experiments.
The cell type dependence of the activities of gPr80 and Nef on MLV-X and HIV-1 were also compared by using a panel of pro-ducer cell lines, which include adherent cell types (Fig. 5B). The requirements of Nef and gPr80 were both high for virions derived from Jurkat and CEM, moderate for viruses produced from 293T and A549, and low for viruses derived from HT1080 and TE671 cells. The requirement of both proteins is therefore similarly determined by the producer cell type. In addition, the absolute infectivity of wt virions generated by the different producer cells is variable (Fig. S2), suggesting that, in addition to gPr80 and Nef, cellular factors are important to modulate the infectivity of MLV and HIV-1.
Finally, to verify whether Nef and gPr80 are required at a similar stage of the virus life cycle, their effect on progression of reverse transcription in infected cells was compared (Fig. 5C). As detected by quantitative real-time PCR, HIV-1Nef- and MLV-Xgg- produced significantly less late RT products than HIV-1wt and MLV-Xwt. gPr80 had a similar effect on the accumulation of HIV-1 RT products (Fig. S3). The low abundance of RT products generated by mutant viruses mirrors their defective infectivity (Fig. S3) and indicates that, like Nef, the activity of gPr80 on MLV and HIV-1 is also manifest during an early step of the infection process.
Discussion
Although the presence of glycosylated gag in gammaretroviruses was found >30 years ago, its function in the context of the retrovirus life cycle has remained enigmatic. In this study, the fortuitous presence of gammaretroviruses in HIV-1 producer cells (49) revealed the ability of glycogag to rescue the infectivity of HIV-1Nef- particles. gPr80 was then found crucial for MLV infectivity. The positive effect of gPr80 on gammaretrovirus replication in vivo and in some cases in vitro has been reported (41–47). Data presented here describe, in addition, the ability of gly-cogag to increase the intrinsic infectivity of retrovirus particles, an activity that shares striking functional similarity with Nef.
Being gPr80, a type II transmembrane protein, the conventional Gag residues are extracellular. However, in this study, the extracellular domain of gPr80 was found to be not strictly required for the activity on infectivity (Fig. 3C), indicating that the Gag residues and, therefore, the glycosylation status of gPr80, is not essential for the infectivity function.
Besides the evidence that glycogag can replace the activity of Nef for the infectivity of HIV-1, a functional similarity between the two proteins is supported by other observations.
The requirement for gPr80 or Nef is similarly determined by the envelope glycoprotein (Fig. 5A). Virions pseudotyped with envelope proteins, such as VSV-G, which require endosomal uptake, could bypass a block which targets Nef- and gPr80-defective virions that fuse directly at the cell membrane (28, 29). Interestingly, the entry of MLV-E has been described to occur via endocytic vesicles in a cell-type dependent manner (52, 53), a property which might explain the target cell-dependent variability of the gPr80 activity observed on MLV-E and the lack of Nef requirement for HIV-1(MLV-E) infecting HT1080 cells.
The requirement for gPr80 or Nef is similarly determined by the producer cell type (Fig. 5B). Both proteins could counteract a cellular condition that equally impairs both HIV-1 and MLV particles and which is differentially present in different cell types. Interestingly, the requirement of both proteins is strongest using producer lymphoid cells. Given the lymphotropic nature of HIV and MLV, the need for Nef or gPr80 activities could be a prerogative of lymphotropic viruses. Of note, consistent with this hypothesis, p12 of HTLV-I, another lymphotropic retrovirus, was also reported to have a Nef-like activity on HIV-1 infectivity (54). Incidentally, the evidence that infectivity of wt MLV and wt HIV-1 varies significantly with different producer cell lines (Fig. 1B and Fig. S2) suggests that additional cell-type specific activities operate to modulate retrovirus infectivity.
Glycogag and Nef have an identical intracellular distribution (Fig. S4). Despite the absence of sequence conservation, the two proteins have therefore similarly developed the ability to target the same cellular compartments, likely to be the TGN and coated pits (55, 56).
MLVgg- and HIV-1Nef- are both defective at an early stage of the infection process (Fig. 5C), which precedes the completion of reverse transcription. Although in the case of HIV-1 a role of Nef during entry into the target cell was ruled out (26, 27, 57), this remains to be established for gPr80. However, glycogag does not alter association of Env with either MLV or HIV-1 particles (Fig. S5). Importantly, the effect of gPr80 on HIV-1 is also manifest during reverse transcription (Fig. S3), further indicating that its activity on HIV-1 resembles that of Nef.
Nef was unable to rescue the infectivity of MLVgg- (Fig. 4C), whereas glycogag not only rescues HIV-1nef-, but can restore it to higher levels than HIV-1WT (Fig. 2B). This evidence could reflect a mechanistic difference. One possibility is that Nef requires the specific interaction with another lentiviral component to which glycogag cannot interact. Because Nef and glycogag are known to be incorporated into virions (58, 59), another possibility is that Nef cannot function as an MLV virion protein. However, abundant Nef was found in MLV particles (Fig. S5) (58), contrasting with its lack of activity on the gammaretrovirus and suggesting that its incorporation into virion is not sufficient for the effect on infectivity.
Interestingly, gPr80 was reported to affect the release of both MLV and HIV-1 by facilitating budding from lipid rafts (48). Intriguingly, Nef has also been reported to favor budding from specialized lipid domains (60–62). Whether this feature of gPr80 is linked to the activity on infectivity described here remains to be established, because a positive effect of gPr80 on MoMLV or HIV-1 release could not be observed in this study.
Nef performs several prominent activities, which include the ability to down-regulate cell surface receptors and to activate cellular kinases. However, Glycogag is unable to down-regulate the HIV-1 receptor CD4, the MLV-E receptor mCat-1 (Fig. S6), and MHC-I complexes (Fig. S6). The effect of glycogag on kinase activation remains to be experimentally assessed. However, those motifs present in Nef that are known to be important for recruiting and activating cellular kinases (13, 63) are not evident within the cytoplasmic tail of glycogag. The similarity between the two proteins could therefore be limited to the activity on infectivity. Despite this limitation, the crucial role of gPr80 for disease progression in infected animals (43, 45–47), the strong selective pressure that ensures its expression during infection in vivo (43, 45, 64), and its requirement for sustained virus replication in animals (41–44) are additional features that highlight a similarity with the Nef function.
In conclusion, a Nef-like requirement for retroviral infectivity is not unique to primate lentiviruses, as shown by the strong functional similarity between Nef and gPr80. Intriguingly, the ORFs of the two proteins have evolved from unrelated genetic regions of divergent retroviruses and do not share any significant homology. The activity on infectivity of Nef and glycogag could therefore be the result of convergent evolution, highlighting a fundamental role in retrovirus biology.
Materials and Methods
Plasmids.
Env- and Nef-defective HIV-1 NL4-3 provirus constructs have been described (32). The MLV provirus construct (MoMLV; accession no. J02255) and its derivatives are based on pNCA (65). Env-defective MLV was generated by introducing a frameshift at a BspEI site in env. A replication-competent MLV-X was generated by replacing MoMLV env in pNCA with that of NZB-9-1. Expression vector for HIV-1HXB2 Env, HIV-1HXB2 Env ΔCT (that encodes a truncated HIV-1 Env that lacks 144 residues of the cytoplasmic domain, used to pseudotype MLV particles), and HIV-1LAINef were described (66). Env from MLV-NZB-9-1 was expressed from a pCDNA-based vector. Vectors expressing Amphotropic (4070-A), ecotropic (MoMLV) Envs, and the vesicular stomatitis virus G protein (pMD.G) have been described (67, 68). Truncations of gPr80 with an N-terminal HA-Tag were cloned into the expression vector PBJ5. Stable cell lines expressing the ecotropic MLV receptor (mCAT-1) were generated by transduction with a pBABE-puro-based vector expressing mCAT-1, followed by puromycin selection.
Virus Production and Infectivity.
Virions capable of a single round of replication were produced by transfecting suspension-growing cells with electroporation and adherent cells with FuGENE-6 (Roche), using env-deficient HIV or MLV proviral DNA and vectors encoding viral Env glycoproteins at a 4:1 ratio. Cotransfections of HIV-1 provirus constructs with pNCA-derived plasmids and HA-gg vectors were performed by keeping a 1:1 plasmid ratio. Virus-containing supernatants were harvested 48 h after transfection. Single-cycle infectivities were determined in triplicate by challenging target cells with serially diluted viruses normalized for reverse transcriptase activity (51, 66). HIV-1 infectivities were revealed by staining infected TZM-bl cells with X-Gal and NP-2 CD4/CXCR4 (National Biological Standards Board) with a mouse monoclonal antibody to HIV-1 p55/p24 (National Biological Standards Board) followed by Alexa-488 conjugated anti-mouse (Invitrogen). MLV infectivity was evaluated by staining infected cells using a goat anti-RLV P30 (Quality Biotech) followed by Alexa-488-conjugated anti-goat (Invitrogen).
Western Blot Analysis.
Cell lysates were analyzed by SDS/PAGE and Western blotting, using goat anti-RLV P30 and anti-RLVgp70 (Quality Biotech), anti-HA antibody HA.11 (Covance), and anti-β-actin (Sigma-Aldrich).
Quantification of Reverse Transcription Products.
To minimize contamination with plasmid DNA, HIV-1 was produced from JTAg cells infected with replication-competent HIV-1NL4-3 and HIV-1NL4-3/Nef-, and MLV-X and MLV-Xgg- pseudotyped with VSV-G derived from transient transfection of HEK293T cells. Viruses were normalized based on RT activity and incubated with target cells in 6-well plates. Cells were harvested, washed extensively with PBS and total DNA was extracted (Qiagen), quantified, and subjected to real-time PCR. To control for contamination of plasmid DNA, infections for the last time point was also performed in the presence of 40 μM AZT. Primers to detect HIV-1 and MLV late RT products in a SYBR-Green I based reaction were as follows: 5′-GACCCTTTTAGTCAGTGTGGAAA-3′ and 5′-TACTCACCAGTCGCCGCC-3′ (HIV-1) and 5′-TGTCTGTCCGATTGTCTAGTGTCTA-3′ and 5′-AGGCGGGAACTGTTTTAGGT-3′ (MLV). Absolute quantification was obtained with a standard curve prepared with known amounts of provirus-containing plasmids.
Supplementary Material
Acknowledgments
I am indebted to Jeremy Luban for his great support. Many thanks to Heinrich Gottlinger (University of Massachusetts Medical School), Yasu Takeuchi (University College London), Lorraine Albritton (University of Tennessee), Peter Cherepanov (Imperial College), and Daniel Pinschewer (University of Geneva) for valuable suggestions and reagents and Myra McClure for her support. I thank the Centre for AIDS Reagents, National Biological Standards Board (Potters Bar, UK), and National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, for cell lines and for antibodies. This work was supported by European Commission FP7 Marie Curie Intra-European Fellowships for Career Development, grant no. 237265.
Footnotes
The author declares no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001554107/-/DCSupplemental.
References
- 1.Kestler HW, 3rd, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 2.Deacon NJ, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 3.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 4.Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 5.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: Requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz O, Maréchal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 7.Bell I, et al. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J Gen Virol. 1998;79:2717–2727. doi: 10.1099/0022-1317-79-11-2717. [DOI] [PubMed] [Google Scholar]
- 8.Münch J, et al. T-cell receptor:CD3 down-regulation is a selected in vivo function of simian immunodeficiency virus Nef but is not sufficient for effective viral replication in rhesus macaques. J Virol. 2002;76:12360–12364. doi: 10.1128/JVI.76.23.12360-12364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindler M, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 10.Baur AS, et al. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1:373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 11.Schrager JA, Marsh JW. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc Natl Acad Sci USA. 1999;96:8167–8172. doi: 10.1073/pnas.96.14.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14:763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 13.Saksela K, Cheng G, Baltimore D. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14:484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renkema GH, Manninen A, Mann DA, Harris M, Saksela K. Identification of the Nef-associated kinase as p21-activated kinase 2. Curr Biol. 1999;9:1407–1410. doi: 10.1016/s0960-9822(00)80086-x. [DOI] [PubMed] [Google Scholar]
- 15.Sawai ET, et al. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fackler OT, Luo W, Geyer M, Alberts AS, Peterlin BM. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol Cell. 1999;3:729–739. doi: 10.1016/s1097-2765(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 17.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene WC. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature. 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 18.Chowers MY, et al. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. The human immunodeficiency virus-1 nef gene product: A positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 21.Chowers MY, Pandori MW, Spina CA, Richman DD, Guatelli JC. The growth advantage conferred by HIV-1 nef is determined at the level of viral DNA formation and is independent of CD4 downregulation. Virology. 1995;212:451–457. doi: 10.1006/viro.1995.1502. [DOI] [PubMed] [Google Scholar]
- 22.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldsmith MA, Warmerdam MT, Atchison RE, Miller MD, Greene WC. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandori MW, et al. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz O, Maréchal V, Danos O, Heard JM. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavrois M, Neidleman J, Yonemoto W, Fenard D, Greene WC. HIV-1 virion fusion assay: Uncoating not required and no effect of Nef on fusion. Virology. 2004;328:36–44. doi: 10.1016/j.virol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Campbell EM, Nunez R, Hope TJ. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chazal N, Singer G, Aiken C, Hammarskjöld ML, Rekosh D. Human immuno-deficiency virus type 1 particles pseudotyped with envelope proteins that fuse at low pH no longer require Nef for optimal infectivity. J Virol. 2001;75:4014–4018. doi: 10.1128/JVI.75.8.4014-4018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fackler OT, et al. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology. 2006;351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 31.Laguette N, Benichou S, Basmaciogullari S. Human immunodeficiency virus type 1 Nef incorporation into virions does not increase infectivity. J Virol. 2009;83:1093–1104. doi: 10.1128/JVI.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pizzato M, et al. Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef. Proc Natl Acad Sci USA. 2007;104:6812–6817. doi: 10.1073/pnas.0607622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bresnahan PA, et al. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 34.Craig HM, Pandori MW, Guatelli JC. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards SA, Fan H. gag-Related polyproteins of Moloney murine leukemia virus: Evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol. 1979;30:551–563. doi: 10.1128/jvi.30.2.551-563.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans LH, Dresler S, Kabat D. Synthesis and glycosylation of polyprotein precursors to the internal core proteins of Friend murine leukemia virus. J Virol. 1977;24:865–874. doi: 10.1128/jvi.24.3.865-874.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neil JC, Smart JE, Hayman MJ, Jarrett O. Polypeptides of feline leukemia virus: A glycosylated gag-related protein is released into culture fluids. Virology. 1980;105:250–253. doi: 10.1016/0042-6822(80)90173-7. [DOI] [PubMed] [Google Scholar]
- 38.Schultz AM, Rabin EH, Oroszlan S. Post-translational modification of Rauscher leukemia virus precursor polyproteins encoded by the gag gene. J Virol. 1979;30:255–266. doi: 10.1128/jvi.30.1.255-266.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prats AC, De Billy G, Wang P, Darlix JL. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 40.Fujisawa R, McAtee FJ, Zirbel JH, Portis JL. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: Identification of differences in processing in vitro and in vivo. J Virol. 1997;71:5355–5360. doi: 10.1128/jvi.71.7.5355-5360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartzberg P, Colicelli J, Goff SP. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J Virol. 1983;46:538–546. doi: 10.1128/jvi.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goff SP, Lobel LI. Mutants of murine leukemia viruses and retroviral replication. Biochim Biophys Acta. 1987;907:93–123. doi: 10.1016/0304-419x(87)90001-1. [DOI] [PubMed] [Google Scholar]
- 43.Chun R, Fan H. Recovery of glycosylated gag virus from mice infected with a glycosylated gag-negative mutant of Moloney murine leukemia virus. J Biomed Sci. 1994;1:218–223. doi: 10.1007/BF02253305. [DOI] [PubMed] [Google Scholar]
- 44.Fan H, Chute H, Chao E, Feuerman M. Construction and characterization of Moloney murine leukemia virus mutants unable to synthesize glycosylated gag polyprotein. Proc Natl Acad Sci USA. 1983;80:5965–5969. doi: 10.1073/pnas.80.19.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbin A, Prats AC, Darlix JL, Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol. 1994;68:3857–3867. doi: 10.1128/jvi.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujisawa R, McAtee FJ, Wehrly K, Portis JL. The neuroinvasiveness of a murine retrovirus is influenced by a dileucine-containing sequence in the cytoplasmic tail of glycosylated Gag. J Virol. 1998;72:5619–5625. doi: 10.1128/jvi.72.7.5619-5625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Münk C, et al. 10A1-MuLV but not the related amphotropic 4070A MuLV is highly neurovirulent: Importance of sequences upstream of the structural Gag coding region. Virology. 2003;313:44–55. doi: 10.1016/s0042-6822(03)00210-1. [DOI] [PubMed] [Google Scholar]
- 48.Nitta T, Kuznetsov Y, McPherson A, Fan H. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc Natl Acad Sci USA. 2010;107:1190–1195. doi: 10.1073/pnas.0908660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol. 2008;82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raisch KP, et al. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology. 2003;308:83–91. doi: 10.1016/s0042-6822(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 51.Pizzato M, et al. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods. 2009;156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Kizhatil K, Albritton LM. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClure MO, Sommerfelt MA, Marsh M, Weiss RA. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 54.Tsukahara T, Ratner L. Substitution of HIV Type 1 Nef with HTLV-1 p12. AIDS Res Hum Retroviruses. 2004;20:938–943. doi: 10.1089/0889222042222728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 56.Greenberg ME, Iafrate AJ, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol. 2003;77:10645–10650. doi: 10.1128/JVI.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bukovsky AA, Dorfman T, Weimann A, Göttlinger HG. Nef association with human immunodeficiency virus type 1 virions and cleavage by the viral protease. J Virol. 1997;71:1013–1018. doi: 10.1128/jvi.71.2.1013-1018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welker R, Harris M, Cardel B, Kräusslich HG. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: Analysis of its role in enhancement of viral infectivity. J Virol. 1998;72:8833–8840. doi: 10.1128/jvi.72.11.8833-8840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JK, Kiyokawa E, Verdin E, Trono D. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc Natl Acad Sci USA. 2000;97:394–399. doi: 10.1073/pnas.97.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc Natl Acad Sci USA. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng YH, Plemenitas A, Linnemann T, Fackler OT, Peterlin BM. Nef increases infectivity of HIV via lipid rafts. Curr Biol. 2001;11:875–879. doi: 10.1016/s0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 63.Arold S, et al. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 64.Low A, et al. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J Virol. 2007;81:3685–3692. doi: 10.1128/JVI.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colicelli J, Goff SP. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 66.Pizzato M, Popova E, Göttlinger HG. Nef can enhance the infectivity of receptor-pseudotyped human immunodeficiency virus type 1 particles. J Virol. 2008;82:10811–10819. doi: 10.1128/JVI.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cosset FL, et al. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.