Abstract
Retinoic acid (RA) has several established functions during cardiac development, including actions in the fetal epicardium required for myocardial growth. An open question is if retinoid effects are limited to growth factor stimulation pathway(s) or if additional actions on uncommitted progenitor/stem populations might drive cardiac differentiation. Here we report the dual effects of RA deficiency on cardiac growth factor signaling and progenitor/stem biology using the mouse retinaldehyde dehydrogenase 2 (Raldh2) knockout model. Although early heart defects in Raldh2−/− embryos result from second-heart-field abnormalities, it is unclear whether this role is transient or whether RA has sustained effects on cardiac progenitors. To address this, we used transient maternal RA supplementation to overcome early Raldh2−/− lethality. By embryonic day 11.5–14.5, Raldh2−/− hearts exhibited reduced venticular compact layer outgrowth and altered coronary vessel development. Although reductions in Fgf2 and target pERK levels occurred, no alterations in Wnt/β-catenin expression were observed. Cell proliferation is increased in compact zone myocardium, whereas cardiomyocyte differentiation is reduced, alterations that suggest progenitor defects. We report that the fetal heart contains a reservoir of stem/progenitor cells, which can be isolated by their ability to efflux a fluorescent dye and that retinoid signaling acts on this fetal cardiac side population (SP). Raldh2−/− hearts display increased SP cell numbers, with selective increases in expression of cardiac progenitor cell markers and reduced differentiation marker levels. Hence, although lack of RA signaling increases cardiac SP numbers, simultaneous reductions in Fgf signaling reduce cardiomyocyte differentiation, possibly accounting for long-term defects in myocardial growth.
Keywords: heart, myocardium, retinoids, stem cells
Developmental genetics has aided us in defining how various signaling pathways regulate myocardial growth. Retinoic acid (RA), the active derivative of vitamin A, by activating its nuclear receptors (RARs), regulates key growth factor pathways essential for organ growth in a variety of settings (reviewed in refs. 1 and 2). Additional RA actions on progenitor/stem cells, although demonstrated in cell culture, have yet to be established in living organisms. Local generation of RA involves retinaldehyde dehydrogenase 2 (Raldh2), which catalyzes the second oxidative step in RA biosynthesis in most developing tissues. Raldh2−/− knockout embryos exhibit severe RA deficiency, have hearts that do not loop, and display impaired sinoatrial growth and defective ventricular trabeculation (3, 4). These early cardiac defects originate in a group of multipotent cardiac precursor cells termed the second heart field (5, 6). Although defective RA signaling affects the survival and proper integration of these progenitors into the growing heart tube (5), subsequent lethality due to alterations in hemodynamic blood flow did not allow us to examine other questions pertinent to myocardial growth regulation.
The critical region of fetal cardiac RA production is thought to be the epicardium. Deriving from proepicardium, epicardial progenitors migrate over the surface of the myocardium to cover the growing heart (7). The growth-promoting effect of the epicardium has been established as its surgical removal results in a hypocellularity of the myocardial compact zone, similar to mutations of various epicardially expressed genes. Epicardial signaling stimulates cell proliferation in the compact myocardium. Raldh2 is strongly expressed in the epicardial region. Presently, the growth-promoting effects of epicardial-derived retinoids have been explained by up-regulation of mitogen secretion (8). Epicardial expression of a dominant-negative RAR (9), compound inactivation of RARα/RARγ (10), and absence of RXRα—a heterodimerization partner of RARs (11, 12)—similarly produce myocardial hypoplasia. Data from the RXRα mutants show that both the Fgf/ERK and the Wnt pathways require RXRα signaling. A model was proposed in which these two pathways act combinatorially to control myocardial growth (11, 13). Fgf2 and Fgf9 are two retinoid-induced growth factor genes (11, 14). Disruption of the RA-dependent, epicardial-induced growth factor gradient was proposed to produce a lack of compact zone outgrowth, which in the fetus results in a thin-walled ventricle and lethality by embryonic day (E) 14.
With recent data implicating uncommitted mesenchymal progenitor/stem populations as critical regulators of myocardial growth, here we test the hypothesis that RA affects these populations. By rescuing the lethality of Raldh2−/− mutants through transient maternal RA supplementation, we show that, at fetal stages, the RA-deficient hearts exhibit a hypoplastic ventricular compact zone and reduced Fgf2 expression. Both adult and developing hearts contain reservoirs of stem and progenitor cells characterized by various markers (c-Kit, Sca-1, Isl1) and by their side population (SP) properties (15). Selected by its ability to exclude Hoechst dye 33342, the SP is a subpopulation containing cardiac progenitor cells (16), which, when isolated from postnatal heart, can differentiate into functional cardiomyocytes (17–19). We confirm the presence of a fetal cardiac SP, which under RA deficiency is increased in overall size and in the levels of cardiac progenitor marker expression. These findings suggest additional functions of retinoids in regulating fetal cardiac growth by controlling cardiac SP size.
Results
Formation and Initial Migration of the Epicardium Occurs in the Absence of RA Signaling.
Epicardial progenitors derive from the proepicardial organ, a transitional structure originating as an extension of the septum transversum. Several genes marking the proepicardial organ, including Epicardin and Tbx18, were robustly expressed at E9.5 in Raldh2−/− mutants, indicating that initial steps of epicardial migration had occurred despite the heart tube abnormalities (Fig. S1). TenascinC (TenC) expression, labeling the proepicardial organ and cardiomyoblasts of the outflow tract, confirmed that the specification of both regions can occur independently of RA production (Fig. S1). These data indicate that early steps of proepicardial development can proceed in the absence of RA synthesis.
Short-Term RA Rescue of Raldh2 Mutants Results in a Reduced Ventricular Compact Zone and Prenatal Heart Failure.
We previously showed that a stage-specific, food-based maternal RA supplementation improves heart looping and postpones the lethality of Raldh2−/− embryos until prenatal stages (4, 20). The minimal time interval for this rescue is 24 h between E7.5 and E8.5 (hereafter designated as “short-term” rescue). At E12.5, such rescued mutants have roughly the same size as wild-type (WT) littermates (Fig. 1 A and B), but show specific forelimb and craniofacial abnormalities (20). Blood vessel dilation (Fig. 1B, arrowhead) and pericardial edema were observed, suggesting compromised heart function.
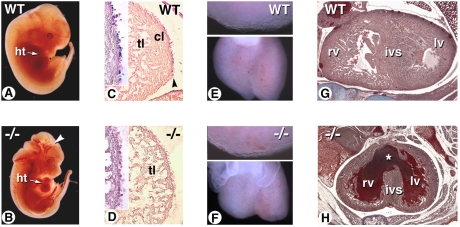
Fig. 1.
Altered cardiac development and reduced ventricular compact zone outgrowth in the RA-rescued Raldh2−/− mutant heart. (A and B) E12.5 WT and mutant embryos given RA at 100 μg/g food from E7.5 to E8.5. Mutant shows abnormal blood stasis in heart cavities and blood vessels (arrowhead). (C and D) Sagittal sections of the ventricular wall of RARE-hsp68-lacZ-transgenic WT (C) and mutant (D) embryos under the same treatment conditions. No lacZ transgene activity is detected in the mutant (D, Inset), and the ventricular compact layer is hypoplastic. (E and F) At E13.5, rounder mutant hearts display signs of fetal cardiomyopathy (F, Lower), yet ventricular epicardial cells remain adherent (E and F, Upper). (G and H) At E18.5, Raldh2−/− fetuses show reduced ventricular outgrowth, blood pooling, and ventricular septal defect (H, asterisk). cl: compact layer; ht: heart; ivs: interventricular septum; lv: left ventricle; rv: right ventricle; tl: trabecular layer.
Histological analysis was performed on E12.5 embryos harboring the RARE-hsp68-lacZ reporter transgene (21) to localize RA-responsive cells (Fig. 1 C and D). In WT embryos, X-gal+ cells were distributed mainly in the epicardial and subepicardial layers of the ventricular wall (Fig. 1C). Raldh2−/− embryos showed almost no X-gal+ cells in these layers (Fig. 1D). Furthermore, they had a reduced myocardial compact layer. By E13.5, the mutant hearts had distinct left and right ventricles with an abnormal rounded shape, suggesting myocardial dilation and heart failure (Fig. 1F). The Raldh2−/− hearts displayed no signs of epicardial detachment (Fig. 1 E and F, Upper), as observed in RXRα mutants (11, 22). Reduced heart function was further evidenced by blood pooling, regions of ventricular hypoplasia, and accompanying reductions in compact zone outgrowth by E18.5 (Fig. 1 G and H).
Epicardial RA Induces Fgf Signaling.
A transmural gradient of expression of several mitogenic factors, including Fgf2 (23), regulates myocyte proliferation in the developing heart. Because Fgf2 expression is reduced by epicardial mutation of RXRα (11), and exogenous RA increases cardiac Fgf9 levels (14), it was not surprising that Raldh2−/− mutation reduced cardiac Fgf signaling at the levels of both Fgf2 and Fgf9 mRNA (Fig. 2G) and Fgf2 protein (Fig. 2F). Immunohistochemistry showed that the Raldh2−/− compact zone is devoid of Fgf2 protein (Fig. 2 A and B, brackets), an alteration partially restored when RA supplementation is extended until E11.5 (Fig. 2C). Fgf signaling acts intracellularly to phosphorylate ERK and AKT (see ref. 24 for a review). The strongest sites of Fgf action can be revealed by immunochemical detection of phosphorylated ERK1/2 (p-ERK1/2) (25) (Fig. 2 D and E). We found p-ERK1/2 to be enriched in the epicardium (Fig. 2 D and E, arrowheads) and proliferative myocardium but reduced by Raldh2 deficiency (Fig. 2E). Western blotting confirmed net reductions in the phosphorylation status of both ERK1/2 and AKT in mutant ventricular tissue (Fig. 2F), changes also observed in RXRα-deficient hearts (13).
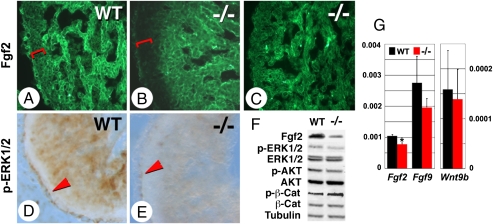
Fig. 2.
Alteration in Fgf signaling in the myocardium of rescued Raldh2−/− mutants. (A–C) Immunostaining for Fgf2 on E12.5 WT and mutant embryos given RA at 100 μg/g food from E7.5 to E8.5 (A and B) or from E7.5 to E11.5 (C). Reductions in Fgf2 in the myocardial compact zone are seen in the short-term rescued mutant (A and B, brackets), which are restored with extended RA supplementation (C). (D and E) Sections immunostained for pERK1/2 show epicardial (arrowheads) and myocardial activation, both of which are reduced in short-term rescued mutants (E). (F) Western blot analysis of Fgf2, pERK1/2, total ERK1/2, pAKT, total AKT, p552 β-catenin, total β-catenin, and tubulin in hearts from E12.5 WT and mutants after short-term rescue. (G) Real time quantitative RT–PCR analysis of Fgf2, Fgf9, and Wnt9b on RNA from E12.5 WT (black bars) and Raldh2−/− (red bars) hearts after short-term RA rescue (relative mRNA levels normalized with Gapdh). Student's unpaired t test: *P < 0.05; **P < 0.01.
In RXRα mutants, several components of the Wnt/β-catenin signaling pathway, in particular, β-catenin and its activator Wnt9b, are reduced (11). Intracellular β-catenin levels reflect activation of the canonical Wnt pathway because upon Wnt ligand binding the β-catenin degradation through GSK3b is inhibited (26). Under Raldh2 deficiency the Wnt9b transcript levels are unaltered (Fig. 2G). Western blot analysis showed unchanged total β-catenin and slightly increased phosphorylated (p552) β-catenin levels in Raldh2−/− hearts (Fig. 2F). Immunohistochemistry showed similar patterns of cytoplasmic vs. nuclear β-catenin in WT and mutant hearts (Fig. S2). Reductions in Wnt signaling produce blunted invasion of epicardial cells into the myocardial space and impaired differentiation into coronary smooth muscle (27), events defective in RXRα mutants as visualized by collagen gel invasion assays (11). Such gel invasion assays performed on E12.5 Raldh2−/− ventricular epicardium tissue explants revealed normal gel penetration (Fig. S3), as expected if Wnt signaling is intact under conditions of RA deficiency.
RA Deficiency Affects Coronary Vessel Growth and Patterning.
The coronary vasculature derives from the epicardium (27 and references therein) and can be affected by direct epicardial contribution to smooth muscle vasculature and indirect signaling via growth factor secretion. Reduced Fgf2 levels produce coronary vessel defects in RXRα mutants (11). The vascular endothelial markers VEGF-R2 (Fig. 3 A and B) and PECAM-1 (Fig. 3 C and D) revealed reduced overall coronary vessel branching over the surface of short-term rescued Raldh2−/− hearts. Interestingly, extending the exogenous RA treatment from E7.5 to E11.5 restored coronary vessel growth and patterning in Raldh2−/− embryos (Fig. 3 E and F). Epicardial-based Fgf signaling triggers fetal coronary growth by stimulating hedgehog signaling and its downstream targets (28). Although levels of expression of the ligand Sonic hedgehog (Shh) were unaltered, reduced expression of the Shh-induced Patched receptor (Ptc1) and two hedgehog targets, Gli1 and Gli3, confirm reduced hedgehog signaling in Raldh2−/− hearts (Fig. 3G).
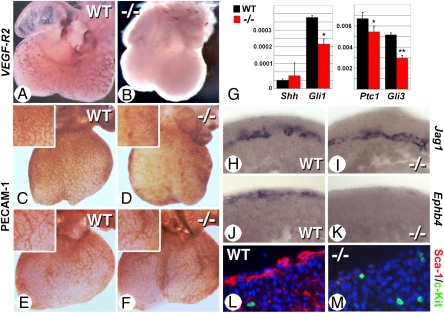
Fig. 3.
Coronary vascular defects in rescued Raldh2−/− mutants. (A–D) Whole-mount in situ hybridization for VEGF-R2 (A and B) and immunostaining for PECAM-1 (C and D) show severe reductions of the developing coronary vascular network in hearts from E12.5 mutants after short-term rescue (B and D: compare with WT in A and C). (E and F) PECAM-1 staining shows a rescue of coronary vascular development in Raldh2−/− mutants (F) when the maternal RA supplementation is extended up to E11.5. (G) RT–PCR analysis of E12.5 hearts shows reduced levels of Gli1, Ptc1, and Gli3 transcripts in short-term rescued mutants, whereas Shh levels are not affected (Student's unpaired t test: *P < 0.05; **P < 0.01). (H–K) In situ hybridization of E14.5 heart sections reveal an almost complete lack of Ephb4-expressing cells in the subepicardial region of the short-term rescued mutants (J and K), whereas Jag1-positive cells are similarly present in deeper myocardial layers (H and I). (L and M) Sca-1 (red) and c-Kit (green) immunofluorescence reveals a marked deficiency in Sca-1+ cells in the short-term rescued mutant, whereas c-Kit+ cells are detected as in WT.
Subepicardial blood vessels represent a venous lineage distinct from arterial intramyocardial vessels. Hedgehog signaling originating from cardiomyocytes is specifically required for subepicardial coronary vein formation, but not for coronary artery formation occurring deeper within the myocardial wall (29). We hypothesized that an epicardial source of RA is required to increase myocardial hedgehog signaling and that its deficiency would specifically affect coronary vein formation. To test this hypothesis, we examined markers of each lineage. Whereas the notch ligand Jagged1 (Jag1) displays endothelial expression restricted to arteries (30), the receptor tyrosine kinase EphB4 marks venous endothelial cells (31), much like the hemangioblast/cardiac progenitor marker Sca-1 (29). Although no alteration of coronary arterial Jag1 expression was observed in E14.5 Raldh2−/− hearts (Fig. 3 H and I), the absence of both markers of coronary veins (Fig. 3 J–M) indicated a distinct requirement for Raldh2 in inducing the coronary venous lineage.
RA Deficiency Affects Differentiation of Cardiomyocyte Precursors.
Several assays were used to examine if ventricular compact layer thinning in the rescued Raldh2−/− mutants might result from diminished proliferation of cardiomyocyte precursors. First, we analyzed the distribution of proliferating cell nuclear antigen on E12.5 vibratome-sectioned hearts and of phospho-histone H3 on E11.5 whole hearts (Fig. S4). Data suggested that RA deficiency allowed both cardiomyocytes and epicardial cells to maintain their growth rate. Further quantification of cardiomyocyte proliferation was performed using double immunofluorescence for MF20 (a cardiomyocyte marker, red) and Ki67 (a mitosis-specific protein, green) on E12.5 sections, and Ki67+ and MF20+ cells were counted on serial sections of the compact zone myocardium. Unexpectedly, Raldh2 deficiency increased the percentage of proliferating (Ki67+) cells among the MF20+ differentiating myocardial population by ≈30% (Fig. 4 A–C).
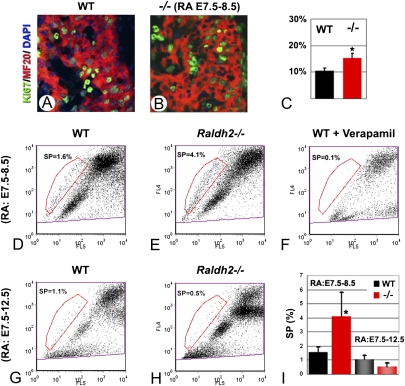
Fig. 4.
RA deficiency impacts on the proliferation of embryonic cardiomyocytes and on the SP. (A–C) Combined immunolabeling for Ki67 (green) and MF20 (red) revealed an increased fraction of proliferating cells among cardiomyocyte progenitors (MF20+) in the short-term rescued mutants. Double-positive cells were counted on serial sections of the ventricular myocardium (C). (D–F) Hoechst 33342 FACS profiles of cell suspensions from hearts of E12.5 WT (D) and Raldh2−/− mutants (E) after short-term RA rescue and WT hearts preincubated in the channel blocker verapamil (F). (G and H) FACS profiles of WT and mutant hearts after an extended RA rescue (E7.5–12.5). (I) Comparison of heart SP cell populations (expressed as percentages of the main populations) under short-term and extended rescue conditions. Ten independent tissue collections were performed, in which nine or more mutant fetuses were obtained.
Because RA accelerates embryonic stem (ES) cell-derived cardiac differentiation (32), we further analyzed myocardial differentiation in the Raldh2−/− mutants. Levels of Nppa, an early marker for working myocardium, which were reduced in the Raldh2−/− trabecular myocardium, indeed suggest an impaired differentiation at an early stage (Fig. S5). This was confirmed by reduced α-sarcomeric actin and cardiac troponin T protein levels in Raldh2 mutants. Extension of RA supplementation until E10.5 or E12.5 partially restored myocardial differentiation in the mutant heart (Fig. S5), confirming that a continuous RA supply was required for cardiomyocyte maturation.
Because Fgf signaling plays a role in inducing the differentiation of cardiac progenitors (33), reduced Fgf signaling might be responsible for differentiation defects observed in Raldh2−/− mutants. To address this possibility, WT and Raldh2−/− hearts were grown in Transwell explant culture for 28 h, and the effect of exogenous Fgf2 administration was tested. As observed in whole embryos, cultured Raldh2−/− hearts displayed reduced cardiomyocyte differentiation (assayed by α-sarcomeric actin expression). Following addition of 200 μg/mL of Fgf2, α-sarcomeric actin was increased in Raldh2−/− hearts (Fig. S6), supporting a role of Fgf signaling in inducing differentiation.
Side Population Changes Indicate that RA Deficiency Increases Cardiac Progenitors.
Recent studies identified a population of multipotent progenitor cells, the SP cells, which are isolated from adult tissues. The unique capacity of stem/progenitors to efflux the Hoechst 33342 dye, a process that requires the action of ABC transporters, provides a characteristic fluorescent-activated cell sorting (FACS) profile of SP cells (16, 34). Adult SP cells have been isolated from multiple tissues including skeletal muscle, bone marrow, liver, heart, and brain and have been shown to be enriched in progenitor/stem cells (reviewed in ref. 15). We performed Hoechst dye exclusion assays on cells isolated from hearts of short-term rescued Raldh2−/− E12.5 mutants and their control littermates. In controls, the fetal cardiac SP was present at a percentage of 1.6% compared with the main population (Fig. 4D, red gated population), with a reproducible profile and fractional ratio consistently seen in over 10 independent collections. Raldh2−/− mutants displayed a 2.3-fold increase in the SP percentage (Fig. 4E). Both WT (Fig. 4F) and Raldh2−/− cardiac SPs were eliminated by addition of the channel blocker verapamil, demonstrating specificity. Extending the period of RA treatment until the day of analysis (E7.5–12.5) produced reductions in the SP fraction in both WT and Raldh2−/− mutants, confirming that the cardiac SP cells were RA responsive (Fig. 4 G–I).
Increased Abcg2 and Altered Cardiac Stem/Differentiation Gene Marker Expression in the Retinoid-Deficient SP.
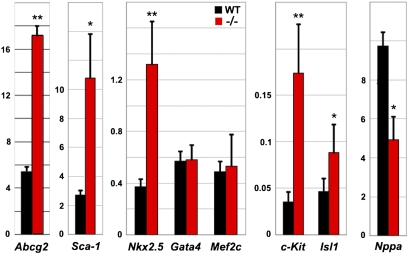
Two ABC transporter superfamily members, Abcg2 and Multidrug resistance (Mdr)1, drive Hoechst dye efflux, and by their differentially enriched expression in stem cells constitute the probable basis of the SP phenotype. Genetic studies indicate that Abcg2 regulates the proliferative capacity and survival of the adult cardiac SP (35). Enriched Abcg2 expression in embryonic and fetal hearts suggests that this transporter also plays a prominent role during cardiac development (18). We observed that Abcg2 levels are ≈100-fold higher than Mdr1 in E12.5 cardiac SP and are elevated in Raldh2−/− mutants (Fig. 5); hence higher amounts of Abcg2 likely account for elevated SP dye efflux due to retinoid deficiency.
Fig. 5.
RA deficiency alters gene expression profiles in heart SP cells. Quantitative RT–PCR analysis of Abcg2, Sca-1, NKx2.5, Gata4, Mef2c, c-Kit, Isl1, and Nppa in SP fractions from WT and Raldh2−/− hearts (black and red bars, respectively) collected at E12.5 after short-term (E7.5–8.5) RA rescue (relative mRNA levels with respect to Gapdh levels). Student's unpaired t test: *P < 0.05; **P < 0.01.
Subpopulations of adult-derived cardiac SP that display elevated levels of cardiac stem cell factors such as Sca-1 (36) have an enhanced ability to form beating cardiomyocytes (19). Although the SP is heterogeneous in composition and requires further characterization (37), the potent role RA plays in inducing differentiation in other stem cell lineages led us to investigate its role in regulating the overall SP transcriptional profile. Several myocytic and progenitor markers were assayed in fetal cardiac SP from WT and Raldh2−/− embryos. Of these, two stem-cell–related surface antigens, Sca-1 (36) and c-Kit (38), both exhibited increased mRNA expression in the Raldh2−/− SP fraction (Fig. 5). Several markers of early cardiogenic progenitors, including Nkx2.5, Gata4, and Mef2c, continue to be expressed in differentiated cardiomyocytes and thus do not discriminate between progenitors and differentiated cardiomyocytes (39). Although Nkx2.5 was increased in Raldh2−/− cardiac SP, Gata4 and Mef2c were not altered in the Raldh2−/− SP fraction (Fig. 5). The homeobox gene Islet1 (Isl1), a marker of the second myocardial lineage, is down-regulated as soon as the cardiac progenitors enter a differentiation program (39, 40). This feature makes it a suitable marker for cardiac precursors. The level of Isl1 was increased in Raldh2−/− FACS-sorted SP cells (Fig. 5). Lower levels of the differentiation marker Nppa were also observed in RA-deficient cardiac SP (Fig. 5). Hence, although the SP is a mixed population of progenitors and differentiated cardiomyocytes (37), these findings suggest that RA can collectively drive SP differentiation, a hypothesis meriting investigation of the RA effects on individual SP cells.
Discussion
Retinoic Acid Deficiency Does Not Fully Phenocopy RXRα Loss of Function.
Defective RAR/RXR signaling reduces cell density in the compact zone myocardial region. This action appears epicardial in origin because both tissue-restricted expression of a dominant-negative RARα transgene (9) and epicardial-specific ablation of RXRα (11) give similar defects. One model is that retinoids regulate cardiomyocyte proliferation by inducing the production of secreted mitogen(s), which mediate the signaling properties of the epicardium (41). Two epicardial targets of RAR/RXR signaling have been deduced using the RXRα mutants. Fgf activity is reduced (11), contributing to deficiencies in compact zone growth and coronary vascular formation (14). Biochemical assays show that deficit in RXRα signaling reduces both the PI3K and the ERK kinase pathways (13). In addition, RXRα mutant epicardial cells have reduced Wnt signaling, rendering the epicardium defective in its cardiac invasive and epithelial-mesenchymal transformation (EMT) ability (11). The Wnt signaling pathway is required for the full contribution of epicardial cells to coronary smooth muscle and coronary fibroblasts (27) and likely is involved in epicardial conversion to cardiomyocytes, although some controversy exists about whether lineage tracing conclusively demonstrates an epicardial origin of cardiomyocytes (42 and references therein).
Our characterization of Raldh2−/− hearts reveals reduced compact zone myocardial density and Fgf signaling, consistent with the RXRα mutant phenotype, but no alterations in Wnt signaling. Phosphorylation of β-catenin at Serine 552 increases its nuclear accumulation, a hallmark of Wnt activation (43). Our Western blot analysis showed slightly increased phosphorylated β-catenin levels in the Raldh2−/− heart; however, immunohistochemistry showed similar β-catenin nuclear localization in Raldh2−/− heart compared with WT. We also observed that increasing the level of Wnt signaling through maternal injection of lithium chloride, which acts by inhibiting β-catenin degradation, does not alter the Raldh2−/− phenotype, further suggesting that this phenotype is not critically Wnt-dependent. Hence although RXRα-dependent signaling was proposed to dually activate β-catenin and Fgf targets, net reductions in Fgf2 and myocardial growth appear less severe in Raldh2 mutants than in RXRα mutants, and Raldh2 inactivation allows canonical Wnt signaling, which manifests as unaltered levels of β-catenin protein. Divergent RAR and RXR requirements for Wnt target signaling may stem from RXRα binding and acting through other members of the nuclear receptor signaling family because RXRs can dimerize with at least 20 other nuclear receptors. Inactivation of PPARγ, one dimerization partner of RXRα, leads to similar ventricular myocardial phenotypes as the RXRα knockout (44), although there are no reports of its effects on coronary vasculature or EMT.
Retinoid-Stimulated Fgf Signaling Drives Fetal Cardiomyocyte Differentiation.
Fgf signaling is clearly implicated in regulating the acquisition of cardiac fate, a role that likely continues in the maintenance of cardiac cell proliferation and control of progenitor differentiation. During early heart specification exogenous Fgf administration expands cardiac field size (45), an action occurring exclusively before cardiomyocyte differentiation (46). Therefore reduced Fgf signaling inhibits cardiomyocyte formation and overall heart size. Mechanistically, Fgf increases recruitment of cells to the arterial pole and drives differentiation to cardiomyocytes (47), an action required for the addition of second-heart-field cardiac progenitors during outflow tract formation (48, 49).
A persistent role of Fgf in influencing ventricular growth, homeostasis, and regeneration at fetal and adult stages (14, 50) may lie in pluripotent epicardial progenitors. Unlike mammalian hearts, injured adult zebrafish hearts can regenerate. During this regeneration both Raldh2 expression and Fgf receptor signaling are increased (50). Although dramatic cardio-regenerative capacities appear to be lost in mammals, selective regenerative roles of the retinoid and Fgf pathways may still exist in rare progenitor populations. Such roles in progenitors may occur in coordination with other RA-regulated signaling pathways, notably Sonic hedgehog (Shh), whose signaling is reduced in Raldh2 mutant hearts.
The Side Population, an RA-Regulated Population.
The heart contains a reservoir of stem and progenitor cells capable of forming functional cardiomyocytes. Extrinsic factors regulating these cells may increase their potential to participate to myocardial regeneration in response to injury and disease (reviewed in ref. 15). The SP phenotype is a universal marker for stem cell activity (51), and one property of transplanted hematopoietic SP cells is their potential to differentiate into muscle (34). Here we report that both endogenous and maternally administered exogenous RA reduce the SP levels in fetal hearts. Our transcriptional analysis of SP cells shows that retinoid deficiency selectively increases the expression of cardiac progenitor/stem cell markers (Isl1, c-Kit, and Sca-1). Reduced levels of the differentiation marker Nppa in the Raldh2−/− SP fraction and global reductions in cardiomyocyte differentiation support a model in which endogenous RA would direct fetal cardiac stem cell differentiation.
As established in embryonic stem cell systems, retinoids act as potent agents driving uncommitted populations to differentiate into predetermined lineages. Such actions are concentration dependent (32). Hence by synthesizing a low-to-intermediate level of RA in the epicardium, Raldh2 increases Fgf/pERK signaling in the compact zone myocardium, growth factor signals established to drive cardiomyocyte differentiation. Consistently, Raldh2 mutants display reduced Fgf/pERK levels and reduced cardiomyocyte differentiation. One isolatable target population is the SP group of cardiac progenitors, which, under conditions of RA deficiency, increases in quantity. We have established that these changes are RA-responsive and can be partially corrected by providing an exogenous supply of RA through maternal supplementation and transplacental transfer.
Concluding Remarks.
A key issue in cardiac regenerative medicine is understanding how developmental pathways are recapitulated when mesenchymal or embryonic stem cells are routed into a cardiac lineage. Understanding the signaling pathways that control differentiation would provide key insights to develop reagents and regimens for enhanced differentiation. Retinoids are one of the signaling pathways acting at several steps of embryonic and fetal heart development and directing cardiomyogenic differentiation. Our data provide a framework for further studies that will assess whether retinoid-dependent modulation of growth factor signaling may be applied to control SP differentiation.
Materials and Methods
The generation of Raldh2 mutant mice (3) and the rescue conditions were previously reported (20, 52). In situ hybridizations were performed as described (53), using an Intavis InSitu Pro robot (procedure available at http://empress.har.mrc.ac.uk/browser/). X-gal assays were performed as described (21). Immunoreactions were performed on whole hearts, 10-μm cryosections, or 100-μm vibratome sections, according to standard protocols (http://www.histochem.net/histochemistry%20protocols%20main.htm). The antibodies used are listed in Table S1.
Flow cytometry of SP cells was performed on E12.5 heart cell suspensions using a modification of standard methods (16). Control and Raldh2−/− embryos were distinguished by their forelimb phenotype (20); each tissue group was pooled and then digested with 0.1 mg/mL Liberase Blendzyme IV (Roche) and 50 μg/mL DNase I (Roche) in HBSS/10 mM Hepes (10 min at 37 °C). Digests were homogenized and filtered with a 40-μm cell strainer into iced HBSS/20% FBS/10 mM Hepes. Digestion was repeated twice; cell suspensions were rinsed and incubated in DMEM/10 mM Hepes/2% FBS containing 5 μg/mL Hoechst 33342 (Sigma-Aldrich) for 1 h at 37 °C, washed, and resuspended in HBSS with 2 μg/mL propidium iodide to distinguish viable cells. Flow cytometry was performed using a MoFlo (Dako Cytomation) cell sorter. Ten independent tissue collections were performed, in which nine or more mutant fetuses were obtained.
Real-time quantitative RT–PCR was performed using SYBR Green Core Reagents (Qiagen) on RNA from three independent pooled groups of 5–10 WT and Raldh2−/− hearts using the RNA mini plus kit (Qiagen). Reverse transcription of 0.5–1 μg RNA was performed using the quantitative PCR MasterMix for the SYBR Green I kit (Eurogentec). SYBR Green incorporation was monitored in real-time with an ABI PRISM 7700 (Applied Biosystems). Primer sequences are shown in Table S2. Relative mRNA abundances were calculated using the comparative threshold cycle method, normalized with Gapdh, and expressed as differences between WT and Raldh2−/−. Final data show means (±SDs) of three or more independent experiments. For SP analysis, WT and Raldh2−/− mRNA was amplified using the Agilent Low RNA Input Linear Amplification Kit.
For epicardial invasion assays, E12.5 ventricular epicardium explants were allowed to form monolayers on 3D gels containing 1% collagen type I (BD Biosciences) for 48 h at 37 °C, 5% CO2 (54). Explanted tissues were removed and gels were incubated for 60 h in M199 medium containing 10% FCS and supplemented with penicillin/streptomycin and glutamine for another 3–5 days (54). E12.5 whole-heart cultures were performed in Transwell inserts (Corning) in BGJb medium (GIBCO-BRL) containing 5% FCS and 20 mg/mL ascorbic acid (Sigma) in the absence or presence of 200 μg/mL recombinant Fgf2 (R&D Systems).
Supplementary Material
Acknowledgments
We thank Drs. J. Rossant for the RARE-lacZ mice and D. Anderson, S. L. Ang, V. Christoffels, R. Harvey, K. Imanaka-Yoshida, A. Kispert, J. Lewis, and E. Olson for providing template plasmids. We thank V. Fraulob and B. Schuhbaur for technical assistance and Dr. S. Tsai for discussions. This work was supported by grants from the American Heart Association (0330265N) and the National Institutes of Health (R01 HL070733) (to K.N.) and funds from the Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Fondation Recherche Médicale (Equipe Fondation pour la Recherche Médicale) (to P.D.), Agence Nationale de la Recherche (ANR-07-MRAR-003), and Association Française contre les Myopathies (AFM 13517) (to S.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910430107/-/DCSupplemental.
References
- 1.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niederreither K, Dollé P. Retinoic acid in development: Towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 3.Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 4.Niederreither K, et al. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- 5.Ryckebusch L, et al. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci USA. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virágh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- 8.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen TH, et al. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 10.Kastner P, et al. Vitamin A deficiency and mutations of RXRalpha, RXRbeta and RARalpha lead to early differentiation of embryonic ventricular cardiomyocytes. Development. 1997;124:4749–4758. doi: 10.1242/dev.124.23.4749. [DOI] [PubMed] [Google Scholar]
- 11.Merki E, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sucov HM, et al. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 13.Kang JO, Sucov HM. Convergent proliferative response and divergent morphogenic pathways induced by epicardial and endocardial signaling in fetal heart development. Mech Dev. 2005;122:57–65. doi: 10.1016/j.mod.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Lavine KJ, et al. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Barile L, Messina E, Giacomello A, Marbán E. Endogenous cardiac stem cells. Prog Cardiovasc Dis. 2007;50:31–48. doi: 10.1016/j.pcad.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hierlihy AM, Seale P, Lobe CG, Rudnicki MA, Megeney LA. The post-natal heart contains a myocardial stem cell population. FEBS Lett. 2002;530:239–243. doi: 10.1016/s0014-5793(02)03477-4. [DOI] [PubMed] [Google Scholar]
- 18.Martin CM, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Pfister O, et al. CD31− but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. doi: 10.1161/01.RES.0000173297.53793.fa. [DOI] [PubMed] [Google Scholar]
- 20.Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Embryonic retinoic acid synthesis is required for forelimb growth and anteroposterior patterning in the mouse. Development. 2002;129:3563–3574. doi: 10.1242/dev.129.15.3563. [DOI] [PubMed] [Google Scholar]
- 21.Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins SJ, Hutson DR, Kubalak SW. Analysis of the proepicardium-epicardium transition during the malformation of the RXRalpha-/- epicardium. Dev Dyn. 2005;233:1091–1101. doi: 10.1002/dvdy.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consigli SA, Joseph-Silverstein J. Immunolocalization of basic fibroblast growth factor during chicken cardiac development. J Cell Physiol. 1991;146:379–385. doi: 10.1002/jcp.1041460307. [DOI] [PubMed] [Google Scholar]
- 24.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 26.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: Regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 27.Zamora M, Männer J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci USA. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavine KJ, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development. 2008;135:3161–3171. doi: 10.1242/dev.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villa N, et al. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Dev. 2001;108:161–164. doi: 10.1016/s0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 31.Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 32.Wobus AM, et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol. 1997;29:1525–1539. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblatt-Velin N, Lepore MG, Cartoni C, Beermann F, Pedrazzini T. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest. 2005;115:1724–1733. doi: 10.1172/JCI23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gussoni E, et al. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 35.Pfister O, et al. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ Res. 2008;103:825–835. doi: 10.1161/CIRCRESAHA.108.174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh H, et al. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann NY Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- 37.Oyama T, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 39.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 40.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Epicardial control of myocardial proliferation and morphogenesis. Pediatr Cardiol. 2009;30:617–625. doi: 10.1007/s00246-009-9391-8. [DOI] [PubMed] [Google Scholar]
- 42.Christoffels VM, et al. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458(7240):E8–9. doi: 10.1038/nature07916. discussion E9–10. [DOI] [PubMed] [Google Scholar]
- 43.He XC, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 45.Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 46.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilagan R, et al. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 49.Park EJ, et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 51.Smalley MJ, Clarke RB. The mammary gland “side population”: A putative stem/progenitor cell marker? J Mammary Gland Biol Neoplasia. 2005;10:37–47. doi: 10.1007/s10911-005-2539-0. [DOI] [PubMed] [Google Scholar]
- 52.Niederreither K, Vermot J, Fraulob V, Chambon P, Dolle P. Retinaldehyde dehydrogenase 2 (RALDH2)- independent patterns of retinoic acid synthesis in the mouse embryo. Proc Natl Acad Sci USA. 2002;99:16111–16116. doi: 10.1073/pnas.252626599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chotteau-Lelièvre A, Dollé P, Gofflot F. Expression analysis of murine genes using in situ hybridization with radioactive and nonradioactively labeled RNA probes. Methods Mol Biol. 2006;326:61–87. doi: 10.1385/1-59745-007-3:61. [DOI] [PubMed] [Google Scholar]
- 54.Dettman RW, Denetclaw W. Ordahl CP, Bristow J., Jr. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.