Abstract
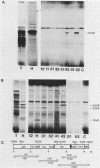
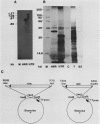
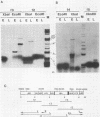
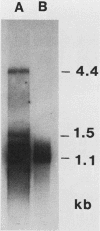
The sequence of the 8,600-base-pair HindIII H fragment, located at the center of the vaccinia virus genome, was determined to analyze several late genes. Seven major complete open reading frames (ORFs) and two that started from or continued into adjacent DNA segments were identified. ORFs were closely spaced and present on both DNA strands. Some adjacent ORFs had oppositely oriented overlapping termination codons or contiguous stop and start codons. Nucleotide compositional analysis indicated that the A-T frequency was consistently lowest in the first codon position. The sizes of the polypeptides predicted from the DNA sequence were compared with those determined by polyacrylamide gel electrophoresis of cell-free translation products of mRNAs selected by hybridization to cloned single-stranded DNA segments or synthesized in vitro by bacteriophage T7 RNA polymerase. Six transcripts that initiated within the HindIII H DNA fragment were detected, and of these, four were synthesized only at late times, one was synthesized only early, and one was synthesized early and late. The sites on the genome corresponding to the 5' ends of the transcripts were located by high-resolution nuclease S1 analysis. For late genes, the transcriptional and translational initiation sites mapped within a few nucleotides of each other, and in each case the sequence TAAATGG occurred at the start of the ORF. The extremely short leader and the absence of A or G in the -3 position, relative to the first nucleotide of the initiation codon, distinguishes the majority of vaccinia virus late genes from eucaryotic and vaccinia virus early genes.
Full text
PDF




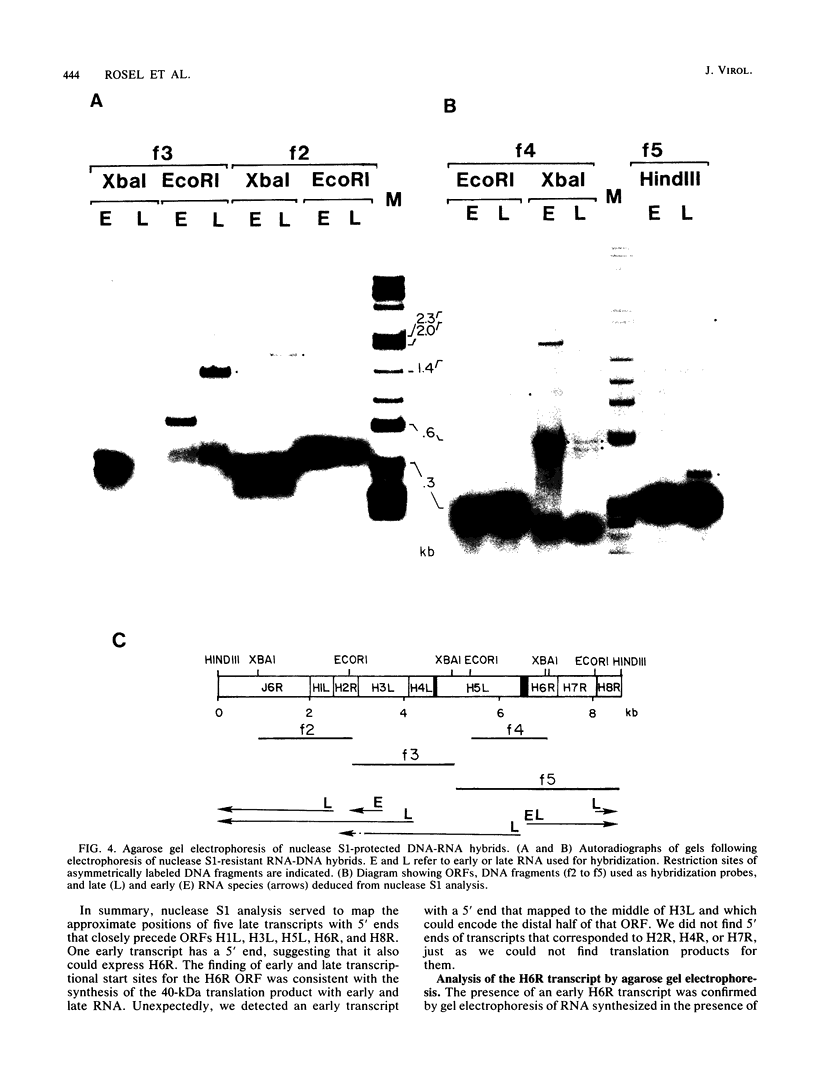

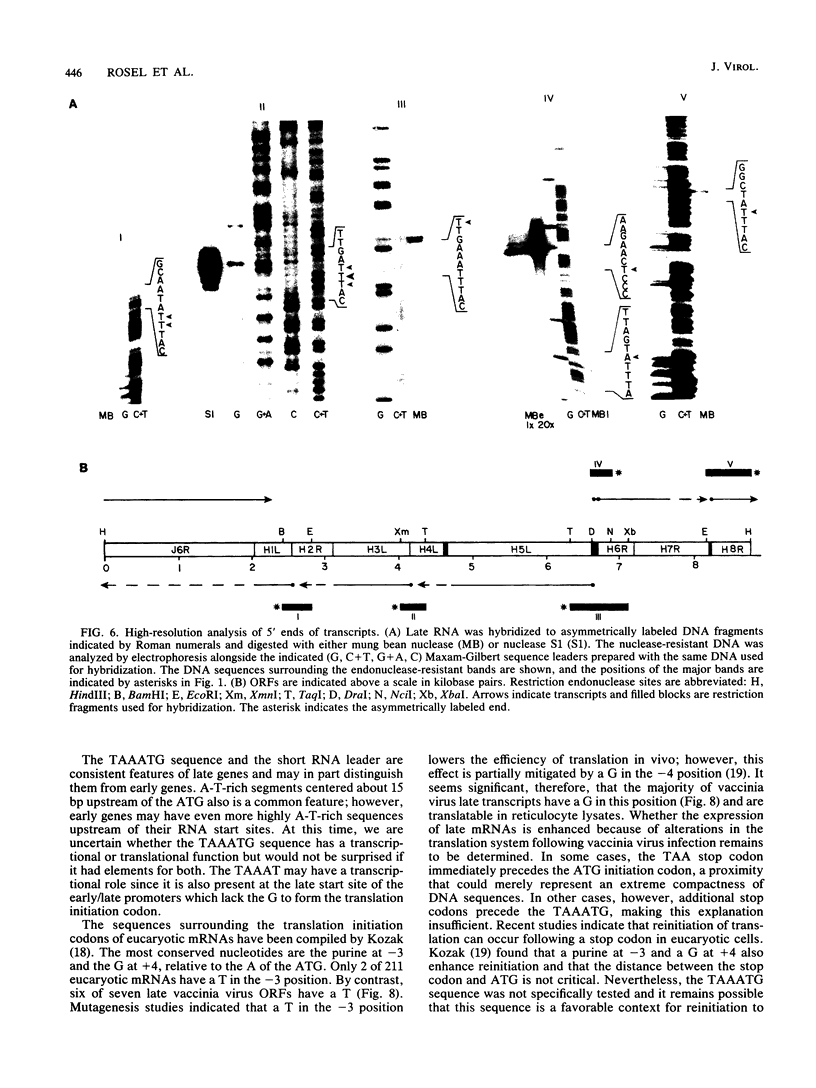
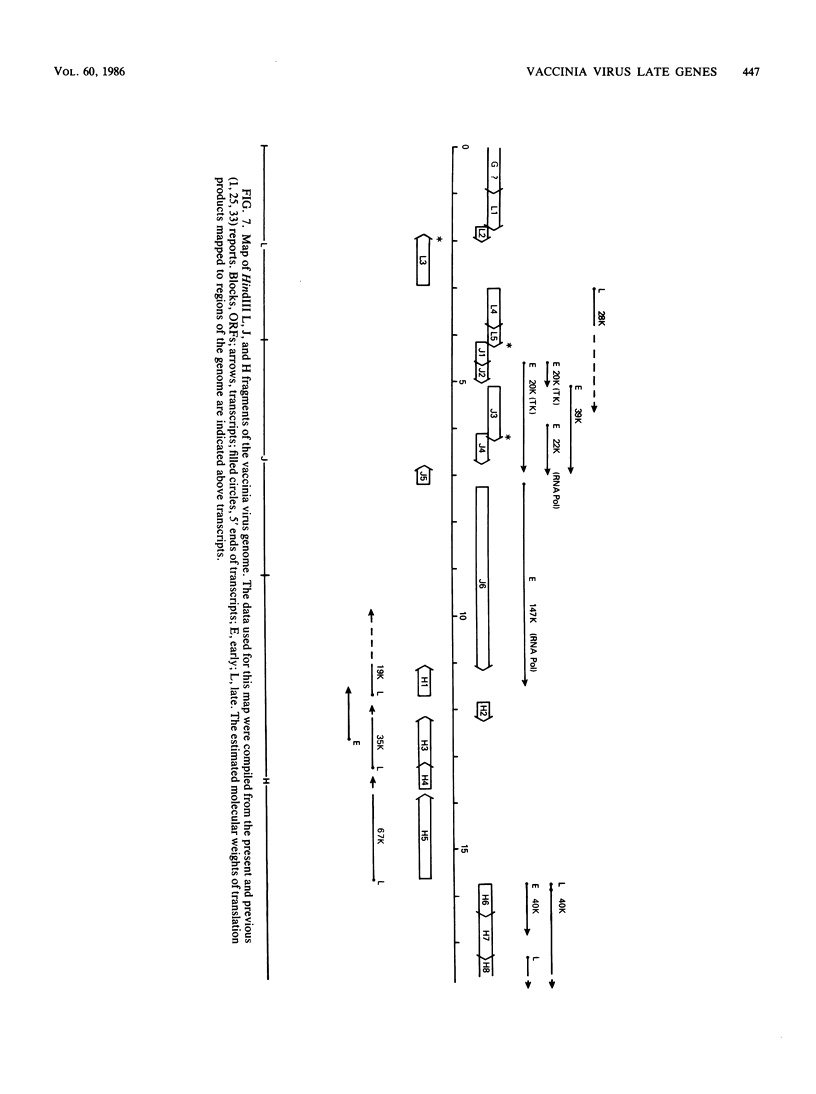
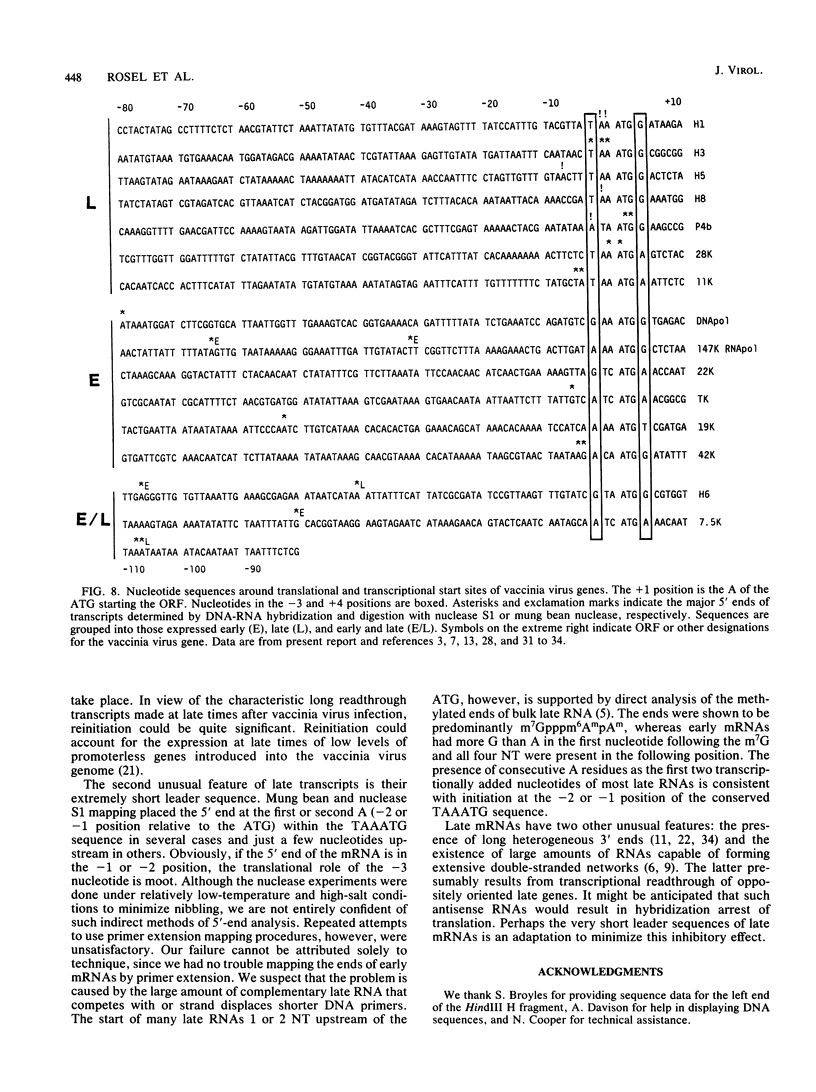

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bajszár G., Wittek R., Weir J. P., Moss B. Vaccinia virus thymidine kinase and neighboring genes: mRNAs and polypeptides of wild-type virus and putative nonsense mutants. J Virol. 1983 Jan;45(1):62–72. doi: 10.1128/jvi.45.1.62-72.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet C., Drillien R., Wittek R. One hundred base pairs of 5' flanking sequence of a vaccinia virus late gene are sufficient to temporally regulate late transcription. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2096–2100. doi: 10.1073/pnas.82.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone R. F., Moss B. Methylated 5'-terminal sequences of vaccinia virus mRNA species made in vivo at early and late times after infection. Virology. 1977 Jun 1;79(1):67–80. doi: 10.1016/0042-6822(77)90335-x. [DOI] [PubMed] [Google Scholar]
- Boone R. F., Parr R. P., Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J Virol. 1979 Apr;30(1):365–374. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci U S A. 1986 May;83(10):3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran M. A., Puckett C., Moss B. In vitro mutagenesis of the promoter region for a vaccinia virus gene: evidence for tandem early and late regulatory signals. J Virol. 1985 Apr;54(1):30–37. doi: 10.1128/jvi.54.1.30-37.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby C., Jurale C., Kates J. R. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J Virol. 1971 Jan;7(1):71–76. doi: 10.1128/jvi.7.1.71-76.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., Moss B. In vitro translation of immediate early, early, and late classes of RNA from vaccinia virus-infected cells. Virology. 1979 Jul 30;96(2):368–380. doi: 10.1016/0042-6822(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J Virol. 1981 Sep;39(3):733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippes F. M. Restriction enzyme mapping of vaccinia virus DNA. J Virol. 1982 Jul;43(1):136–149. doi: 10.1128/jvi.43.1.136-149.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golini F., Kates J. R. A soluble transcription system derived from purified vaccinia virions. J Virol. 1985 Jan;53(1):205–213. doi: 10.1128/jvi.53.1.205-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Roeder R. G. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell. 1980 Nov;22(1 Pt 1):231–242. doi: 10.1016/0092-8674(80)90171-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hruby D. E., Maki R. A., Miller D. B., Ball L. A. Fine structure analysis and nucleotide sequence of the vaccinia virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3411–3415. doi: 10.1073/pnas.80.11.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isle H. B., Venkatesan S., Moss B. Cell-free translation of early and late mRNAs selected by hybridization to cloned DNA fragments derived from the left 14 million to 72 million daltons of the vaccinia virus genome. Virology. 1981 Jul 15;112(1):306–317. doi: 10.1016/0042-6822(81)90636-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mackett M., Smith G. L., Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984 Mar;49(3):857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A., Roberts B. E. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984 Feb;49(2):510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Morrison D. K., Carter J. K., Moyer R. W. Isolation and characterization of monoclonal antibodies directed against two subunits of rabbit poxvirus-associated, DNA-directed RNA polymerase. J Virol. 1985 Sep;55(3):670–680. doi: 10.1128/jvi.55.3.670-680.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucienniczak A., Schroeder E., Zettlmeissl G., Streeck R. E. Nucleotide sequence of a cluster of early and late genes in a conserved segment of the vaccinia virus genome. Nucleic Acids Res. 1985 Feb 11;13(3):985–998. doi: 10.1093/nar/13.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmann G., Moss B. Transcription of vaccinia virus early genes by a template-dependent soluble extract of purified virions. J Virol. 1985 Nov;56(2):349–355. doi: 10.1128/jvi.56.2.349-355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel J., Moss B. Transcriptional and translational mapping and nucleotide sequence analysis of a vaccinia virus gene encoding the precursor of the major core polypeptide 4b. J Virol. 1985 Dec;56(3):830–838. doi: 10.1128/jvi.56.3.830-838.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Slabaugh M. B., Johnson T. L., Mathews C. K. Vaccinia virus induces ribonucleotide reductase in primate cells. J Virol. 1984 Nov;52(2):507–514. doi: 10.1128/jvi.52.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S., Baroudy B. M., Moss B. Distinctive nucleotide sequences adjacent to multiple initiation and termination sites of an early vaccinia virus gene. Cell. 1981 Sep;25(3):805–813. doi: 10.1016/0092-8674(81)90188-4. [DOI] [PubMed] [Google Scholar]
- Venkatesan S., Gershowitz A., Moss B. Complete nucleotide sequences of two adjacent early vaccinia virus genes located within the inverted terminal repetition. J Virol. 1982 Nov;44(2):637–646. doi: 10.1128/jvi.44.2.637-646.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Nucleotide sequence of the vaccinia virus thymidine kinase gene and the nature of spontaneous frameshift mutations. J Virol. 1983 May;46(2):530–537. doi: 10.1128/jvi.46.2.530-537.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Regulation of expression and nucleotide sequence of a late vaccinia virus gene. J Virol. 1984 Sep;51(3):662–669. doi: 10.1128/jvi.51.3.662-669.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Use of a bacterial expression vector to identify the gene encoding a major core protein of vaccinia virus. J Virol. 1985 Nov;56(2):534–540. doi: 10.1128/jvi.56.2.534-540.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G., Hammarskjöld M. L. Isolation of DNA from agarose gels using DEAE-paper. Application to restriction site mapping of adenovirus type 16 DNA. Nucleic Acids Res. 1980 Jan 25;8(2):253–264. doi: 10.1093/nar/8.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Menna A., Schümperli D., Stoffel S., Müller H. K., Wyler R. HindIII and Sst I restriction sites mapped on rabbit poxvirus and vaccinia virus DNA. J Virol. 1977 Sep;23(3):669–678. doi: 10.1128/jvi.23.3.669-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]