Abstract
ATP hydrolysis-dependent molecular machines and motors often drive regulated conformational transformations in cell signaling and gene regulation complexes. Conformational reorganization of a gene regulation complex containing the major variant form of bacterial RNA polymerase (RNAP), Eσ54, requires engagement with its cognate ATP-hydrolyzing activator protein. Importantly, this activated RNAP is essential for a number of adaptive responses, including those required for bacterial pathogenesis. Here we characterize the initial encounter between the enhancer-dependent Eσ54 and its cognate activator AAA+ ATPase protein, before ADP+Pi formation, using a small primed RNA (spRNA) synthesis assay. The results show that in a prehydrolysis state, sufficient activator-dependent rearrangements in Eσ54 have occurred to allow engagement of the RNAP active site with single-stranded promoter DNA to support spRNA synthesis, but not to melt the promoter DNA. This catalytically competent transcription intermediate has similarity with the open promoter complex, in that the RNAP dynamics required for DNA scrunching should be occurring. Significantly, this work highlights that prehydrolysis states of ATPases are functionally important in the molecular transformations they drive.
Keywords: transcription initiation, sigma factors, ATP analogues, DNA melting
Macromolecular motion occurs as a part of almost all major enzyme-driven processes and is essential for the biological function of enzymes and nucleic acids. Molecular machines and motors (often belonging to the P-loop NTPase superfamily) are capable of catalyzing a chemical reaction, capturing the free energy released from this reaction, and using this energy to perform biologically useful mechanical work. Molecular machines are present in all kingdoms of life and play essential roles in diverse biological processes ranging from muscle contraction to neuron development and mitosis to gene expression (1–3). The regulated conformational transformations they drive are achieved through distinctly different ATP- and ADP-bound functional states of such molecular machines and motors (4–6). However, which of the distinct nucleotide-bound states of the ATPases cause conformational change in their target substrates is often unknown (5).
One important ATP-dependent gene regulation complex is that of the bacterial RNA polymerase (RNAP) containing the major variant σ54 factor (Eσ54). Transcription initiation is a multistep process (including the obligatory DNA-melting step) that is subject to tight regulation. RNAP binds to specific promoter DNA sequences, forming the closed complex, and then proceeds to melt the DNA sequences surrounding the transcription start site at +1 to yield an open complex (7–10). In the case of enhancer-dependent Eσ54, open complex formation strictly requires engagement of the closed complex with a specialized ATP-hydrolyzing enhancer binding transcriptional activator protein (a member of the AAA+ superfamily of P-loop ATPases) (11–18), resembling the transcription initiation process of eukaryotic RNAP II (19–22). The promoter complexes formed with activated Eσ54 support numerous crucial adaptive responses, including those required for bacterial pathogenesis and survival under extreme nutritional stress (23). One approach to investigate the action of the activator ATPase involves the use of metal fluoride analogs to capture the different nucleotide-bound states associated with nucleotide binding and hydrolysis (5, 24, 25). These ATP analogs allow us to probe the differential contributions that the presence (and state) of the γ-β-phosphate bond (of ATP) makes to the functionality of ATPases by setting and restricting their nucleotide-bound state. One such nonhydrolyzable ATP analog is ADP-AlF, thought to represent ATP at the point of ATP hydrolysis (26). Using ADP-AlF and the catalytic AAA+ domain of the archetypal activator ATPase phage shock protein F (PspF1–275), we can “trap” Eσ54 and study the properties of this trapped complex (Eσ54-PspF1–275:ADP-AlF) (11, 24–28). Recent cryoelectron microscopy structural studies of this trapped complex demonstrated that activator binding to σ54 reconfigures Eσ54 to (i) help align the +1 promoter DNA with the RNAP active site and (ii) begin to make the DNA-binding cleft of RNAP available (11). However, the precise outcomes of the encounters between Eσ54 and the activator ATPase before ADP+Pi formation and release remain unknown.
Here we report that a prehydrolysis state (i.e., a conformational state associated with ATP binding in which the γ-β-phosphate bond is intact) of the activator ATPase (here using the transition-state analog ADP-AlF) is sufficient to reconfigure Eσ54 such that it can engage single-stranded promoter DNA, resulting in a catalytically competent transcription intermediate complex. However, because the interaction between Eσ54 and ADP-AlF–bound activator ATPase does not lead to DNA melting, we suggest that the obligatory step of DNA melting is associated with ADP+Pi formation.
Results
ADP-AlF Supports Activation of RNAP.
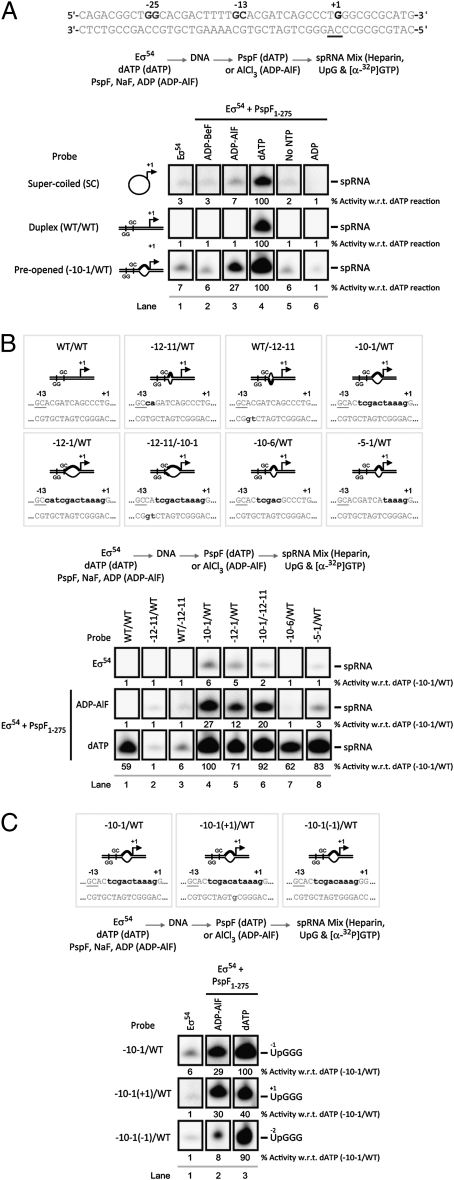
The ability of Eσ54 to form transcriptionally competent open complexes was assessed using abortive initiation assays performed on the Sinorhizobium meliloti nifH promoter (where the sequence of the transcription start site is TGGGC: positions −1 to +4 on the nontemplate strand). In the presence of the dinucleotide primer UpG, radiolabeled GTP, and cold dATP (required by the activator ATPase to form Eσ54 open complexes), a small (dinucleotide) primed RNA product UpGGG is formed. Given that we are artificially stalling transcription from this complex (by only adding GTP), we have renamed this initiation assay a small primed RNA (spRNA) assay (Materials and Methods). Using this spRNA assay, we initially studied the ability of Eσ54 to produce short RNA products in response to PspF1–275 incubated with either dATP or ADP-AlF. Strikingly, in the presence of ADP-AlF, PspF1–275 supported synthesis of an spRNA product identical to that formed in the presence of dATP, when the promoter DNA templates contained a region of mismatched DNA from −10 to −1 (with respect to the +1 site)—thereby mimicking the state of DNA in the open complex (17)—but not with DNA templates where the −10 to −1 region is base-paired (Fig. 1A). Where formed, the majority of the spRNA product was released from RNAP (Fig. S1A). Importantly, the spRNA product was dependent on the presence of ADP (to form the ADP-AlF trapped complex; Fig. S1B) and the initiating dinucleotide primer (either UpG or GpG—identical to the requirements for the dATP reactions; Fig. S1C). Interestingly, no spRNA synthesis (above background levels) was observed in the presence of ADP-AlF from fully double-stranded linear or supercoiled templates, in contrast to the full hydrolysis (dATP) reactions (Fig. 1A).
Fig. 1.
ADP-AlF supports spRNA synthesis on preopened promoter templates. Denaturing gels showing (A) that synthesis of the spRNA product (UpGGG) occurs in the presence of ADP-AlF (lane 3) only with the preopened (−10–1/WT) probe, contrasting results obtained with dATP (lane 4). Control reactions with: no activator ATPase (lane 1); an alternative nonhydrolyzable ATP analog (lane 2, ADP-BeF); no nucleotide, demonstrating the activator is not hydrolyzing GTP from the spRNA mix to form competent complexes (lane 6, No NTP); and no trapping reagents (lane 6, ADP). (B) The template requirements for spRNA synthesis for both ADP-AlF- and dATP-dependent complexes. (C) The organization of the RNAP active site in the presence of ADP-AlF is more restricted because it cannot use the single-base-pair deletion [−10–1(−1)/WT] probe. In A–C, the reaction schematic is as illustrated. The nucleotide sequence of the S. meliloti nifH promoter with the consensus GG and GC and regions of mismatch (in bold) are depicted. The level of spRNA synthesis was quantified and expressed as a percentage of the spRNA synthesized in the dATP reaction.
These data indicate that within the ADP-AlF complex (i) the conformational changes in Eσ54 required to provide access of single-stranded DNA to the RNAP active site have occurred and (ii) RNAP is catalytically active (in terms of phosphodiester-bond formation), ultimately resulting in a transcriptionally active intermediate state. This implies that ADP+Pi formation and/or release are required for DNA opening—potentially coupling changes in DNA structure with changes in accessibility of the RNAP active site to single-stranded DNA.
Control reactions, where we measured the ability of Eσ54 to synthesize the spRNA product in the absence of nucleotide (Fig. 1A, no NTP), in the absence of trapping reagents (Fig. 1A, ADP), and with a different nonhydrolyzable ATP analog (thought to represent the ground-state form of ATP; ADP-BeF) (24, 25), demonstrated that the spRNA product obtained in the presence of ADP-AlF is trapping-condition-specific [given that similar amounts of activator are cross-linked under both trapping conditions, which suggests the difference observed in spRNA synthesis between the two ATP analogs is not a simple binding issue (24)] and requires a precise nucleotide-dependent conformation of the activator ATPase. Further control reactions demonstrated that formation of the spRNA product requires a form of RNAP that is catalytically active (Fig. S1D) and that spRNA formation under ADP-AlF conditions cannot be readily accounted for by the activator hydrolyzing GTP from the reaction mix (less than 10% of the GTP is hydrolyzed in the presence of ADP-AlF compared with 60% in the dATP reactions; Fig. S1E), and therefore is a property of the trapped complex.
Using the antibiotic myxopyronin, which binds to a specific site in RNAP termed the switch region and completely inhibits Eσ70 transcription (7, 29), we demonstrated that the spRNA products formed by Eσ54 in the presence of either dATP or ADP-AlF are proportionally reduced (Fig. S1F). We note that Eσ54 is not as sensitive to myxopyronin as Eσ70, suggesting that the route to open complex formation may be different in Eσ54 compared with Eσ70—particularly in DNA interactions downstream of the transcription start site mediated by the switch regions of RNAP in controlling clamp closure (7, 29, 30). These data suggest that in the presence of either ADP-AlF or dATP, RNAP is functioning in a similar mechanistic manner.
Template Requirements in spRNA Synthesis.
We next considered the precise template requirements for ADP-AlF–dependent spRNA product formation. Eσ54 closed complexes are maintained in a transcriptionally silent state by repressive interactions that σ54 makes with a −12 fork junction structure (formed when Eσ54 binds promoter DNA) (31–33). These repressive interactions are a target for the reorganization activity of the activator ATPase. We measured the contribution of DNA sequences immediately downstream of the consensus promoter GC element (Fig. 1B) and observed that in the presence of ADP-AlF (and dATP), spRNA products were not detected from templates harboring two-base-pair mismatches (lanes 2 and 3) (27, 31). When these mismatch regions were extended, to encompass positions −10 to −1 (lanes 5 and 6), the ADP-AlF-dependent spRNA product was now observed. The set of templates tested supported formation of a stable ADP-AlF trapped complex (scored as a resolvable complex by native gel analysis; Fig. S2A), but spRNA synthesis was only detected from a subset. Interestingly, the amount of spRNA product obtained was lower in reactions when the nontemplate strand was mismatched (at positions −12 and −11), suggesting that the −12 promoter element still restricts the functionality or range of ADP-AlF-dependent changes in Eσ54 complexes.
Thus far, our data indicate that the DNA sequence between positions −10 and −1 in single-stranded form is required for the ADP-AlF-dependent activity of the Eσ54 complex (Fig. 1 A and B). When we shortened the DNA bubble by five bases on each strand, we failed to detect spRNA products above background levels in the presence of ADP-AlF (Fig. 1B). These observations again indicate that, in contrast to dATP hydrolysis, ADP-AlF-induced changes in Eσ54 are not sufficient to permit normal or full DNA melting. To study the contribution of the preopened −10 to −1 sequence further, we shortened or lengthened the bubble by a single base on each strand, thereby altering the relative position of the conventional transcription start site within the RNAP active site. Truncating the bubble (to yield a nine-base melted-out element) resulted in the loss of ADP-AlF-dependent spRNA product formation only when the UpG primer was used (Fig. 1C), not with GpG (Fig. S2B) and not in the dATP reactions (compare lanes 2 and 3). Therefore, a single-base-length change has a primer-specific effect on the activity of Eσ54 complexes that is dependent on ADP-AlF, suggesting that ADP-AlF does not support the exact same range of changes in the relationship between promoter DNA and Eσ54 as dATP hydrolysis can. However, the common utilization of many different templates by Eσ54 in the presence of ADP-AlF and dATP suggests a large overlap in the nature of all activator-dependent reorganizations of RNAP.
Differential Roles of the DNA Strands in spRNA Synthesis.
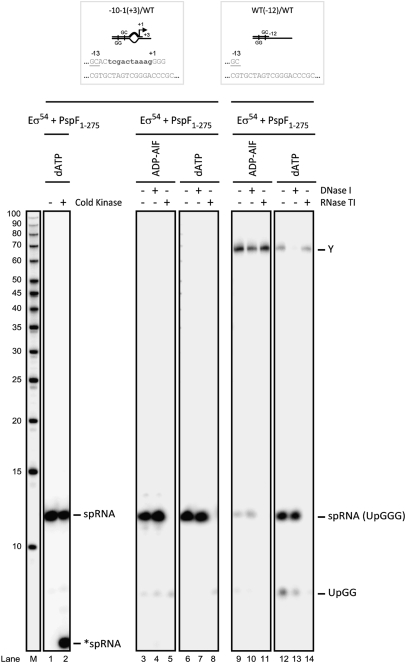
In open complexes, RNAP makes interactions with double-stranded DNA downstream of the melted-out region (from +1 to +20), via the mobile modules of RNAP (namely the β lobes, β′ jaw, and β′ clamp domains), to stabilize the open complex (8). We therefore examined the contribution of sequences downstream of the +3 position in spRNA synthesis. Truncating the template DNA strand at +3 eliminated all above background spRNA synthesis (even when trapped complexes were still capable of forming; Fig. S3), suggesting RNAP interactions downstream of +3 on the template strand are important. We do note, however, the formation of a slower-migrating nucleic acid product containing [α-32P]GTP which does not require the primer (labeled product X, discussed below; see Fig. S3C). In contrast, truncation of the nontemplate strand at +3 did not effect ADP-AlF- or dATP-dependent spRNA product formation, in accordance with the UpG- and/or GpG-specific priming of spRNA from the −1 or +1 sites, respectively (Fig. 2 and Fig. S3A). Clearly nontemplate strand interactions downstream of +3 are relatively unimportant for spRNA synthesis.
Fig. 2.
ADP-AlF–dependent spRNA synthesis requires the nontemplate strand −12 to +3 sequence. Denaturing gels demonstrating that truncated templates give rise to a slower-migrating species: product Y (compared with the RNA marker, lane M; USB). Notably, the spRNA product (with a 5′-OH) migrated slower than a 10-base RNA marker (5′-32P-labeled), but cold kinasing the 5′ end (Materials and Methods) resulted in the expected spRNA migration (lane 2, *spRNA). A further control illustrates that the spRNA product is sensitive to RNase TI but not DNase I; however, product Y was sensitive to DNase I and was subsequently identified as nonspecific incorporation of [α-32P]GTP on one DNA strand. The truncated probes used in this study with the consensus GC and regions of mismatch in bold are indicated.
Given that the presence of single-stranded DNA between the −10 to −1 site is critical for formation of the ADP-AlF-dependent spRNA product, we then addressed the contribution of these sequences by truncating the DNA strand at the −12 site. As a control, we truncated the template strand at −12 and, as expected, spRNA products were no longer detected from such templates (Fig. S3A). When we truncated the nontemplate strand at the −12 site, we observed that spRNA products were only formed in the full ATP hydrolysis event (in the dATP reactions) (Fig. 2, compare lanes 9 and 12; Fig. S3A). In the presence of ADP-AlF, we observed formation of one unusually long-length species (Fig. 2, labeled product Y). This product is not UpG-dependent, and appears to be the result of nonspecific incorporation of [α-32P]GTP via extension of one DNA strand (34). These data imply that a single-stranded region of the nontemplate strand between positions −12 and +3 is critical for formation of the ADP-AlF-dependent spRNA product. In marked contrast, because truncating the nontemplate strand at position −12 did not prevent spRNA product formation in the dATP reactions, we suggest that steps in activation associated with full ATP hydrolysis (i.e., ADP+Pi formation and release) may direct open complex formation independent of the nontemplate strand. As the additional contributions of the activator ATPase in DNA delivery into RNAP are absent under trapping (ADP-AlF) conditions, interactions with the nontemplate strand assume greater importance in these complexes.
Contribution of the Activator to spRNA Synthesis.
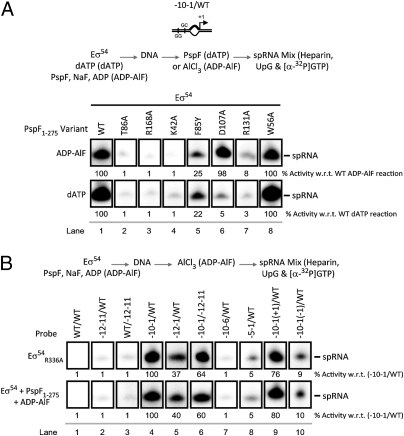
To dissect the promoter activation steps that are specifically attributable to nucleotide hydrolysis, we investigated determinants in the AAA+ domain of the activator ATPase that are specifically required for ADP-AlF-dependent (compared with dATP) spRNA synthesis. Strikingly, spRNA products were observed with the Walker B mutant D107A (Fig. S4A), which is severely deficient for ATP (and dATP) hydrolysis but binds ATP effectively (35), in the presence of ADP-AlF, but not dATP (Fig. 3A). These data indicate that the basis of the activation defect with hydrolyzable nucleotide in the D107 mutant lies in establishing a specific nucleotide-dependent productive binding interaction with the target Eσ54 promoter complex, further indicating that ATP hydrolysis is not required for spRNA synthesis. We also note that ADP-AlF-dependent stable complex formation (scored as a resolvable complex by native gel analysis; Fig. S4B) between D107A and Eσ54-DNA was not required for spRNA product formation, because such complexes were not observed with the D107A variant. Further, the ADP-AlF-dependent spRNA-stimulating activity of the D107A variant argues that binding of a prehydrolysis state of ATP (by the wild-type activator ATPase) generates transient interactions with Eσ54 that are sufficient for the primary remodeling activity of the closed complex. This is consistent with the findings that in the presence of ATPγS (a slowly hydrolyzable ATP analog), spRNA synthesis is only observed with preopened templates (36). Recall that the ATP ground-state analog ADP-BeF, which supports formation of trapped complexes between the activator ATPase and Eσ54-DNA (24), failed in spRNA product formation (above the background level obtained with Eσ54 alone; Fig. 1A). We infer that the conformational changes in Eσ54 needed for spRNA product formation are more complete when the transition-state analog (ADP-AlF) rather than the ground-state analog (ADP-BeF) is used.
Fig. 3.
Protein components that contribute to spRNA synthesis. Denaturing gels showing (A) the effect of specific substitutions in the activator ATPase on ADP-AlF- and dATP–dependent spRNA synthesis. Substitutions correspond to (i) impaired σ54 interactions: T86A, F85Y, and R131A, (ii) impaired nucleotide binding: K42A, (iii) impaired R-finger functionality: R168A, (iv) reduced ATPase activity: D107A, and (v) a wild-type-like control: W56A. The locations of these residues are indicated in Fig. S4A. Results demonstrate that D107A forms more productive interactions with Eσ54 in the presence of ADP-AlF than dATP. (B) The activator bypass mutant, R336A, phenocopies the ADP-AlF state in a template-specific manner. The reaction schematic is shown above the gels. The level of spRNA synthesis has been quantified and expressed as a percentage of the indicated reaction.
Importantly, control reactions demonstrate that single amino acid substitutions in the σ54-contacting L1 loop (T86A and F85Y; Fig. S4A) resulted in the loss (for T86A) or severe reduction (for F85Y) of ADP-AlF- and dATP-dependent spRNA product formation (Fig. 3A), consistent with a total (for T86A) or partial (for F85Y) loss of binding to the target Eσ54-DNA complex (Fig. S4B) (37, 38). Similarly, the absence of a functional R-finger (R168A) or impaired nucleotide binding site (K42A),caused a general loss in spRNA product formation (in the presence of both ADP-AlF and dATP). The L2 loop variant R131A, which poorly hydrolyzes ATP and is compromised in its ability to interact with σ54 (due to disrupted communication with the σ54-interacting L1 loop), also fails with ADP-AlF and dATP (39). However, mutations which do not impact on the ATPase- or σ54-interacting activities of the activator (such as W56A) (40) have no effect on spRNA production in the presence of either ADP-AlF or dATP.
Taken together, these results suggest that (i) ADP-AlF- and dATP-dependent activities require similar determinants within the activator ATPase for spRNA synthesis and (ii) that ADP-AlF stabilizes the otherwise transient ATP prehydrolysis state of the activator ATPase (and hence the interactions with Eσ54) that naturally occurs upon open complex formation.
Phenocopy of ADP-AlF by a σ54 Variant.
To further explore the activator ATPase-dependent actions giving rise to spRNA synthesis, we examined the properties of a σ54 variant, R336A (in the context of Eσ54), which is capable of transcribing in vitro in the absence of an activator ATPase and its nucleotide-dependent action (termed activator bypass transcription) (41–43). Strikingly, the R336A variant (which was specifically chosen because the activator ATPase-interacting domain remained intact) showed the exact same template specificities evident in ADP-AlF-dependent spRNA synthesis—where distinctions between dATP- and ADP-AlF-dependent spRNA synthesis were apparent (Fig. 3B). Notably, like ADP-AlF and unlike the dATP reactions, the R336A form of σ54 failed to form the spRNA product on the −12 truncated nontemplate strand probe and instead favored the unusual long-length species (Fig. S4C). Like ADP-AlF, R336A displayed a stricter dependence upon template structure than observed with dATP, for example in not supporting use of a template where the conventional +1 site was moved closer to the −12 promoter element (Fig. 3B). The phenocopy of ADP-AlF action by the R336A variant establishes that much of the action of the ADP-AlF-dependent activator ATPase is established via its effects on σ54.
Discussion
Our understanding of how promoter specificity factors contribute to RNAP functionality is rapidly increasing. For Eσ54, a large-scale translational movement of σ54 relative to core RNAP has been proposed as part of the activation mechanism that facilitates the delivery of the +1 template DNA to the RNAP active site (11). Our data now establish that a prehydrolysis state of an AAA+ ATPase (here ADP-AlF) is capable of productively coordinating the specific reorganization of its cognate substrate (here σ54) to control RNAP activity. The ability of ADP-AlF to support spRNA synthesis is comparable to results obtained using ATPγS, which supported a role for the ATP-bound form of the activator ATPase (36). The outcome of this nucleotide-dependent reorganization event is a catalytically competent transcription intermediate that appears to be fully active, in terms of the RNAP dynamics required for the abortive initiation DNA-scrunching mechanism to occur (44–46), and so at this level is comparable to the open complex. The binding energies of the component parts of the trapped complex are sufficient to support the transformations in RNAP structure required for RNA synthesis activity, but not to melt the DNA, indicating that any thermodynamically unfavorable steps in the transition from the closed to the open promoter complex are likely to be closely associated with the DNA-opening event.
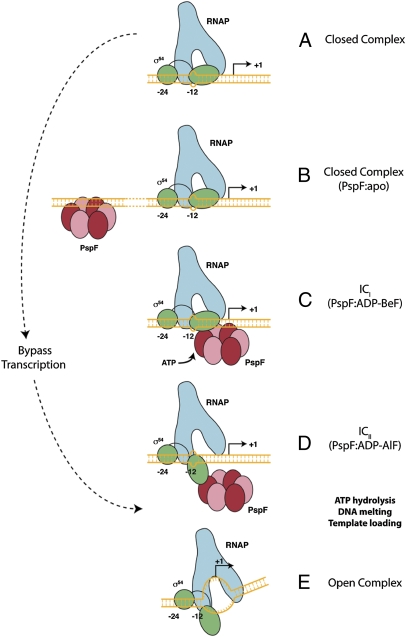
This study revealed that depending on the nucleotide-bound state of the activator ATPase, different templates changed the outcome—in terms of spRNA synthesis. Comparing outcomes with dATP to those with ADP-AlF, or the activator bypass σ54 (R336A), suggests that ADP+Pi formation and/or release greatly influences (i) template utilization, as evidenced by the ability to use the −2 position as a start site with dATP but not with ADP-AlF (Fig. 1C), and (ii) DNA melting, as demonstrated by the absence of spRNA synthesis from supercoiled or fully duplexed linear templates in the presence of ADP-AlF (Fig. 1A). Each of these separate observations can be rationalized if the ATPase-driven activation pathway involves at least two distinct conformational states of the RNAP-DNA complex when progressing from the closed to the open complex (Fig. 4), one in which the active site of RNAP becomes accessible (as in the ADP-AlF trapped complexes; Fig. 4D) and one (or more) in which melted DNA is created and delivered to the active site (Fig. 4E).
Fig. 4.
Schematic representation of the proposed pathway to open complex formation by Eσ54. (A) The closed complex with inhibitory −12 fork junction structure is potentially engaged by the apo-form of the activator ATPase (here PspF) binding to upstream DNA enhancer sites (B). ICi (C) and ICii (D) represent two different ATP-bound states of the activator ATPase, with ADP-BeF being closer to the ground state than ADP-AlF. Importantly, in (D), the RNAP active site is functional and accessible to single-stranded DNA. We infer that ADP+Pi formation and/or release are required to establish the open complex (E). The properties of the ADP-BeF- (C) and ADP-AlF–dependent (D) complexes are distinct, emphasizing the importance of prehydrolysis states for substrate engagement. These discrete nucleotide-bound states are suggested to reflect different states of the activator ATPase in relation to being poised for hydrolysis—a view supported by recent crystallographic studies of a Rho-RNA complex in which individual catalytic (i.e., ATP hydrolysis) sites were shown to be distinct, some closer to the hydrolysis state than others and each with different modes of RNA binding (5).
Although the study here is restricted to a type-member-specialized transcriptional activator, we suggest other ATP-dependent transformations driven by molecular machines will have a large component of their remodeling activity associated with the ATP-bound state. Full remodeling is likely to require mixed nucleotide-bound states, potentially accessible with real-time single-molecule studies (47).
Materials and Methods
Proteins.
Escherichia coli core RNAP was purchased from Epicentre Biotechnologies (Cambio). Klebsiella pneumoniae σ54 and E. coli PspF1–275 were purified as described (35). The S. meliloti nifH DNA probes (MWG Operon) were 32P-labeled (where appropriate) and annealed to the complementary strand as described (48).
Small Primed RNA Assays.
Small primed RNA assays (a form of abortive initiation assay) were performed in STA buffer (25 mM Tris-acetate, pH 8.0, 8 mM Mg-acetate, 10 mM KCl, and 3.5% wt/vol PEG 6000) in a 10-μL reaction volume containing 100 nM Eσ54 (reconstituted using a 1:4 ratio of E:σ54) and 20 nM promoter DNA probe, which was initially incubated at 37 °C for 5 min to form Eσ54-DNA complexes. ADP-AlF trapped complexes were formed in the presence of 5 μM PspF1–275 (wild-type or variants), 5 mM NaF, and 1 mM ADP and “trapping” was initiated by adding 0.2 mM AlCl3 (24, 26). Open complexes (dATP reactions) were formed in the presence of 5 μM PspF1–275 (wild-type or variants) and 4 mM dATP. Control reactions contained (i) no activator (Eσ54 alone), (ii) ADP-BeF (0.2 mM BeCl2 is used instead of AlCl3), (iii) no nucleotide (Eσ54 and PspF1–275), or (iv) no trapping reagents (Eσ54, PspF1–275, and ADP, but no NaF or AlCl3). All reactions were incubated for 10 min at 37 °C (as per reaction schematic; see Fig. 1A). Synthesis of spRNA (UpGGG) was initiated by adding a mix containing 100 μg/mL heparin, 0.5 mM UpG, and 4 μCi [α-32P]GTP and incubated for 20 min at 37 °C. The reaction was quenched by addition of loading buffer and analyzed on a 20% denaturing gel, and visualized and quantified using a Fuji FLA-5000 PhosphorImager. All experiments were minimally performed in triplicate. Specific modifications to the general spRNA protocol are outlined below.
Analysis of spRNA products by native gel.
A 2-μL sample of the spRNA reaction was analyzed using a 4.5% native (nondenaturing) gel run at 100 V for 55 min. The gels were dried and the protein-DNA complexes were visualized and quantified using an FLA-5000 PhosphorImager.
Analysis of spRNA products by thin-layer chromatography.
A 2-μL sample of the spRNA reaction was diluted with 40 μL of 2 M formic acid and [α-32P]GDP separated from [α-32P]GTP by thin-layer chromatography. The relative ratios of the products were quantified by an FLA-5000 PhosphorImager.
Myxopyronin inhibition assays.
Myxopyronin (2 μM final) was added to the reaction as indicated (see reaction schematic; see Fig. S1F). All reactions were analyzed as described above.
DNase I or RNase TI cleavage.
Where appropriate, either 1 U (final) DNase I (Roche) or 10 U (final) RNase TI (Fermentas) was added to the spRNA reaction (after spRNA synthesis) and the reaction was incubated at 37 °C for 10 min to initiate cleavage. The reaction was quenched by addition of loading buffer and the reactions were analyzed as described above.
Cold kinasing reactions.
Half the spRNA reaction (5 μL) was kinased at 37 °C for 30 min using 1 U (final) T4 PNK (USB), 1 μL of 10× reaction buffer, and 1 μL of 1 mM ATP. The reaction was quenched by addition of loading buffer and the reactions were analyzed as described above.
Native Gel Mobility Shift Assays.
Native gel mobility shift assays were conducted in STA buffer in a total reaction volume of 10-μL containing 100 nM Eσ54 (reconstituted using a 1:4 ratio of E:σ54) and 20 nM 32P-labeled probe, which was incubated for 5 min at 37 °C. Trapped complexes were formed as described above. Reactions were analyzed using a 4.5% native (nondenaturing) gel run at 100 V for 55 min. The gels were dried and protein-DNA complexes were visualized and quantified using an FLA-5000 PhosphorImager. These experiments were minimally performed in triplicate.
Supplementary Material
Acknowledgments
We thank M. Jovanovic and N. Zhang for proteins and the members of the M.B. laboratory and D. Bose, E. James, and C. Engl for their comments on the manuscript. This work was supported by the Wellcome Trust (Grant 084599/Z/07/Z to M.B.) and the Biotechnology and Biological Sciences Research Council (Grant BB/G001278/1 to M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001188107/-/DCSupplemental.
References
- 1.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure—diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 3.Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol. 2008;69:1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 5.Thomsen ND, Berger JM. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker PA, Sallai L. The AAA+ superfamily—A myriad of motions. Curr Opin Struct Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Belogurov GA, et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 9.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 10.Young BA, Gruber TM, Gross CA. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 11.Bose D, et al. Organization of an activator-bound RNA polymerase holoenzyme. Mol Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Carlo S, et al. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SY, et al. Regulation of the transcriptional activator NtrC1: Structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popham DL, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 15.Sallai L, Tucker PA. Crystal structure of the central and C-terminal domain of the σ(54)-activator ZraR. J Struct Biol. 2005;151:160–170. doi: 10.1016/j.jsb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Tintut Y, Wang JT, Gralla JD. A novel bacterial transcription cycle involving σ 54. Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 17.Wedel A, Weiss DS, Popham D, Dröge P, Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 18.Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 19.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 20.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 21.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative σ factors and their roles in bacterial virulence. Microbiol Mol Biol Rev. 2005;69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrows PC, Joly N, Nixon BT, Buck M. Comparative analysis of activator-Eσ54 complexes formed with nucleotide-metal fluoride analogues. Nucleic Acids Res. 2009;37:5138–5150. doi: 10.1093/nar/gkp541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, et al. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, σ54. Structure. 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaney M, et al. Binding of transcriptional activators to σ 54 in the presence of the transition state analog ADP-aluminum fluoride: Insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burrows PC, Severinov K, Buck M, Wigneshweraraj SR. Reorganisation of an RNA polymerase-promoter DNA complex for DNA melting. EMBO J. 2004;23:4253–4263. doi: 10.1038/sj.emboj.7600406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappas M, et al. Structural insights into the activity of enhancer-binding proteins. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay J, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannon WV, Gallegos MT, Buck M. Isomerization of a binary σ-promoter DNA complex by transcription activators. Nat Struct Biol. 2000;7:594–601. doi: 10.1038/76830. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Lew CM, Gralla JD. Promoter opening by σ(54) and σ(70) RNA polymerases: σ factor-directed alterations in the mechanism and tightness of control. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krupp G. Unusual promoter-independent transcription reactions with bacteriophage RNA polymerases. Nucleic Acids Res. 1989;17:3023–3036. doi: 10.1093/nar/17.8.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joly N, Rappas M, Wigneshweraraj SR, Zhang X, Buck M. Coupling nucleotide hydrolysis to transcription activation performance in a bacterial enhancer binding protein. Mol Microbiol. 2007;66:583–595. doi: 10.1111/j.1365-2958.2007.05901.x. [DOI] [PubMed] [Google Scholar]
- 36.Burrows PC, et al. Coupling σ factor conformation to RNA polymerase reorganisation for DNA melting. J Mol Biol. 2009;387:306–319. doi: 10.1016/j.jmb.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordes P, et al. The ATP hydrolyzing transcription activator phage shock protein F of Escherichia coli: Identifying a surface that binds σ 54. Proc Natl Acad Sci USA. 2003;100:2278–2283. doi: 10.1073/pnas.0537525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N, et al. The role of the conserved phenylalanine in the σ54-interacting GAFTGA motif of bacterial enhancer binding proteins. Nucleic Acids Res. 2009;37:5981–5992. doi: 10.1093/nar/gkp658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrows PC, et al. Functional roles of the pre-sensor I insertion sequence in an AAA+ bacterial enhancer binding protein. Mol Microbiol. 2009;73:519–533. doi: 10.1111/j.1365-2958.2009.06744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elderkin S, Bordes P, Jones S, Rappas M, Buck M. Molecular determinants for PspA-mediated repression of the AAA transcriptional activator PspF. J Bacteriol. 2005;187:3238–3248. doi: 10.1128/JB.187.9.3238-3248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaney M, Buck M. The σ 54 DNA-binding domain includes a determinant of enhancer responsiveness. Mol Microbiol. 1999;33:1200–1209. doi: 10.1046/j.1365-2958.1999.01566.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang JT, Syed A, Gralla JD. Multiple pathways to bypass the enhancer requirement of σ 54 RNA polymerase: Roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JT, Syed A, Hsieh M, Gralla JD. Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: Role of an NH2-terminal leucine patch in σ 54. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 44.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz A, et al. A stepwise 2′-hydroxyl activation mechanism for the bacterial transcription termination factor Rho helicase. Nat Struct Mol Biol. 2009;16:1309–1316. doi: 10.1038/nsmb.1711. [DOI] [PubMed] [Google Scholar]
- 48.Wigneshweraraj SR, et al. Enhancer-dependent transcription by bacterial RNA polymerase: The β subunit downstream lobe is used by σ 54 during open promoter complex formation. Methods Enzymol. 2003;370:646–657. doi: 10.1016/S0076-6879(03)70053-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.