Abstract
Changes in brain cholesterol homeostasis are associated with multiple diseases, such as Alzheimer’s and Huntington’s, however, controversy persists as to whether adult neurons produce their own cholesterol, or if it is outsourced to astrocytes. To address this issue, we analyzed 25 genes most immediately involved in cholesterol homeostasis from in situ data provided by the Allen Mouse Brain Atlas. We compared the relative mRNA expression in the pyramidal and granule layers, populated with neurons, with the rest of the hippocampus which is populated with neuronal processes and glia. Comparing the expression of the individual genes to markers for neurons and astrocytes, we found that cholesterol homeostasis genes are preferentially targeted to neuronal layers. Therefore, changes in gene expression levels might affect neuronal populations directly.
Keywords: systems biology, shuttle hypothesis, Allen brain mouse atlas, glia, Alzheimer’s disease
The shuttle hypothesis of brain cholesterol homeostasis suggests that adult neurons outsource cholesterol synthesis to astrocytes (Nieweg et al., 2009; Pfrieger, 2003). However, an in situ hybridization study of adult mouse brain tissue demonstrated the expression of Hmgcr and Dhcr7, the rate-limiting and final enzymes in cholesterol biosynthesis, respectively, within cortical, hippocampal, and basal forebrain-cholinergic neurons (Korade et al., 2007). Moreover, recent reports have concluded that neuronal expression of the SREB1 Hmgcs gene and SREB1 protein is necessary for cholesterol production (Ong et al., 2000; Swanson et al., 1988). Overall, these studies partially suggest that cholesterol is produced in adult neurons, however, cholesterol production requires multiple steps (Figure 1), and it is therefore necessary to analyze the expression of all genes involved in its production.
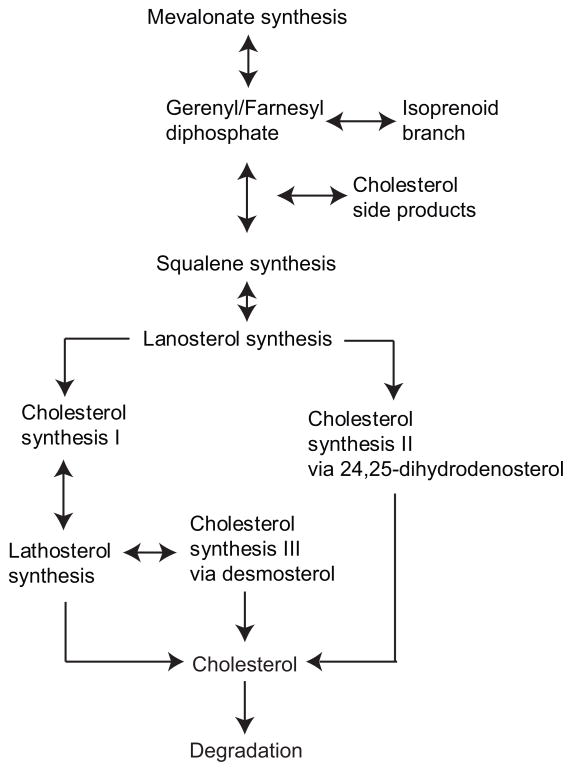
Figure 1.
Schematic diagram of cholesterol anabolism divided in a branching process that starts with mevalonate synthesis and includes the rate limiting enzyme, HMGCR. Subsequent to the mevalonate pathway, isoprenoid side-products and squalene are generated. The post squalene portion commits to sterol synthesis and leads to lanosterol production. The pathway then takes two routes: cholesterol synthesis I and II. Both pathways result in the production of cholesterol but differ in the specific enzymatic steps and intermediate metabolites of the pathways. Cholesterol synthesis pathway I is further subdivided into cholesterol synthesis III that is most prominent during early brain development.
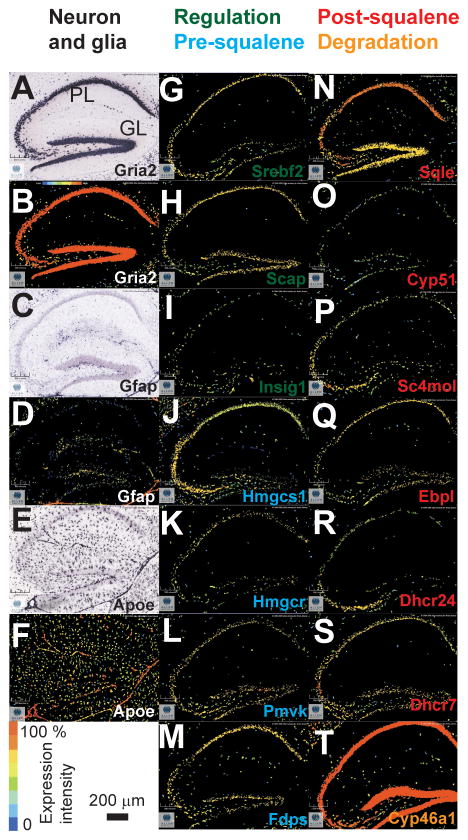
Using the Allen Mouse Brain Atlas (AMBA, http://www.brain-map.org/) we assembled the first complete in situ cholesterogenic map in the adult hippocampus. We concentrated on the hippocampus since cholesterol homeostasis is greatly affected by diseases such as Alzheimer’s in this region (Blalock et al., 2004). In Figure 2 the localization of neuronal and astrocytic specific biomarkers is provided as are 14 examples of genes specific in cholesterol homeostasis in the adult mouse hippocampus. We also show the in situ hybridization of Grai2, a neuron specific marker of AMPA receptor production in Figure 2A (Geiger et al., 1995); the dark puncta in the image are clustered in the pyramidal and granule cell layers. Likewise, the corresponding pseudo-colored image of Gria2 is provided in Figure 2B. The expression intensity is centered in the pyramidal and granule cell layers, indicated by the maximum intensity color (see scale bar). A similar analysis of glial fibrillary acidic protein (Gfap), an astrocyte specific marker is presented in Figures 2C–D. As opposed to the results of Gria2, Gfap shows little to no dark puncta in the pyramidal or granule cell layers of the hippocampus. Figures 2E–F demonstrate the expression of apolipoprotein E (Apoe), another astrocyte specific marker involved in the production of lipid packets used for shuttling and collecting excess cholesterol from the extra-cellular fluid (Boyles et al., 1985). The dark puncta in Figure 2E are spread non-specifically throughout the hippocampus and surrounding tissue. In both astrocyte specific markers the pyramidal and granule cell layers are indistinguishable from the surrounding areas. The images shown in Figures 2A–F establish the criteria for distinguishing a neuron or astrocyte specific pattern to analyze all genes involved in cholesterol homeostasis.
Figure 2.
Localization of cholesterol homeostasis mRNAs in the adult mouse hippocampus. A: in situ hybridization of Gria2. B: pseudocolor expression patterns obtained from in situ hybridization in A. C: in situ hybridaztion of Gfap, an astrocyte marker. D: pseudocolor expression for C. E–F: same as in C–D for Apoe, another astrocyte marker. G–I: cholesterol genes involved in regulation (green) and pre-squalene biosynthesis (blue). N–T: cholesterol genes involved in post-squalene biosynthesis (red) and degradation (orange). All cholesterol markers retain a neuron specific distribution pattern seen in pyramidal (PL) and granule layer (GL) as seen in images A–B. All data collected from the adult mouse hippocampus completed by the Allen Institute for Brain Science.
Figures 2G–T show examples of genes present in regulation, biosynthesis, and degradation of cholesterol in the hippocampus, and Figures 2G–I show the expression of three regulatory genes of cholesterol homeostasis; Srebf2, Scap, and Insig1. The images indicate that the signal for each gene is centered in the pyramidal and granule cell layers. We separated the biosynthesis component of cholesterol homeostasis into the pre- and post-squalene segments. While pre-squalene expression can be associated with other metabolic processes, post-squalene gene expression is exclusive to cholesterol production (see Figure 1). In Figures 2J–M, we show four genes involved in the pre-squalene segment: Hmgcs1, Hmgcr, Pmvk, and Fdps. The distribution patterns of the pre-squalene genes unlike the expression patterns of Gfap or Apoe retain a robust neuron specific distribution. The data presented in Figures 2J–M ultimately shows that regulatory and pre-squalene biosynthesis genes are targeted to neuronal layers in the hippocampus.
Figures 2N–S show expression patterns of six post-squalene genes: Sqle, Cyp51, Sc4mol, Ebpl, Dhcr24, and Dhcr7. Notably, the signal for these six genes retains a neuron specific distribution pattern in the pyramidal and granule cell layers, as do all other genes in the post-squalene segment. Figure 2T shows Cyp46a1 which is involved in degradation, as the final common pathway of all brain cholesterol catabolism and exit to the blood. Additionally, the intensity values in Figure 2T are nearly exact to the ones observed for the neuron biomarker (Gria2, Figure 2B). Overall, the expression patterns of the genes shown in Figure 2 suggest that adult neurons express all necessary genes for regulation, synthesis, and degradation of cholesterol.
Since the analysis presented in Figure 2 suggests an almost exclusive neuronal specificity of cholesterol related genes, we decided to further quantify this possibility. Specifically, we developed a targeting factor measurement that quantified the amount of expression signal in the pyramidal and granule cell layers, normalized by the area occupied by these structures. We calculated the targeting to these layers using the following formula:
| 1 |
Where Rf is the relative fluorescence given by the ratio of the integrated expression value in the pyramidal/granule cell layers (fpyr) divided by the total fluorescence in the region (Ftotal):
| 2 |
Ra was the relative area occupied by the pyramidal/granular layer (apyr) over the entire hippocampus (Aarea):
| 3 |
To perform the analysis, we implanted a program in Matlab (Mathworks, Natick, MA) that could read TIFF images downloaded from the AMBA, manually delineate the pyramidal and granule cell layers and compute the ratios.
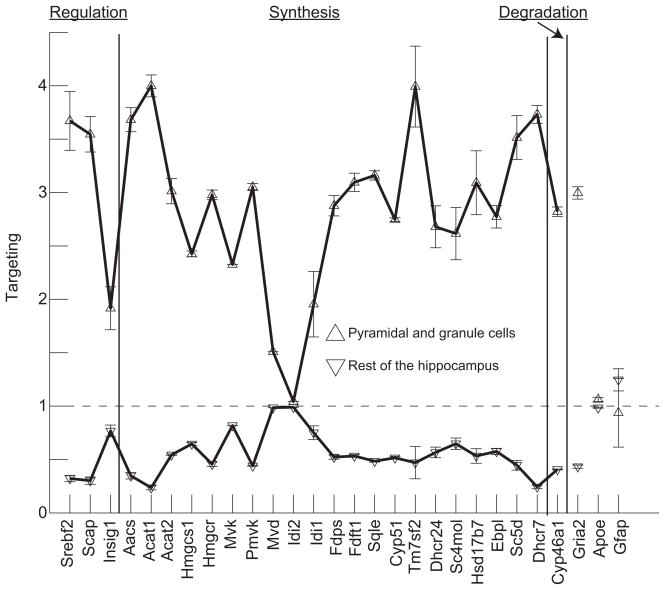
Using the analysis outlined in equations 1–3 we studied the targeting of 25 genes involved in cholesterol homeostasis to the neuronal and extra-neuronal regions in the hippocampus. These genes participate in the core reactions of cholesterol metabolism (Horton et al., 2003) (see also superpathway of cholesterol biosynthesis in http://biocyc.org/ and steroid biosynthesis in http://www.genome.jp/kegg). At time of analysis Nsdhl, another gene involved in cholesterol metabolism, was not reported in the AMBA. Cyp27b1, Ch25h, involved in cholesterol and 7-dehydrocholesterol degradation, showed no expression; while Srebf1, Lbr and Lss showed minimal expression in the hippocampus (not shown). Figure 3 shows two plots, the data represented with triangles pointing up corresponds to targeting to the pyramidal and granule cell layers; while the data plotted with triangles pointing down correspond to an identical analysis performed in the extra-neuronal regions of the hippocampus (all areas except the pyramidal and granule cell layers). A targeting value of one means no specificity in the gene expression in the area of interest with respect to the entire hippocampus. The larger the targeting value, the more gene expression is preferentially localized within the studied area. The error bars correspond to the standard error of the mean calculated from at least three slices obtained from different animals, except in the case of Mvd and Mvk where only one image was available in the AMBA. The targeting analysis is consistent with the anatomical distribution of cells in the hippocampus since the targeting of Gria, a neuron specific marker, shows a high targeting value; while two astrocyte markers (Apoe and Gfap) show a targeting value close to one.
Figure 3.
Cholesterol homeostasis genes are preferentially expressed in neuron specific layers. The plots show two curves, the one with triangles pointing up is for the targeting factor calculated for the pyramidal and granule cell layers, the plot with triangles pointing down is for the targeting factor for all the areas of the hippocampus, except the pyramidal and granule cell layers. A targeting value of one means no specificity in the gene expression in the area of interest with respect to the entire hippocampus. The average targeting of the genes suggests that they are neuron specific since a neuronal exclusive gene (Gria2) has the same targeting factor. Error bars are for the standard error of the mean calculated for slices obtained from 3–4 different animals, except in the case of Mvd and Mvk in which there was only one slice available in the AMBA database.
The plots shown in Figure 3 demonstrate that the targeting of cholesterol genes correlates with neuronal expression but not with astrocytes. The neuronal specific plot has an average targeting value almost identical to Gria2. Independently of the focus of the analysis in neuronal or extra-neuronal regions, the targeting of astrocyte markers suggests and almost homogeneous distribution of these cells in the hippocampus. Never-the-less, the extra-neuronal analysis shows that the targeting of almost all genes again corresponded with the targeting value of Gria2; suggesting that such expression was inside neuronal processes. In both cases, the expression of Idi2 was close to one, implying no specific targeting of this gene to neuronal or extra-neuronal areas. Thus, the analyses shown in Figures 2 and 3 suggest that all but one (Idi2) of the genes related to cholesterol production homeostasis are preferentially targeted to neuron specific layers in the hippocampus. Therefore, changes in gene expression levels related to cholesterol production might reflect neuronal populations when brain tissue homogenates are used, as in microarray studies.
Overall our analysis, of in situ hybridization data from the AMBA, shows a complete expression map of cholesterol homeostasis markers in the hippocampus. Partial experimental evidence shows that Hmgcr and Dhrc7, the rate limiting and final enzymes of cholesterol production, are expressed within hippocampal neurons (Korade et al., 2007). Our analyses complete this study by showing that all cholesterol homeostasis markers target neurons much more than glia. From the combined analysis we conclude that cholesterol markers retain a neuron specific distribution pattern, showing that adult neurons retain the molecular components necessary for cholesterol homeostasis.
The shuttle hypothesis suggests that cholesterol synthesis in adult neurons is outsourced to glia. Further, it implies that neurons abandon the cholesterol synthesis pathway, beginning with mevalonate. However, our results show that neurons in the pyramidal and granule cell layers express much higher levels of mevalonate pathway genes than glia. The locally produced isoprenoids from the mevalonate pathway are essential for neuronal survival (Michikawa and Yanagisawa, 1999; Tanaka et al., 2000). Glia derived cholesterol is needed for axon maintenance through myelination (Edgar and Nave, 2009; Griffiths et al., 1998). We found that the localization of myelin via expression of the proteolipid protein gene (plp1) was nearly absent from neuron specific layers but found heavily elsewhere in the hippocampus (not shown), discarding the possibility that myelin was contributing to our neuronal layer measurements. Our analysis shows that adult hippocampal neurons express all known genes required for cholesterol homeostasis, thus we infer that cholesterol is locally produced in neurons. However, glia also produces cholesterol (Edgar and Nave, 2009), providing the possibility that neuronal and glia cholesterol pools are differentially regulated. For example, during diseased states neurons may down-regulate the mevalonate pathway, thus lowering isoprenoid production and therefore activating outsourcing of cholesterol synthesis to glia. The shift in localization of Cyp46a1 from neurons to glia in severe Alzheimer’s disease (Papassotiropoulos et al., 2003) indeed suggests this.
Acknowledgments
TRAC-UTSA; NSF, 0932339; NIH, G12 RR13646-08; NIH, MBRS-RISE; San Antonio Comparative Biology of Aging Center, 5K07AG025063; NIH, R01AG026151-03; and Alzheimer’s Association, ZEN-07-59500.
References
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101(7):2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest. 1985;76(4):1501–13. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JM, Nave KA. The role of CNS glia in preserving axon function. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15(1):193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, et al. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280(5369):1610–3. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100(21):12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Mi Z, Portugal C, Schor NF. Expression and p75 neurotrophin receptor dependence of cholesterol synthetic enzymes in adult mouse brain. Neurobiol Aging. 2007;28(10):1522–31. doi: 10.1016/j.neurobiolaging.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Michikawa M, Yanagisawa K. Apolipoprotein E4 isoform-specific actions on neuronal cells in culture. Mechanisms of Ageing and Development. 1999;107(3):233–243. doi: 10.1016/s0047-6374(98)00134-1. [DOI] [PubMed] [Google Scholar]
- Nieweg K, Schaller H, Pfrieger FW. Marked differences in cholesterol synthesis between neurons and glial cells from postnatal rats. J Neurochem. 2009;109(1):125–34. doi: 10.1111/j.1471-4159.2009.05917.x. [DOI] [PubMed] [Google Scholar]
- Ong WY, Hu CY, Soh YP, Lim TM, Pentchev PG, Patel SC. Neuronal localization of sterol regulatory element binding protein-1 in the rodent and primate brain: a light and electron microscopic immunocytochemical study. Neuroscience. 2000;97(1):143–153. doi: 10.1016/s0306-4522(00)00031-2. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Streffer JR, Tsolaki M, Schmid S, Thal D, Nicosia F, Iakovidou V, Maddalena A, Lutjohann D, Ghebremedhin E, et al. Increased brain beta-amyloid load, phosphorylated tau, and risk of Alzheimer disease associated with an intronic CYP46 polymorphism. Arch Neurol. 2003;60(1):29–35. doi: 10.1001/archneur.60.1.29. [DOI] [PubMed] [Google Scholar]
- Pfrieger FW. Outsourcing in the brain: do neurons depend on cholesterol delivery by astrocytes? Bioessays. 2003;25(1):72–8. doi: 10.1002/bies.10195. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Simmons DM, Hofmann SL, Goldstein JL, Brown MS. Localization of mRNA for low density lipoprotein receptor and a cholesterol synthetic enzyme in rabbit nervous system by in situ hybridization. Proc Natl Acad Sci U S A. 1988;85(24):9821–5. doi: 10.1073/pnas.85.24.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tatsuno I, Uchida D, Moroo I, Morio H, Nakamura S, Noguchi Y, Yasuda T, Kitagawa M, Saito Y, et al. Geranylgeranyl-Pyrophosphate, an Isoprenoid of Mevalonate Cascade, Is a Critical Compound for Rat Primary Cultured Cortical Neurons to Protect the Cell Death Induced by 3-Hydroxy-3-Methylglutaryl-CoA Reductase Inhibition. J Neurosci. 2000;20(8):2852–2859. doi: 10.1523/JNEUROSCI.20-08-02852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]