Abstract
Several genes contained in the Francisella pathogenicity island (FPI) encode proteins needed for intracellular growth and virulence of Francisella tularensis. The pdpA gene is the first cistron in the larger of the two operons found in the FPI. In this work we studied the intracellular growth phenotype of a Francisella novicida mutant in the pdpA gene. The ΔpdpA strain was capable of a small amount of intracellular replication but, unlike wild-type F. novicida, remained associated with the lysosomal marker LAMP-1, suggesting that PdpA is necessary for progression from the early phagosome phase of infection. Strains with in cis complementation of the ΔpdpA lesion showed a restoration of intracellular growth to wild-type levels. Infection of macrophages with the ΔpdpA mutant generated a host-cell mRNA profile distinct from that generated by infection with wild-type F. novicida. The transcriptional response of the host macrophage indicates that PdpA functions directly or indirectly to suppress macrophage ability to signal via growth factors, cytokines and adhesion ligands.
INTRODUCTION
Francisella tularensis is a facultative intracellular pathogen that is able to grow in a number of cell types, and is often found in infected tissues within cells of the monocytic cell lineage (Anthony et al., 1991; Bosio & Dow, 2005; Conlan & North, 1992; Ellis et al., 2002; Hall et al., 2007). In vitro studies have shown that F. tularensis subverts the normal endocytic pathway of host macrophages and grows rapidly within these cells (Santic et al., 2005a). Approximately 20 min after macrophage engulfment, the Francisella-containing phagosome acquires the early endosomal markers EEA-1 and Rab5 (Clemens et al., 2004). The phagosome subsequently gains late endosomal markers such as Rab7, CD63, LAMP-1 and LAMP-2 (Clemens et al., 2004; Santic et al., 2005a). This late endosome-like compartment acquires the proton vacuolar ATPase pump and becomes transiently acidified but does not associate with lysosomal markers, such as cathepsin D (Clemens et al., 2004; Santic et al., 2005a, 2008). Francisella is thus able to prevent phagosomal–lysosomal fusion, and within 4 h of uptake actively breaks down the phagosomal membrane in order to escape into the host-cell cytosol and replicate. The infective process eventually leads to host-cell death, whereby the bacteria are freed to infect neighbouring cells.
All Francisella species contain at least one copy of a gene cluster known as the Francisella pathogenicity island (FPI). A number of genes, particularly within the FPI, have been shown necessary for phagosomal escape and intracellular replication; however, most of these studies have not investigated the nature of the gene products nor provided any evidence to support a mechanism of action of the gene products (Bonquist et al., 2008; de Bruin et al., 2007; Nano et al., 2004; Santic et al., 2005b, 2008). Studies of mutants of Francisella novicida that have knockouts of genes encoding phosphatases suggest that one or more of them play a role in phagosome membrane degradation. Since the acid phosphatase, AcpA, also has lipase activity, it may play an important role in degrading the membrane (Mohapatra et al., 2007a, 2008; Reilly et al., 1996).
Microarray analysis has recently been employed in a variety of ways to study Francisella infection. The majority of these studies have used microarray technology to profile the transcriptional response of Francisella-infected immune cells (Andersson et al., 2006a, b; Butchar et al., 2008; Paranavitana et al., 2008). Some have used the microarray-based studies to identify virulence genes (Weiss et al., 2007) or study the control of virulence gene expression by regulators such as pmrA, mglA and sspA (Charity et al., 2007; Mohapatra et al., 2007b; Sammons-Jackson et al., 2008). Microarray analysis has not hitherto been used to profile the change in host-cell responses to Francisella strains with mutations in virulence genes.
The pdpA gene is one of the largest in the FPI and is located at the beginning of a putative operon containing the pdpB, vgrG and dotU genes. A previously described gene replacement mutant in the pdpA gene of F. novicida exhibited impaired intracellular replication and avirulence in mice; however, the substitution of the erythromycin resistance (EmR) cassette for pdpA has since been shown by us (unpublished data) to have polarity effects on genes downstream of pdpA. Because many of these downstream genes affect intracellular growth, it is possible that the altered intracellular growth phenotype of the ΔpdpA : : EmR mutant was due to the suppressed expression of multiple FPI-encoded proteins. In a companion paper (Schmerk et al., 2009) we describe a non-polar, F. novicida, pdpA deletion mutant and show that it is highly attenuated for virulence. In this paper we examine the intracellular growth phenotype of the ΔpdpA mutant, and the effect of the deletion of pdpA on macrophage gene expression response to F. novicida infection.
METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. F. novicida strains were grown using trypticase soy agar or broth supplemented with 0.1 % (w/v) cysteine (TSAC, TSBC).When necessary, 15 μg kanamycin ml−1 was added to the growth medium.
Table 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference |
|---|---|---|
| U112 | Wild-type F. novicida | ATCC 15482 |
| JLO | U112 with deletion in FTN_1390, where SKX vector inserts; identical growth and virulence with respect to U112 | Ludu et al. (2008b) |
| GB2 | U112 with point mutation in global virulence regulator, mglA | Baron & Nano (1998) |
| ΔpdpA | JLO with a deletion of pdpA | This study |
| ΔpdpA/SKX : : pdpA | ΔpdpA complemented with the integrating pJL-SKX : : pdpA construct | This study |
| ΔiglC | JLO with a deletion of iglC | de Bruin et al. (2007) |
Intracellular growth assays.
Bone marrow cells were isolated from the femurs of male BALB/c mice and seeded in 96-well cell culture plates at a density of 3×105 cells per well. Cells were incubated for 1 week in complete Dulbecco's Modified Eagle Medium (cDMEM) supplemented with 10 % fetal bovine serum, 2 mM l-glutamine, 1 % MEM nonessential amino acids, 10 mM HEPES buffer solution and 10 % conditioned L929 cell supernatant. After 7 days the macrophages were infected with F. novicida strains at an m.o.i. of 50 : 1 (bacteria : macrophage). The monolayers were incubated at 37 °C, 5 % CO2 for 1 h to allow uptake of the bacteria and then washed five times using sterile PBS. Infected macrophages were lysed with 0.1 % (w/v) deoxycholic acid at 0, 24 and 48 h post-infection. Lysates were serially diluted in PBS containing 0.1 % (w/v) gelatin and plated on TSAC for enumeration.
Experiments using the J774A.1 murine macrophage cell line were carried out in a manner similar to those used for bone marrow-derived macrophages (BMDM). Cells were seeded in 96-well cell culture plates at a density of 5×104 cells per well and allowed to adhere overnight. F. novicida strains were added to the wells at an m.o.i. of 50 : 1 and the infection was carried out as described above. Infected macrophages were lysed with 0.1 % deoxycholic acid at 0, 24 and 48 h post-infection.
Real-time PCR assays.
J774A.1 murine macrophage infections were performed as described above, except that infections were performed in 25 cm2 tissue culture flasks. mRNA was isolated from the macrophage cell line 12 h after infection using PureLink Micro-to-Midi total RNA purification system (Invitrogen) according to the manufacturer's protocols. Quantitative real-time (qRT-PCR) data were generated using the RT2 Profiler PCR array mouse Signal Transduction PathwayFinder (SuperArray Bioscience). qRT-PCR was performed with 1.5 μg total RNA according to the manufacturer's protocol using the Stratagene MX4000 thermocycler.
Immunofluorescence and LAMP-1 association.
J774A.1 macrophages were seeded on 22 mm glass coverslips in 6-well tissue culture plates at a density of 1×106 cells per well and allowed to adhere overnight. Cells were chilled on ice for 5 min and bacteria were added to each well at an m.o.i. of 500 : 1, after which the cells were chilled for an additional 10 min. Cells were immediately warmed in a 37 °C water bath for 3 min to synchronize bacterial uptake and then incubated at 37 °C, 5 % CO2 for an additional 20 min. Wells were washed five times in PBS and fresh medium was added. At the appropriate time points the coverslips were removed and rinsed in PBS. The coverslips were then fixed in 2 % (w/v) paraformaldehyde, 1 % (w/v) sucrose, PBS for 20 min at room temperature followed by immersion in ice-cold methanol for 10 min at −20 °C. Coverslips were blocked in PBS containing 5 % lamb serum for 30 min. Rabbit polyclonal anti-F. novicida (Nano, 1988) and rat monoclonal anti-LAMP-1 (DSHB, University of Iowa) antibodies were incubated with the coverslips overnight at 4 °C at a dilution of 1 : 1000. Coverslips were then washed three times with PBS and incubated for 2 h with goat anti-rabbit Alexafluor568 and goat anti-rat Alexafluor488 secondary antibodies (Molecular Probes) at a dilution of 1 : 1500. The coverslips were incubated for 5 min with Hoescht 33258 (Molecular Probes) to label the DNA. Cells were imaged with a Leica DMIREZ inverted fluorescent microscope using a 100× oil immersion lens. Using Openlab 5.1 software, multiple-channel Z stacks were captured and deconvoluted in order to score macrophage LAMP-1 association with the bacterial strains.
Graphing and statistics.
The Prism GraphPad v4.03 software was used to generate graphs and to calculate the appropriate statistical values, including standard deviation, standard error of the mean and P-values. For P-values the Student t-test or two-way ANOVA were used where appropriate.
RESULTS
Intracellular growth of pdpA mutants
Our previous work had shown that a gene replacement mutation in pdpA resulted in a strain defective for intramacrophage growth (Nano et al., 2004). We have since learned that this and other replacement mutations generated polarity effects; hence we examined the phenotype of the newly created ΔpdpA strain (Schmerk et al., 2009), which has a non-polar deletion mutation. Infection of the ΔpdpA mutant in mouse BMDM resulted in limited growth that showed an initial increase in F. novicida numbers followed by a decline in numbers (Fig. 1a). Macrophages infected with the ΔpdpA mutant appeared healthy throughout the infection, displaying no signs of cytotoxicity or cell-rounding. Genetic complementation, but not a mock complementation (data not shown) with the pJL-SKX vector, restored nearly complete growth in the macrophages (Fig. 1a). Very similar results were observed when J774A.1 macrophages were infected with the ΔpdpA mutant and its genetic complement (Fig. 1b). While statistical analysis showed that the growth of the ΔpdpA mutant is clearly different from both the wild-type strain and from the complemented strain, this analysis also showed that the growth of the wild-type and complemented strains was different, indicating that the wild-type phenotype was not fully restored by genetic complementation. The influence of an exogenous promoter in the pJL-SKX : : pdpA vector may account for these differences.
Fig. 1.
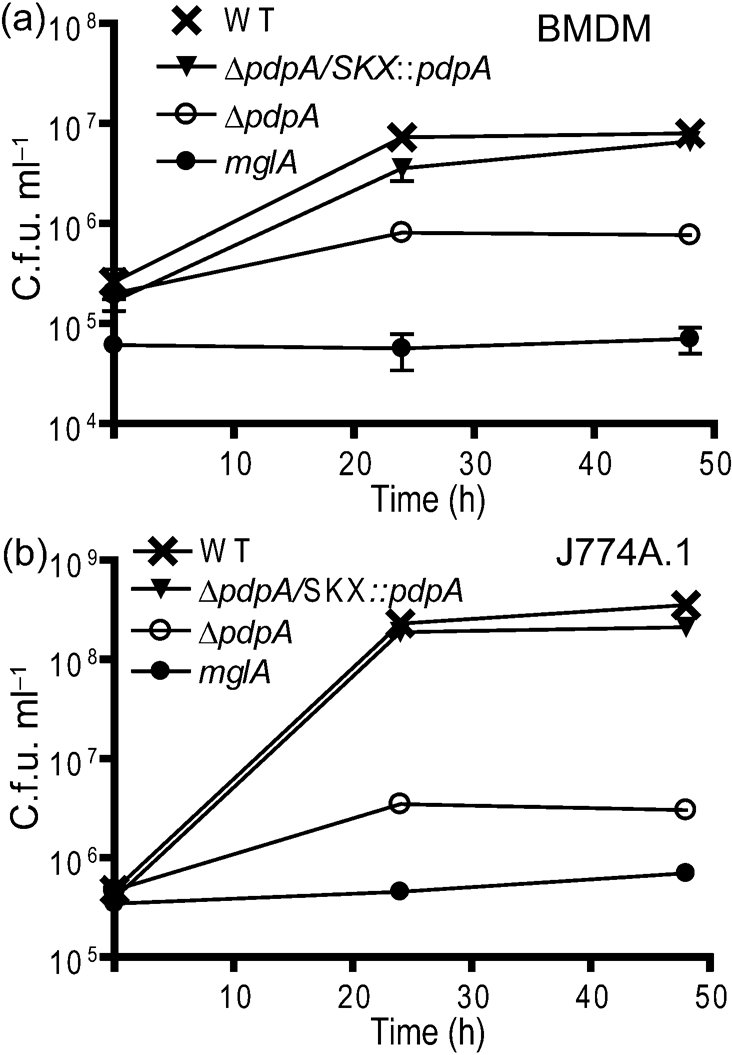
Intracellular growth of ΔpdpA mutants. The ΔpdpA mutant was able to replicate in BMDM (a) and J774A.1 cells (b) within the first 24 h of infection. Bacterial numbers decreased after 48 h and macrophages exhibited no signs of cytotoxicity. Complementation of the ΔpdpA deletion restored the wild-type (WT) growth phenotype. The ΔmglA mutant was included as a control as it is unable to replicate within macrophages. All data points in both panels are representative of three replicates and each experiment was performed in triplicate. Two-way ANOVA was used to calculate the significance of the differences in the growth curves between pairs of strains. For BMDM: WT vs ΔpdpA, P<0.00l; WT vs ΔpdpA/SKX : : pdpA, P=0.0261; and for ΔpdpA vs ΔpdpA/SKX : : pdpA, P=0.0028. For J774A.1 macrophages: WT vs ΔpdpA, P=<0.001; WT vs ΔpdpA/SKX : : pdpA, P=0.006; and for ΔpdpA vs ΔpdpA/SKX : : pdpA, P=<0.001.
LAMP-1 association of the pdpA mutant
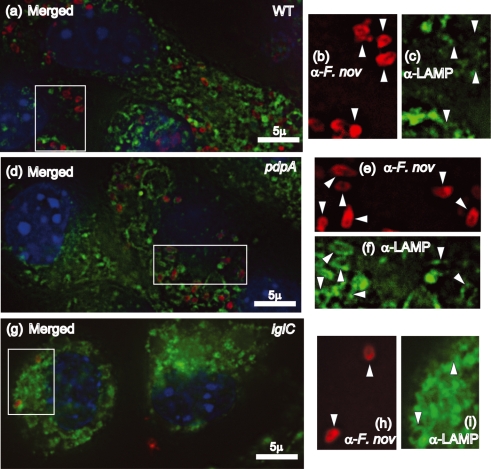
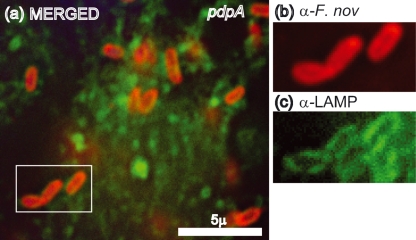
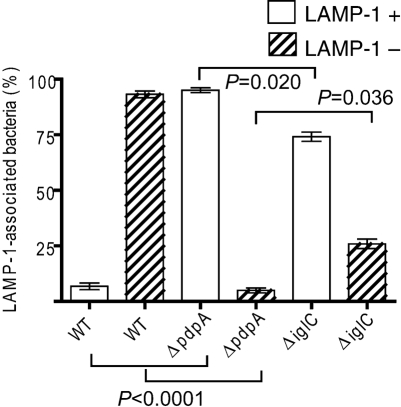
There is now a substantial body of literature that documents the escape of Francisella from LAMP-1-laden phagosomal vesicles, and the failure of some FPI mutants to escape at wild-type levels. In order to evaluate the intracellular trafficking of the ΔpdpA mutant, we infected mouse macrophage-like cell line J774A.1 cells, and examined the co-localization of LAMP-1 with wild-type F. novicida, the ΔpdpA mutant, and a ΔiglC mutant. F. tularensis and F. novicida mutants in iglC have been shown by others to be defective for phagosome escape (Bonquist et al., 2008; Lindgren et al., 2004; Santic et al., 2005b). Our experimental results using deconvoluting fluorescence microscopy showed a clear separation of wild-type F. novicida and LAMP-1 localization at 12 h post-infection (Fig. 2a–c). In contrast, the ΔpdpA mutant (Fig. 2d–f) and the ΔiglC mutant (Fig. 2g–i) showed close association of F. novicida and LAMP-1 localization. A close examination of the micrographs revealed that individual cells of ΔpdpA (Fig. 2d–f) and ΔiglC (Fig. 2g–i) were surrounded by a LAMP-1-laden structure that took the shape of the bacterial cells. However, this was not seen in the images of wild-type F. novicida (Fig. 2a–c). Genetic complementation with pJL-SKX : : pdpA restored LAMP-1 association to wild-type levels (data not shown). The clear association of ΔpdpA cells within a LAMP-1-loaded structure continued even after 19 h infection (Fig. 3a–c). A quantitative analysis of F. novicida-LAMP-1 association showed that less than 10 % of the wild-type F. novicida was LAMP-1 associated 12 h post-infection while 92 % of ΔpdpA and 74 % of the ΔiglC strain were LAMP-1 associated (Fig. 4).
Fig. 2.
The ΔpdpA mutant replicates in macrophages but remains LAMP-1 associated. J774A.1 macrophages adhered to glass coverslips were infected with F. novicida U112 (a–c), ΔpdpA (d–f), or ΔiglC (g–i) at an m.o.i. of 500 : 1. The colours in merged panels a, d and g correspond to Hoechst DNA (blue), anti-LAMP-1 (green), and anti-F. novicida (red). Panels b, e and h represent staining with anti-F. novicida antibody, and panels c, f and i represent staining with anti-LAMP-1. The arrowheads indicate areas of LAMP-1 staining that correspond to bacterial location.
Fig. 3.
The ΔpdpA mutant remains LAMP-1 associated late in infection. The association of ΔpdpA with LAMP-1 continues after 19 h infection (a). This localization is clearly seen in the corresponding enlarged images of the boxed region stained with only anti-F. novicida (b) or anti-LAMP-1 (c).
Fig. 4.
Association of F. novicida mutants with LAMP-1. Bacterial association with LAMP-1 was scored at 12 h post-infection by counting a minimum of 100 F. novicida cells for each strain. The Student's t-test was used to examine the significance of the differences between strains of the LAMP-1 positive or negative F. novicida cells and some of these P-values are shown on the graph. The P-values for wild-type (WT) F. novicida vs the ΔiglC strain are (LAMP-1 positive), 0.0001; and (LAMP-1 negative), 0.0024. The experiment was performed in triplicate.
Effect of the deletion of pdpA on host-cell mRNA responses
As an approach to discern a possible role of PpdA in host-cell processes, we compared the effect of infection of macrophages with wild-type and the ΔpdpA deletion strain on host-cell mRNA levels of selected signalling pathways. A summary from a representative set of experiments of the mRNA levels that were affected is shown in Table 2 and the complete dataset is presented in Supplementary Tables S1–S3 (available with the online version of this paper). Each of the columns showing the fold changes represents the results of separate experiments in which the mRNA levels were measured from sets of macrophage cultures subjected to two different infections or an infection and a mock infection. In addition, all experimental runs were duplicated. When the mRNA levels following infection with the ΔpdpA strain are compared to those following infection with the wild-type strain there are 28 mRNA species that are elevated and four that are depressed (Table 2, column 3) at least twofold. Importantly, the experimental fold differences seen for infections with the ΔpdpA vs the wild-type strain (column 3) are reflected by a comparison between the results of a wild-type infection vs uninfected macrophages (column 5) and the results of the mRNA levels measured in ΔpdpA infections vs uninfected macrophages (column 7). Of the 32 RNAs shown in Table 2, 13 encode ligands, five encode receptors, six encode pathway components and eight encode effector molecules. Of the ten most strongly upregulated messages, eight are transcripts from ligand-encoding genes.
Table 2.
Changes in mRNA levels of J744 macrophage-like cells following infection with F. novicida strains
| mRNA | Corresponding protein | ΔpdpA infection vs wild-type infection | Wild-type infection vs uninfected J774A.1 | ΔpdpA infection vs uninfected J774A.1 | |||
|---|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | Fold change | P-value | ||
| Lep | Leptin | 454 | <0.001 | 0.41 | 0.282 | 184.82 | <0.001 |
| Igfbp3 | Insulin-like growth factor-binding protein 3 | 387 | <0.001 | 2.03 | 0.142 | 786.88 | 0.001 |
| Wnt1 | Wingless-related MMTV integration site 1 | 305 | <0.001 | 0.57 | 0.607 | 174.37 | <0.001 |
| Ccl20 | Chemokine (C-C) ligand 20 | 303 | <0.001 | 1.47 | 0.285 | 446.34 | <0.001 |
| Wnt2 | Wingless-related MMTV integration site 1 | 191 | <0.001 | 1.65 | 0.397 | 315.17 | <0.001 |
| Birc1a | Bacloviral I AP repeat-containing 1 | 115 | <0.001 | 2.79 | 0.213 | 82.25 | 0.0167 |
| Csf2 | Colony stimulating factor 2 | 110 | <0.001 | 5.91 | 0.033 | 648.07 | <0.001 |
| Fgf4 | Fibroblast growth factor 4 | 82 | 0.014 | 1.30 | 0.702 | 105.86 | 0.0095 |
| Lef1 | Lymphoid enhancer binding factor1 | 76 | <0.001 | 1 | N/A | 75.90 | <0.001 |
| Cyp19a1 | Cytochrome P450, family 19, subfamily a, polypeptide 1 | 68 | 0.02 | 1.86 | 0.334 | 126.06 | 0.0084 |
| Sele | Selectin, endothelian cell | 50 | <0.001 | 1.48 | 0.580 | 74.54 | <0.001 |
| Tmepai | Transmembrane, prostate androgen-induced RNA | 45 | 0.04 | 2.9 | 0.215 | 131.23 | 0.007 |
| Hhip | Hedgehog-interacting protein | 21 | <0.001 | 4.21 | 0.033 | 86.70 | <0.001 |
| Wisp1 | WNT inducible pathway protein 1 | 8.8* | 0.001* | 6.06* | 0.011* | 19.62 | 0.0348 |
| Hk2 | Hexokinase | 0.11 | <0.001 | 1.01 | 0.9817 | 0.12 | 0.0015 |
| Fasl | Fas (TNF super family receptor) | 8.5 | 0.08 | 0.7 | 0.767 | 5.91 | 0.0092 |
| Cxcl1 | Chemokine (C-x-C) ligand 1 | 7.5 | <0.001 | 1.62 | 0.341 | 12.18 | 0.002 |
| Igfbp4 | Insulin-like growth factor-binding protein 4 | 6.8 | 0.004 | 0.08* | 0.0013* | 0.56* | 0.058* |
| Lta | Lymphotoxin A | 6.3 | <0.001 | 0.81 | 0.489 | 5.11* | <0.001* |
| Il2ra | Interleukin 2 receptor α | 6.1 | 0.002 | 0.72 | 0.613 | 4.40* | 0.044* |
| Il1a | Interleukin 1α | 4.9 | 0.035 | 2.73 | 0.0211 | 13.38 | 0.001 |
| Ccnd1 | Cyclin D1 | 3.7 | <0.001 | 0.28 | 0.002 | 1.02* | 0.093* |
| Mdm2 | Transformed mouse 3T3 cell double minute 2 | 0.3 | 0.002 | 3.22 | 0.0088 | 0.97 | 0.901 |
| Tert | Telomerase reverse transcriptase | 3.3 | <0.001 | 0.58* | 0.053* | 1.92 | 0.008 |
| Trp53 | Transformation-related protein 53 | 3.2 | <0.001 | 0.54 | 0.019 | 1.72 | 0.021 |
| Nos2 | Nitric oxide synthase 2 inducible, macrophage | 0.31 | 0.002 | 11.3* | <0.001* | 3.49* | 0.004* |
| Ikbkb | Inhibitor of kappaB kinase β | 2.8 | 0.004 | 0.79* | 0.252* | 2.24* | 0.009* |
| FasN | Fatty acid synthase | 0.38 | 0.16 | 0.23 | 0.028 | 0.09 | 0.001 |
| Il4ra | Interleukin 4 receptor α | 2.54 | 0.002 | 0.52* | 0.026* | 1.32* | 0.22* |
| Tfrc | Transferrin receptor | 2.4 | <0.001 | 0.82* | 0.047* | 19.3* | 0.026* |
| Nab2 | Ngfi-A-binding protein | 2.3 | <0.001 | 0.81* | 1.38* | 1.85* | 0.001* |
| Cdk2 | Cyclin-dependent kinase 2 | 2.1 | <0.001 | 0.45* | 0.001 | 0.98 | 0.877 |
*Indicates that a value represents the mean of four samples rather than five in an experimental run.
DISCUSSION
Like many intracellular pathogens, the ability of F. tularensis to interfere with the endocytic pathway of its host cell is vital in the pathogen's ability to cause disease. Although it is clear that infecting bacteria are able to break down the phagocytic membrane prior to lysosomal fusion, it is not clear what virulence factors are responsible for this process. It is apparent that many of the FPI-encoded proteins are required for Francisella intracellular growth and virulence, although some of the pathogenicity island proteins are required only for the latter (Ludu et al., 2008a). There is also substantial bioinformatic and biochemical evidence that the FPI encodes proteins that make up a type VI secretion apparatus or a macromolecular structure related to the type VI secretion system (T6SS) (Bingle et al., 2008; de Bruin et al., 2007; Ludu et al., 2008a). Although we and others have adopted the hypothesis that an FPI-encoded secretion system is responsible for transport of virulence factors that modulate host-cell functions, there is, at present, little evidence for this model, and no experimental results that precisely define the role of any FPI-encoded protein.
This study represents the first detailed analysis of a non-polar pdpA deletion mutant and the role of PdpA in intracellular growth. Although the ΔpdpA mutant is able to replicate, albeit minimally, in both BMDM and J774A.1 macrophages, this replication ceases after 24 h and bacterial burdens begin to decrease. This growth phenotype differs from that of a previously studied polar allelic replacement mutant that was completely impaired in its ability to replicate within macrophages (Nano et al., 2004). Recent work by Chong et al. (2008) reveals that polarity effects are also observed in the ΔiglC : : Em mutant, with levels of IglD expression being drastically reduced in this strain. These authors suggest that such observations require the re-evaluation of conclusions drawn from previous studies using the ΔiglC : : Em mutant. This agrees with our findings regarding the ΔpdpA : : Em mutant and emphasizes the importance of creating non-polar mutants for analysis, especially in the FPI, where the full expression of several transcriptionally linked genes appears to be required for intracellular growth. Statistical analysis indicated that genetic complementation of the pdpA deletion did not fully restore intracellular growth to wild-type levels. The incomplete complementation may have resulted from the recombinant pdpA gene being located at an ectopic location within the chromosome, leading to subtle alterations in pdpA expression.
Other researchers have shown that mutation of some FPI genes leads to Francisella strains that fail to escape the phagosome (Bonquist et al., 2008; Lindgren et al., 2004; Santic et al., 2005b). Like this work, these studies are attempts to surmise the function of different FPI-encoded proteins by examining the intramacrophage phenotype of Francisella strains with lesions in FPI genes. The deletion mutant of pdpA made in this work behaves similarly to mutants in iglC and iglD, in that all the mutants are impaired in their intracellular replication and have an increased association with the LAMP-1 lysosomal marker at time points when wild-type Francisella has escaped the phagosome (Bonquist et al., 2008). Since the FPI appears to encode a T6SS, the current challenge is to decipher which FPI genes are needed for secretion and which, if any, encode secreted effector proteins that interact with host-cell components. We do not believe that PdpA is a structural component of the T6SS because the protein does not seem to play a part in the secretion of IglC (Ludu et al., 2008a, and data not shown), and bioinformatic analysis fails to show any similarity of PdpA with known T6SS components in other bacteria.
Without knowing the nature of PdpA, and whether its mode of action has a direct or indirect effect on host-cell function, it is difficult to attribute a biological response directly to its expression. Nevertheless, our comparison of host-cell mRNA responses to infection by wild-type F. novicida and to the ΔpdpA mutant may contribute to the development of hypotheses about the role of PdpA and its interaction with host-cell components. Our examination of the levels of selected host-cell transcripts shows that the absence of PdpA in F. novicida results in significantly different host-cell responses to infection compared to an infection with wild-type F. novicida. That the absence of PdpA results in higher levels of mRNA for genes encoding ligands suggests that one of PdpA's functions is to suppress macrophage ability to signal via growth factors, cytokines and adhesion ligands. Our inference from these limited data is that PdpA plays a role in suppressing the infected macrophage's ability to recruit and stimulate other immune cells. However, these results can only be considered as clues to the role of PdpA in the intracellular parasitism by F. novicida, and considerably more work is needed to define the role and mode of action of PdpA.
Acknowledgments
This work was supported by grants 5R01 AI056212-02 from the National Institute of Allergy and Infectious Diseases and MOP 89812 from the Canadian Institutes of Health Research to F. E. N.
Abbreviations
BMDM, bone-marrow-derived macrophages
FPI, Francisella pathogenicity island
T6SS, type VI secretion system
Footnotes
Three supplementary tables are available with the online version of this paper.
References
- Andersson, H., Hartmanova, B., Kuolee, R., Ryden, P., Conlan, W., Chen, W. & Sjostedt, A. (2006a). Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J Med Microbiol 55, 263–271. [DOI] [PubMed] [Google Scholar]
- Andersson, H., Hartmanova, B., Ryden, P., Noppa, L., Naslund, L. & Sjostedt, A. (2006b). A microarray analysis of the murine macrophage response to infection with Francisella tularensis LVS. J Med Microbiol 55, 1023–1033. [DOI] [PubMed] [Google Scholar]
- Anthony, L. D., Burke, R. D. & Nano, F. E. (1991). Growth of Francisella spp. in rodent macrophages. Infect Immun 59, 3291–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, G. S. & Nano, F. E. (1998). MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol 29, 247–259. [DOI] [PubMed] [Google Scholar]
- Bingle, L. E., Bailey, C. M. & Pallen, M. J. (2008). Type VI secretion: a beginner's guide. Curr Opin Microbiol 11, 3–8. [DOI] [PubMed] [Google Scholar]
- Bonquist, L., Lindgren, H., Golovliov, I., Guina, T. & Sjostedt, A. (2008). The MglA and Igl proteins contribute to the modulation of Francisella tularensis LVS-containing phagosomes in murine macrophages. Infect Immun 76, 3502–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio, C. M. & Dow, S. W. (2005). Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J Immunol 175, 6792–6801. [DOI] [PubMed] [Google Scholar]
- Butchar, J. P., Cremer, T. J., Clay, C. D., Gavrilin, M. A., Wewers, M. D., Marsh, C. B., Schlesinger, L. S. & Tridandapani, S. (2008). Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS One 3, e2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity, J. C., Costante-Hamm, M. M., Balon, E. L., Boyd, D. H., Rubin, E. J. & Dove, S. L. (2007). Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog 3, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, A., Wehrly, T. D., Nair, V., Fischer, E. R., Barker, J. R., Klose, K. E. & Celli, J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun 76, 5488–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, D. L., Lee, B. Y. & Horwitz, M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun 72, 3204–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan, J. W. & North, R. J. (1992). Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect Immun 60, 5164–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin, O. M., Ludu, J. S. & Nano, F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Oyston, P. C., Green, M. & Titball, R. W. (2002). Tularemia. Clin Microbiol Rev 15, 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J. D., Craven, R. R., Fuller, J. R., Pickles, R. J. & Kawula, T. H. (2007). Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect Immun 75, 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, H., Golovliov, I., Baranov, V., Ernst, R. K., Telepnev, M. & Sjostedt, A. (2004). Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol 53, 953–958. [DOI] [PubMed] [Google Scholar]
- Ludu, J. S., de Bruin, O. M., Duplantis, B. N., Schmerk, C. L., Chou, A. Y., Elkins, K. L. & Nano, F. E. (2008a). The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J Bacteriol 190, 4584–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludu, J. S., Nix, E. B., Duplantis, B. N., de Bruin, O. M., Gallagher, L. A., Hawley, L. M. & Nano, F. E. (2008b). Genetic elements for selection, deletion mutagenesis and complementation in Francisella spp. FEMS Microbiol Lett 278, 86–93. [DOI] [PubMed] [Google Scholar]
- Mohapatra, N. P., Balagopal, A., Soni, S., Schlesinger, L. S. & Gunn, J. S. (2007a). AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect Immun 75, 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra, N. P., Soni, S., Bell, B. L., Warren, R., Ernst, R. K., Muszynski, A., Carlson, R. W. & Gunn, J. S. (2007b). Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun 75, 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra, N. P., Soni, S., Reilly, T. J., Liu, J., Klose, K. E. & Gunn, J. S. (2008). The combined deletion of four Francisella acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect Immun 76, 3690–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano, F. E. (1988). Identification of a heat-modifiable protein of Francisella tularensis and molecular cloning of the encoding gene. Microb Pathog 5, 109–119. [DOI] [PubMed] [Google Scholar]
- Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., Ludu, J. S., Letendre, G. W., Meierovics, A. I. & other authors (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 186, 6430–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranavitana, C., Pittman, P. R., Velauthapillai, M., Zelazowska, E. & Dasilva, L. (2008). Transcriptional profiling of Francisella tularensis infected peripheral blood mononuclear cells: a predictive tool for tularemia. FEMS Immunol Med Microbiol 54, 92–103. [DOI] [PubMed] [Google Scholar]
- Reilly, T. J., Baron, G. S., Nano, F. E. & Kuhlenschmidt, M. S. (1996). Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem 271, 10973–10983. [DOI] [PubMed] [Google Scholar]
- Sammons-Jackson, W. L., McClelland, K., Manch-Citron, J. N., Metzger, D. W., Bakshi, C. S., Garcia, E., Rasley, A. & Anderson, B. E. (2008). Generation and characterization of an attenuated mutant in a response regulator gene of Francisella tularensis live vaccine strain (LVS). DNA Cell Biol 27, 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic, M., Molmeret, M. & Abu Kwaik, Y. (2005a). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol 7, 957–967. [DOI] [PubMed] [Google Scholar]
- Santic, M., Molmeret, M., Klose, K. E., Jones, S. & Kwaik, Y. A. (2005b). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol 7, 969–979. [DOI] [PubMed] [Google Scholar]
- Santic, M., Asare, R., Skrobonja, I., Jones, S. & Abu Kwaik, Y. (2008). Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun 76, 2671–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk, C. L., Duplantis, B. N., Wang, D., Burke, R. D., Chou, A. Y., Elkins, K. L., Ludu, J. S. & Nano, F. E. (2009). Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D. S., Brotcke, A., Henry, T., Margolis, J. J., Chan, K. & Monack, D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104, 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]