Abstract
Alternative sigma factor 54 (RpoN) is an important regulator of stress resistance and virulence genes in many bacterial species. In this study, we report on the gene expression alterations that follow rpoN inactivation in Escherichia coli O157 : H7 strain Sakai (SakairpoN : : kan), and the influence of RpoN on the acid resistance phenotype. Microarray gene expression profiling revealed the differential expression of 103 genes in SakairpoN : : kan relative to Sakai. This included the growth-phase-dependent upregulation of genes required for glutamate-dependent acid resistance (GDAR) (gadA, gadB, gadC and gadE), and the downregulation of locus of enterocyte effacement (LEE) genes, which encode a type III secretion system. Upregulation of gad genes in SakairpoN : : kan during exponential growth correlated with increased GDAR and survival in a model stomach system. Complementation of SakairpoN : : kan with a cloned version of rpoN restored acid susceptibility. Genes involved in GDAR regulation, including rpoS (sigma factor 38) and gadE (acid-responsive regulator), were shown to be required for the survival of SakairpoN : : kan by the GDAR mechanism. This study describes the contribution of rpoN to acid resistance and GDAR gene regulation, and reveals RpoN to be an important regulator of stress resistance and virulence genes in E. coli O157 : H7.
INTRODUCTION
Escherichia coli O157 : H7 is an important aetiological agent of food-borne diarrhoea, bloody diarrhoea and haemolytic uraemic syndrome (Mead & Griffin, 1998; Mead et al., 1999). Infection with E. coli O157 : H7 is characterized by a dramatic intestinal histopathology termed attaching and effacing (A/E) lesions. These lesions result from the intimate adherence of E. coli O157 : H7 to host intestinal cells, forming cup-like pedestals on which the bacteria are intimately perched (Donnenberg & Whittam, 2001). The determinants of the A/E phenotype are encoded on a 35.6 kb laterally acquired pathogenicity island termed the locus of enterocyte effacement (LEE) (Perna et al., 1998). The LEE encodes a type III secretion apparatus, which is required for the translocation of virulence factors (effectors) from E. coli O157 : H7 into host enterocytes (Perna et al., 1998).
For successful transmission, E. coli O157 : H7 is dependent on the expression of acid resistance, which allows for passage through the acid barrier of the stomach and low oral infectious dose (Chart, 2000; Teunis et al., 2004). The E. coli acid resistance phenotype is characterized by protracted survival under conditions of extreme acid stress. More specifically, cells are considered acid resistant if survival following 2 h of acid challenge (pH 2.0–2.5) exceeds 10 % of initial cell densities (Gorden & Small, 1993). These conditions are representative of the retention time of food in the stomach, and the pH of stomach gastric acid. There are three major systems of acid resistance in E. coli O157 : H7: arginine- and glutamate-dependent systems, and an oxidative system (Foster, 2004). Of these three, the glutamate-dependent acid resistance (GDAR) system is believed to provide the highest protection from acid stress. GDAR requires exogenous glutamate, glutamate decarboxylase (GadA and GadB isoforms), and an antiporter (GadC), which exchanges the decarboxylation product, γ-aminobutyrate, for fresh glutamate (Castanie-Cornet et al., 1999). Regulation of the GDAR system is multifactorial, and partly includes alternative sigma factor 38 (RpoS), the nucleoid structuring protein H-NS, the AraC/XylS-like transcriptional regulators GadX and GadW, and the LuxR-like regulator GadE (De Biase et al., 1999; Ma et al., 2002, 2003; Masuda & Church, 2003; Tramonti et al., 2002).
Alternative sigma factor 54 (RpoN) is one of seven RNA polymerase sigma subunits in E. coli required for promoter-initiated transcription. RpoN plays a major role in the response of E. coli to nitrogen-limiting conditions. Under such conditions, RpoN directs the transcription of at least 14 E. coli operons/regulators in the nitrogen regulatory (Ntr) response (Reitzer & Schneider, 2001). RpoN also plays a role in various stress response mechanisms of E. coli. For instance, the E. coli phage-shock operon (psp) is driven by an RpoN-dependent promoter and protects E. coli from alkaline pH during stationary-phase growth (Model et al., 1997). In addition, transposon insertion in rpoN has been shown to result in elevated resistance to the DNA gyrase inhibitor novobiocin in E. coli (Jovanovic et al., 1999). RpoN has also been shown to be important for stress resistance in bacteria other than E. coli. For example, inactivation of rpoN in Listeria monocytogenes affects its ability to grow under osmotic stress (Okada et al., 2006), and leads to increased resistance to certain bacteriocins in L. monocytogenes and Enterococcus faecalis (Dalet et al., 2000; Robichon et al., 1997). In addition, RpoN has been determined to regulate virulence gene expression. This includes type III secretion in Pseudomonas aeruginosa, and O-antigen expression in Salmonella enterica (Bittner et al., 2002; Matz et al., 2008). Lastly, RpoN has been reported to co-regulate various bacterial virulence genes in conjunction with RpoS (Bittner et al., 2004; Yang et al., 2005).
This study investigated the RpoN regulon in the 1996 Osaka Japan outbreak E. coli O157 : H7 strain Sakai (Michino et al., 1999), using gene expression microarray analysis. Our results revealed RpoN to be a new regulator of the LEE, and of acid-resistance genes of the GDAR system. The effect of RpoN on the phenotypic expression of acid resistance, the regulation of GDAR genes, and the role for GDAR system regulators in RpoN-dependent repression of acid resistance was also examined.
METHODS
Bacterial strains, genetic manipulations, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Isogenic and polygenic mutants were constructed using the λ-Red recombinase-assisted one-step inactivation method adapted for enterohaemorrhagic E. coli (Murphy & Campellone, 2003). Briefly, primers P1 and P2, which amplify a kanamycin (Kan) resistance cassette from plasmid pKD4 (Datsenko & Wanner, 2000), were constructed with 40 bp oligonucleotide 5′ extensions, which were homologous to the up- and downstream intergenic regions of genes targeted for inactivation (see Supplementary Table S1, available with the online version of this paper, for details of all primers). These primers were used to generate a PCR product, which was then electroporated (2.5 kV, 5.6 ms) using a MicroPulser electroporator (Bio-Rad) into a Red-recombinase-producing background of E. coli O157 : H7 strain Sakai, as described previously (Kailasan Vanaja et al., 2009). Transformants were selected on Luria–Bertani (LB) 1.5 % (w/v) agar plates with Kan (20 μg ml−1). Removal of Kan cassettes was performed by introducing pCP20, encoding a Flp recombinase, into isogenic backgrounds by electroporation following the method of Datsenko & Wanner (2000). Polygenic mutants were constructed by inactivating rpoN in ΔrpoS and ΔgadE isogenic backgrounds to produce SakaiΔrpoS rpoN : : kan and SakaiΔgadE rpoN : : kan. SakairpoN : : kan was complemented in trans with the wild-type Sakai rpoN ORF. This was generated by PCR with TaKaRa LA Taq polymerase, and cloned into pCR2.1 (Invitrogen) to produce SakairpoN : : kan pCR2.1(rpoN+). All genetic constructs were confirmed by PCR and restriction mapping. In addition, impaired growth and glutamine auxotrophy for rpoN mutants (Reitzer et al., 1987) were confirmed in DMEM-MOPS (Supplementary Fig. S1), and by the absence of growth in minimal medium without glutamine, respectively. Diminished catalase activity and glycogen storage of rpoS mutants was confirmed for SakaiΔrpoS rpoN : : kan according to previously designed methods (Bohannon et al., 1991; Hengge-Aronis & Fischer, 1992).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source/reference |
|---|---|---|
| E. coli O157 : H7 | ||
| TW08264 | WT strain Sakai | Michino et al. (1999) |
| EcJR-8 | SakairpoN : : kan | This study |
| EcJR-5 | SakairpoN : : kan pCR2.1(rpoN+) | This study |
| EcJR-6 | SakaiΔgadE rpoN : : kan | This study |
| EcJR-9 | SakaiΔrpoS rpoN : : kan | This study |
| TW04863 | WT strain 93-111 | STEC Center, Mich. State Univ. |
| EcJR-10 | 93-111rpoN : : kan | This study |
| TW14359 | WT | STEC Center, Mich. State Univ. |
| EcJR-11 | TW14359rpoN : : kan | This study |
| TW15901 | Sakai pKM208 | Kailasan Vanaja et al. (2009) |
| TW15902 | SakaiΔgadE | Kailasan Vanaja et al. (2009) |
| Plasmids | ||
| pCR2.1 | TOPO cloning vector | Invitrogen |
| pCR2.1(rpoN+) | pCR2.1 containing rpoN | This study |
| pKM208 | Red-recombinase expression vector | Datsenko & Wanner (2000) |
| pKD4 | Template plasmid for Kan cassette | Datsenko & Wanner (2000) |
| pCP20 | Flp recombinase expression vector | Datsenko & Wanner (2000) |
For all experiments, LB starter cultures were inoculated with single colonies and grown to an OD600 of 0.5 at 37 °C (180 r.p.m.). These cultures were then used to inoculate (0.5 %, v/v, inoculum) 50 mM MOPS-buffered Dulbecco's Modified Eagle's Medium containing 4 g glucose l−1 (pH 7.4) (DMEM-MOPS), LB containing 50 mM MOPS (pH 7.4), LB containing 50 mM MOPS and 4 g glucose l−1 (LBG-MOPS) (pH 7.4), or LB with 4 g glucose l−1 (LBG) (pH 7.4), and grown overnight at 37 °C (180 r.p.m.). DMEM is a rich but defined medium, which contains glutamine to support the growth of rpoN mutants, which are auxotrophic for glutamine. These overnight cultures were then used to inoculate various growth media (initial inoculum OD600≈0.05) for all experiments. The growth of DMEM-MOPS cultures utilized for RNA isolation, DNA microarray analysis and quantitative real-time PCR (qRT-PCR) experiments were monitored by taking OD600 readings at 1 h intervals for 8 h (Supplementary Fig. S1).
DNA microarrays.
Microarray analysis was performed to determine transcriptome alterations
that occur in E. coli O157 : H7 Sakai as a result of rpoN inactivation in exponential- and early stationary-phase cultures.
DNA microarrays consisted of 6088 spotted 70-mer oligonucleotides (E. coli Oligo set version 1.0.1, Qiagen) representing ORFs from E. coli O157 : H7 strains EDL-933 and Sakai, and K-12 strain
MG1655. Over 1700 ORFs were specific to E. coli O157 : H7,
and included 85 ORFs from pO157 and three ORFs from pOSAK1. DMEM-MOPS cultures (n=4) were sampled during exponential (OD600
0.5) and early stationary-phase growth (OD600 1.8),
and RNA was extracted as described previously (Bergholz et al., 2007b). Amino-allyl
labelling and cDNA synthesis, cy-dye coupling and hybridization conditions
followed previously described methods (Bergholz et al., 2007b). Image files (TIFF)
of 16 hybridized microarray slides (four slides per strain, per phase
of growth) were generated using an Axon 4000B scanner (Molecular
Devices), and analysed using GenePix Pro software (Molecular Devices,
version 6.0). The resulting microarray intensity data were log2-transformed, and normalized using the lowess algorithm in maanova version 0.98-8 (R version 2.2.1). Statistical analysis
was performed using a significance analysis of microarrays (SAM, MultiExperiment
Viewer, version 4.0) (Tusher et al., 2001) unpaired contrast, available through the
TM4 software package (JCVI). A d-statistic (Tusher et al., 2001) was
calculated for each gene based on repeated permutations, and a false discovery
rate (FDR) of 0.05 was used to assign a critical cutoff for significance.
Gene expression ratios (R) were calculated for each significant
gene using  , and were
considered biologically significant using a cutoff of 0.5≥R≥2.0.
Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was also used
on select microarray datasets of gene clusters that shared similar biological
function/regulation. Normalized microarray intensity data files have been
deposited in the Gene Expression Omnibus (GSE17467) through NCBI.
, and were
considered biologically significant using a cutoff of 0.5≥R≥2.0.
Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was also used
on select microarray datasets of gene clusters that shared similar biological
function/regulation. Normalized microarray intensity data files have been
deposited in the Gene Expression Omnibus (GSE17467) through NCBI.
qRT-PCR.
For microarray validation, DMEM-MOPS cultures (n=3)
were sampled for RNA extraction, as described above. Sampling was also performed
as described above at OD600 0.2, 0.4, 0.6, 0.8 and 1.2, to examine
the expression of GDAR genes in Sakai and SakairpoN : : kan during exponential into transition-phase growth. cDNA was prepared
from 1 μg RNA samples using iScript Select cDNA synthesis (Bio-Rad)
and qRT-PCR was performed using the iQ5 system (Bio-Rad) with the
primers and probes listed in Supplementary Table S1. For gadA and gadB, TaqMan probes were developed and optimized according to established
protocols (Bergholz et al.,
2007a) and for all remaining qRT-PCR targets, SYBR chemistries
and reaction conditions were used as previously described (Bergholz et al., 2007b). Cycle
threshold (Ct) data were normalized to rrsA and normalized cycle threshold values (ΔCt) were transformed using 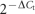 /10−6 (Livak & Schmittgen, 2001), and reported as arbitrary
gene expression units (EU) or as the gene expression ratio of SakairpoN : : kan/Sakai. In keeping with
microarray analysis, a twofold cutoff was used for gene expression ratios
to denote biologically significant changes in expression. Differences in mean
EU between Sakai and SakairpoN : : kan
for individually analysed genes were compared using Welch's t-test (R).
/10−6 (Livak & Schmittgen, 2001), and reported as arbitrary
gene expression units (EU) or as the gene expression ratio of SakairpoN : : kan/Sakai. In keeping with
microarray analysis, a twofold cutoff was used for gene expression ratios
to denote biologically significant changes in expression. Differences in mean
EU between Sakai and SakairpoN : : kan
for individually analysed genes were compared using Welch's t-test (R).
Assessment of GDAR.
GDAR was determined as previously described (Large et al., 2005) with slight modification. DMEM-MOPS, LB-MOPS, LBG-MOPS or LBG cultures were initially grown to the OD600 described below, before using these cultures (≈50 μl culture volume) to inoculate 10 ml (10 : 1 flask to medium ratio) minimal EG medium (73 mM K2HPO4, 10 mM sodium citrate, 0.8 mM MgSO4, 5.7 mM glutamate and 4 g glucose l−1, pH 2.0) to reach a final density of 106 c.f.u. ml−1. To determine the effects of glutamate on the survival of SakairpoN : : kan by the GDAR mechanism, EG medium was also prepared without glutamate addition. GDAR was tested with Sakai and SakairpoN : : kan cultures grown to OD600 0.5, 0.75, 1.0 and 3.5; all remaining strains were tested at OD600 0.5 only. Samples were drawn from EG cultures (180 r.p.m., 37 °C) at 0.5 h intervals and diluted before adding aliquots of the dilutions to LB agar, followed by incubation at 37 °C for 18–20 h before determining c.f.u. ml−1. Survival rates (ΔV) were extrapolated from plate count data using linear regression analysis (R), and were reported as the log10 c.f.u. decrease per h (log c.f.u. h−1). The strength of linearity was estimated by the correlation coefficient (r2), which exceeded 0.85 (85 %) for all analyses. Differences in mean ΔV between wild-type and mutant derivative strains were examined statistically using Welch's t-test.
Assessment of acid resistance in a model stomach system.
Acid resistance in a model stomach system was determined as previously described (Bergholz & Whittam, 2007), with the following adaptations. DMEM-MOPS cultures were grown to OD600 0.5 before using these cultures to inoculate 106 c.f.u. ml−1 into simulated gastric fluid (pH 2) mixed with 30 g Gerber Turkey baby food using a Stomacher 400C (Seward). Aliquots were recovered from this system at 1 h intervals for a total of 2 h for plate counts on LB agar. Mean counts for Sakai and SakairpoN : : kan were then compared using Welch's t-test.
RESULTS
Gene expression profile of rpoN-inactivated E. coli O157 : H7 strain Sakai
We used microarray analysis to examine the impact of rpoN inactivation in E. coli O157 : H7 Sakai (SakairpoN : : kan) on global gene expression during exponential- and early stationary-phase growth in DMEM-MOPS. Inactivation of rpoN resulted in the differential expression of 103 ORFs (Table 2). This altered expression was growth phase dependent, with 95 ORFs being differentially expressed in exponential phase, and 19 in early stationary phase. Fifty-four exponential-phase ORFs were upregulated in SakairpoN : : kan, whereas all 19 ORFs altered in early stationary phase were downregulated. Only 11 ORFs were found to be differentially expressed in both growth phases. Overall, our microarray analysis identified 74 ORFs that have not been shown previously to be regulated by RpoN in E. coli (Table 2). The majority of differentially expressed ORFs (43/103) represented genes involved in nitrogen assimilation, as well as genes involved in the metabolism and transport of carbohydrates and inorganic compounds. As rpoN is required for stimulation of the nitrogen regulatory response in nitrogen-limiting media, such as DMEM (Reitzer & Schneider, 2001), genes involved in nitrogen uptake and assimilation were downregulated in SakairpoN : : kan relative to Sakai. These included glnALG, glnHPQ, glnK-amtB, gltIJKL, yhdWXYZ, astCADBE and nac (Table 2). These operons and genes are involved in glutamine biosynthesis, glutamine transport, ammonia transport, glutamate transport, ABC-type transport, arginine metabolism and nitrogen assimilation, respectively.
Table 2.
Differentially expressed genes in SakairpoN : : kan compared to Sakai
| ECs no.* | Gene | Function | Expression ratio SakairpoN : : kan/Sakai† | |
|---|---|---|---|---|
| Expo | Stat | |||
| Hypothetical and miscellaneous | ||||
| 0069 | yabI | Membrane-associated protein | 0.20 | – |
| 0781 | ybgS | Homeobox protein | 7.73 | – |
| 0982 | ycaC | Predicted hydrolase | 9.24 | – |
| 1152 | yccJ | Unknown | 3.24 | – |
| 1683 | ycgB | Putative sporulation protein | 5.59 | – |
| 1695 | yihV | Unknown | – | 0.05 |
| 1768‡ | Sp9-encoded protein | 4.21 | – | |
| 2316 | tus | DNA-replication protein | 3.63 | – |
| 2430 | ydiZ | Unknown | 3.88 | – |
| 2546 | yebV | Unknown | 20.0 | – |
| 2547 | yebW | Unknown | 5.85 | – |
| 2662 | fliC | Flagellin | 0.25 | – |
| 2692 | yodD | Unknown | 6.94 | – |
| 3154 | elaB | Unknown | 12.1 | – |
| 3413‡ | Unknown | 5.32 | – | |
| 4037 | yhbQ | Predicted endonucleases | 4.44 | – |
| 4291‡ | Unknown | 8.59 | – | |
| 4323 | yhhT | Inner-membrane protein | 0.07 | 0.30 |
| 4363 | yhiM | Unknown | 6.48 | – |
| 4699 | yifE | Unknown | 0.12 | – |
| 4737 | yzcX | Unknown | 5.81 | – |
| 4745 | yigE | Unknown | 2.65 | – |
| 4801‡ | Unknown | 3.17 | – | |
| Stress resistance | ||||
| 1829 | yciE | Unknown | 6.00 | – |
| 1830 | yciF | Structural protein | 13.5 | – |
| 1831 | yciG | Unknown | 9.39 | – |
| 1881§ | pspA | Phage-shock protein A | 0.28 | 0.04 |
| 1882§ | pspB | Phage-shock protein B | – | 0.14 |
| 1883§ | pspC | Phage-shock protein C | – | 0.34 |
| 1884§ | pspD | Phage-shock protein D | – | 0.36 |
| 1885§ | pspE | Phage-shock protein E | – | 0.49 |
| 2097 | gadC | Acid-sensitivity protein (XasA) | 5.28 | – |
| 2098 | gadB | Glutamate decarboxylase isozyme | 4.07 | – |
| 2604 | otsA | Trehalose-6-phosphate synthase | 4.46 | – |
| 2605 | otsB | Trehalose-6-phosphate phosphatase | 2.82 | – |
| 3186 | yfcG | Glutathione S-transferase | 3.23 | – |
| 3533 | ygaM | Unknown | 6.62 | – |
| 4377 | slp | Starvation lipoprotein | 3.29 | – |
| 4392 | gadE | Acid-responsive regulator | 3.83 | – |
| 4396 | gadX | Glutamate decarboxylase activator | 3.31 | – |
| 4397 | gadA | Glutamate decarboxylase isozyme | 3.49 | – |
| 5586 | ecnB | Bacteriolytic enterocidin B | 7.47 | – |
| Transport and metabolism | ||||
| 0504§ | glnK | Nitrogen regulatory protein P-II 2 | 0.01 | 0.02 |
| 0505§ | amtB | High-affinity ammonium transporter | 0.08 | 0.12 |
| 0538 | ybaS | Glutaminase | 5.03 | – |
| 0693§ | gltJ | Glutamate transporter, permease | 0.19 | – |
| 0694§ | gltI | Glutamate transporter, periplasmic | 0.08 | 0.07 |
| 0887§ | glnQ | ATP-binding protein for Gln transporter | 0.26 | – |
| 0888§ | glnP | Permease protein of Gln transporter | – | 0.30 |
| 0889§ | glnH | Periplasmic protein for Gln transporter | 0.07 | 0.09 |
| 1254§ | rutE | Nitroreductase | 0.19 | – |
| 1255§ | rutD | Putative acetyltransferase | 0.24 | – |
| 1711 | ychM | Sulfate permease | 5.73 | – |
| 1722 | chaB | Cation transport regulator | 7.70 | – |
| 1879 | puuE | GABA-aminotransferase | 4.06 | – |
| 1921 | abgB | p-Aminobenzoyl-glutamate hydrolase | – | 0.22 |
| 1922 | abgA | Putative aminohydrolase | – | 0.06 |
| 2082 | adhP | Ethanol dehydrogenase | 7.50 | – |
| 2091§ | ddpA | Putative d-Ala-d-Ala transport protein | 0.09 | – |
| 2092§ | ddpX | Putative d-Ala-d-Ala dipeptidase | 0.09 | – |
| 2103 | ydeN | Predicted sulfatase | 0.12 | – |
| 2451§ | astB | Succinylarginine dihydrolase | 0.25 | – |
| 2452§ | astD | Succinylglutamic semialdehyde dehydrolase | 0.15 | – |
| 2453§ | astA | Arginine succinyltransferase | 0.21 | – |
| 2454§ | astC | 0.20 | – | |
| 2650 | pgsA | Phosphatidylglycerophosphate synthetase | 2.93 | – |
| 2784§ | nac | Nitrogen assimilation control protein | 0.03 | 0.08 |
| 2900 | Fructose bisphosphate aldolase | 7.65 | – | |
| 3058 | yeiC | Predicted kinase | 0.27 | |
| 3192§ | hisQ | Permease for histidine transport | 0.28 | – |
| 3193§ | hisJ | Periplasmic protein for histidine transport | 0.15 | – |
| 3327 | tktB | Transketolase | 8.50 | – |
| 3425 | tadA | tRNA-specific adenosine deaminase | 5.13 | – |
| 3689 | ygdQ | TerC-like transport protein | 0.06 | 0.09 |
| 4141§ | yhdW | Periplasmic-binding protein | 0.38 | – |
| 4142§ | yhdX | Transport system permease protein | 0.46 | – |
| 4144§ | yhdZ | ATP-binding protein | 0.29 | – |
| 4448 | xylA | Xylose isomerase | 0.17 | – |
| 4490 | yibO | Phosphoglyceromutase | 7.18 | – |
| 4492 | yibQ | 2.48 | – | |
| 4734 | hemD | Uroporphyrinogen-III synthetase | 2.54 | – |
| 4735 | hemC | Porphobilinogen deaminase | 2.07 | – |
| 4790§ | glnG | Glutamine utilization response regulator | 0.15 | – |
| 4791§ | glnL | Glutamine utilization sensor kinase | 0.08 | 0.34 |
| 4792§ | glnA | Glutamine synthetase | 0.08 | 0.07 |
| Transcription and translation | ||||
| 1902 | tyrR | Transcriptional dual regulator | 6.36 | – |
| 2032 | rimL | Ribosomal-serine N-acetyltransferase | 4.59 | – |
| 2084 | rpsV | 30S ribosomal protein S22 | 6.34 | – |
| 2783 | cbl | CysB-like regulator of cys operon | 0.09 | 0.18 |
| 3136 | yfaX | Putative regulator | 0.12 | – |
| 3403 | hcaR | Transcriptional activator of hca cluster | 2.75 | – |
| 4440 | yiaG | Transcriptional regulator | 7.66 | – |
| 4484 | yibK | rRNA methylase | 4.51 | – |
| 4867 | metJ | Methionine transcriptional repressor | 2.40 | – |
| Pathogenesis | ||||
| 0780 | zitB | Zinc transport/adhesin | 0.36 | – |
| 3704‡ | yqeI | ETT2 sensory transducer | 3.91 | – |
| 4551‡ | orf29 | LEE4 type III secretion protein | 0.33 | – |
| 4554‡ | espB | LEE4 translocon protein | 0.29 | – |
| 4560‡ | cesT | LEE5 type III secretion chaperone | 0.17 | – |
| 4561‡ | tir | LEE5 translocated intimin receptor | 0.26 | – |
| 4563‡ | cesF | LEE3 type III secretion chaperone | 0.32 | – |
| 4491 | envC | Murien hydrolase | 4.31 | – |
| 4571‡ | espZ | LEE2 type III secretion protein | 0.24 | – |
*Locus tag (ECs no.) for E. coli O157 : H7 strain Sakai (GenBank no. BA000007).
†Expression ratio for exponential (Expo) and early stationary (Stat)
phase cultures of SakairpoN : : kan
and Sakai determined as  . –,
No significant differential expression between SakairpoN : : kan and Sakai.
. –,
No significant differential expression between SakairpoN : : kan and Sakai.
‡Gene is encoded within an E. coli O157 : H7-specific sequence.
§Gene previously shown to be regulated by RpoN.
RpoN regulates the expression of stress resistance genes
Nineteen ORFs determined to be differentially expressed in SakairpoN : : kan by microarray analysis encoded gene products associated with stress resistance (Table 2). Interestingly, 14/19 stress-resistance genes were upregulated in exponential-phase SakairpoN : : kan cultures. These included numerous genes shown to be induced upon exposure to osmotic stress, such as bacteriolytic enterocidin B (ecnB), which is activated under high-osmolarity conditions (Bishop et al., 1998); otsBA, which encodes the enzymes trehalose-6-phosphate phosphatase and trehalose-6-phosphate synthase, involved in metabolism of the osmoprotectant trehalose (Giaever et al., 1988; Hengge-Aronis et al., 1991); and yciGFE, encoding predicted or conserved proteins, the function of which is unknown (Weber et al., 2005) (Table 2). In addition, yfcG (Dong et al., 2007), which encodes a glutathione S-transferase (Wadington et al., 2009) required for hydrogen peroxide resistance (Kanai et al., 2006), was upregulated in SakairpoN : : kan. Two genes induced upon exposure to moderate acid stress, ygaM and slp (Tucker et al., 2002; Weber et al., 2005), were also upregulated in SakairpoN : : kan in exponential phase. As expected, the phage-shock operon, encoded by pspABCDE and transcriptionally regulated by RpoN (Weiner et al., 1991), was downregulated in stationary-phase SakairpoN : : kan cultures (Table 2). E. coli psp mutants demonstrate poor survival in stationary-phase alkaline cultures, reduced biofilm formation, and deficiencies in proton-motive force and protein secretion (Darwin, 2005). Many of these stress-resistance-associated genes (ecnB, otsBA, yciGFE, yfcG and ygaM) have been previously shown to be regulated by RpoS (Bishop et al., 1998; Giaever et al., 1988; Hengge-Aronis et al., 1991; Kanai et al., 2006; Tucker et al., 2002; Weber et al., 2005).
RpoN positively regulates LEE gene expression
Microarray analysis using significance analysis of microarrays (SAM) identified 11 E. coli O157 : H7-specific ORFs to be differentially expressed in exponential-phase SakairpoN : : kan compared to Sakai, which included six downregulated ORFs encoded on the LEE (Table 2). The five remaining E. coli O157 : H7-specific genes encoded an E. coli type III secretion system 2 sensory transducer (yeqI), a prophage-borne gene (ECs1768), and three genes encoding hypothetical proteins: ECs3413, ECs4291 and ECs4801. The LEE genes downregulated in exponential-phase SakairpoN : : kan encoded a protein required for the translocation of type III secretion effectors (espZ) and a secreted structural component of the translocation apparatus (espB); the translocated intimin receptor (tir) and a chaperone required for proper secretion of Tir (cesT); a chaperone (cesF) required for proper folding of EspF; and orf29, which has no known function (reviewed by Crawford et al., 2002). The LEE is composed of 41 ORFs, and is essential to the pathogenesis of E. coli O157 : H7. We therefore utilized gene set enrichment analysis (GSEA) to determine if the expression of other LEE-encoded genes was also altered by rpoN inactivation. GSEA identified 28 genes encoded throughout the LEE to be significantly downregulated in SakairpoN : : kan, with a normalized enrichment score (NES) of −2.8 (FDR q value=0.0; Fig. 1). This set included those genes determined to be differentially expressed by SAM alone. Collectively, the results of SAM and GSEA analysis of microarray data indicate that genes within all five of the LEE operons, including the LEE regulator grlA, were downregulated in exponential-phase cultures of SakairpoN : : kan relative to Sakai (Table 2, Fig. 1).
Fig. 1.
Differential expression of LEE genes by GSEA analysis in SakairpoN : : kan compared to Sakai. The fold change in gene expression is plotted against gene or locus ID (ECs no.) for all five LEE operons. Filled symbols represent genes determined to be significantly altered in expression by GSEA. All genes significant by SAM were also significant by GSEA. For orientation, names for selected LEE genes are provided to the immediate right of their respective plots.
Validation of microarray data using qRT-PCR
qRT-PCR was used to independently validate gene expression differences for selected GDAR and LEE genes (Table 3). The expression of GDAR genes gadC and gadE and of LEE genes cesT, espA, espB, espZ, ler and tir, during exponential growth as determined by qRT-PCR was concordant with microarray expression results. Gene expression ratios from microarray and qRT-PCR analyses were found to be tightly correlated (r2=0.86). The gadC gene, however, was substantially more upregulated in SakairpoN : : kan as determined by qRT-PCR than by microarray analysis. In addition, espA was determined to be significantly downregulated in SakairpoN : : kan by qRT-PCR analysis, but not by microarray analysis (Table 3). These disparities may reflect reported differences in sensitivity between the two techniques (Canales et al., 2006).
Table 3.
Validation of microarray genes by qRT-PCR
| Gene | Expression ratio (SakairpoN : : kan/Sakai)* Expo. (OD600=0.5) | |
|---|---|---|
| qRT-PCR | MA† | |
| cesT | 0.16±0.03‡ | 0.17 |
| espA | 0.29±0.06 | – |
| espB | 0.14±0.03 | 0.29 |
| espZ | 0.20±0.05 | 0.24 |
| ler | – | – |
| tir | 0.15±0.04 | 0.26 |
| gadC | 33.3±13.0 | 5.28 |
| gadE | 4.00±1.00 | 3.83 |
*Normalized gene expression ratios for SakairpoN : : kan/Sakai during exponential (Expo.) phase growth in DMEM-MOPS.
Gene expression ratio was calculated as 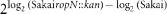 . –, Not significantly altered in expression.
. –, Not significantly altered in expression.
†MA is gene expression ratio for microarray data from Supplementary Table S2.
‡95 % confidence interval on the mean (n=3).
Negative regulation of gad genes by RpoN
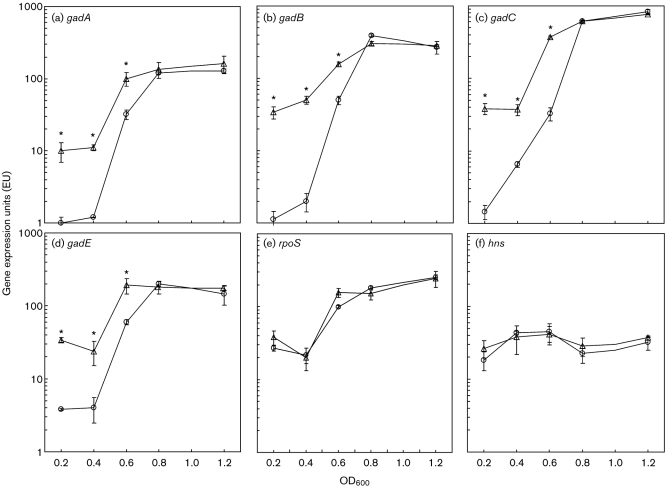
Genes essential to the GDAR system, gadA, gadB and gadC, and genes encoding factors that regulate the GDAR system, gadE and gadX, were all upregulated in exponential-phase SakairpoN : : kan, as determined by microarray analysis, but were not differentially expressed in early stationary phase (Table 2). Since gad genes were upregulated in SakairpoN : : kan, and the GDAR system is essential for full expression of the acid-resistance phenotype of E. coli O157 : H7, we decided to compare the expression of these genes in SakairpoN : : kan and Sakai at various OD600 values during the exponential (OD600 0.2–0.8) and transition (OD600 >0.8) phases of growth (Fig. 2). Transcriptional profiling of gadA, gadB, gadC and gadE by qRT-PCR revealed a significant upregulation of these genes in both SakairpoN : : kan and Sakai during exponential-phase growth (Fig. 2a–d). All of these genes, however, were significantly more upregulated in SakairpoN : : kan compared to Sakai (P<0.01) (Fig. 2). Upon entry into transition-phase growth, the expression of these four genes in both SakairpoN : : kan and Sakai did not differ significantly (Fig. 2).
Fig. 2.
Growth-phase analysis of gad genes, rpoS and hns expression by qRT-PCR. ○, Sakai; △, SakairpoN : : kan. Error bars represent sem (n=3), and the asterisks denote significance by Welch's t-test (P<0.05).
RpoS positively influences gad transcription during stationary-phase growth (Ma et al., 2003), and H-NS represses these genes during exponential-phase growth (Giangrossi et al., 2005; Tramonti et al., 2006). Since the differential expression of gad genes in SakairpoN : : kan was growth phase dependent, we wanted to determine if this was the result of altered rpoS or hns expression. Analysis by qRT-PCR, however, revealed no significant differences in the expression of rpoS or hns between SakairpoN : : kan and Sakai (Fig. 2e, f). Therefore, despite the fact that RpoS and H-NS control the expression of gad genes, and RpoS regulates many of the RpoN-regulated stress-resistance genes discussed above, its does not appear that altered rpoS or hns expression contributes to the differential expression of gad genes in SakairpoN : : kan.
Effects of rpoN inactivation on acid resistance
Since gad gene expression was upregulated in SakairpoN : : kan, we hypothesized that acid resistance would also be altered in SakairpoN : : kan. GDAR was tested in exponential- (OD600 0.5) and stationary- (OD600 3.5) phase DMEM-MOPS cultures of Sakai and SakairpoN : : kan. Following 2 h acid exposure in EG medium, plate counts for exponential-phase SakairpoN : : kan were substantially greater than for Sakai, at 9.5×103 c.f.u. ml−1 and 10 c.f.u. ml−1, respectively (n=3). Plate counts for stationary-phase SakairpoN : : kan and Sakai did not differ, at 5.7×105 c.f.u. ml−1 and 5.5×105 c.f.u. ml−1, respectively. Correspondingly, the survival rate (ΔV) of acid-challenged SakairpoN : : kan grown to exponential phase in DMEM-MOPS, LBG-MOPS and LB-MOPS was significantly improved, at ΔV=−1.49, −1.73 and −0.82 log c.f.u. h−1, when compared to Sakai, at ΔV=−4.48, −4.39 and −3.23 log c.f.u. h−1 (P<0.001). Since GDAR was altered in SakairpoN : : kan, we wanted to determine if rpoN inactivation in other genetically distinct strains of E. coli O157 : H7 also altered GDAR. As expected, exponential-phase DMEM-MOPS cultures of E. coli O157 : H7 strains 93-111 and TW14359 did not survive well when inoculated into EG medium, at ΔV=−4.45 and −4.06 log c.f.u. h−1, respectively. However, like SakairpoN : : kan, ΔV values for 93-111rpoN : : kan and TW14359rpoN : : kan were significantly improved, at −1.49 and −2.04 log c.f.u. h−1 (P≤0.034). Therefore, the inactivation of rpoN leads to increased acid resistance in E. coli O157 : H7 by the GDAR system and rpoN allows for increased susceptibility to acid during exponential growth. Interestingly, examination of GDAR using DMEM-MOPS culture inocula of Sakai and SakairpoN : : kan grown to OD600 0.5, 0.75 and 1.0, when gad gene expression was greatest (Fig. 2), did not show significantly improved survival rates of either strain (data not shown). Complementation of SakairpoN : : kan with a cloned version of the rpoN structural sequence to produce SakairpoN : : kan pCR2.1(rpoN+) reconstituted wild-type levels of acid susceptibility, at ΔV=−4.11 log c.f.u. h−1, in exponential cultures of the complemented strain grown in DMEM-MOPS.
Since both RpoS and GadE act as regulators of GDAR, and many RpoS-regulated stress-resistance genes are altered following rpoN inactivation, we wanted to determine if inactivation of these genes in the SakairpoN : : kan background would affect the expression of GDAR during exponential growth. Interestingly, GDAR in DMEM-MOPS, LBG-MOPS and LB-MOPS cultures was abolished in exponential-phase culture inocula of SakaiΔgadE rpoN : : kan and SakaiΔrpoS rpoN : : kan, where <10 c.f.u. ml−1 of both polygenic mutant backgrounds could be recovered following 0.5 h acid exposure in EG medium.
The effect of glutamate and different culture media on acid resistance in SakairpoN : : kan was also examined. Glutamate is required for acid resistance by the GDAR mechanism and, as predicted, removal of glutamate from EG medium negatively affected the survival of exponential DMEM-MOPS culture inoculums of SakairpoN : : kan, where <10 c.f.u. ml−1 were recovered following 0.5 h acid exposure. The survival of Sakai and SakairpoN : : kan grown in LBG-MOPS compared to DMEM-MOPS, when inoculated into EG medium (pH 2), did not differ significantly. Growth in LB-MOPS, which does not contain glucose, did, however, significantly improve survival rates in EG medium for both Sakai and SakairpoN : : kan (ΔV=−3.23 and −0.82 log c.f.u. h−1, respectively), compared to survival rates when grown in LBG-MOPS (ΔV=−4.39 and −1.73 log c.f.u. h−1, respectively, P<0.001) and DMEM-MOPS (ΔV=−4.48 and −1.49 log c.f.u. h−1 respectively, P<0.001).
The effect of rpoN inactivation on survival in a model stomach system was also determined. The model stomach is a complex in vitro system, containing multiple stressors such as bile salts, food matrix and acid, designed to emulate the complex in vivo stress conditions of the stomach (Bergholz & Whittam, 2007). We therefore examined cultures of SakairpoN : : kan and Sakai grown to exponential phase (OD600 0.5) in DMEM-MOPS, for survival in the model stomach system. Plate counts for SakairpoN : : kan were significantly higher after 1 h (P=0.004) and 2 h (P<0.001) in the model stomach, whereas initial counts did not differ. After 1 h in the model stomach system, plate counts averaged 1.2×102 c.f.u. ml−1 for Sakai, and 4.5×105 c.f.u. ml−1 for SakairpoN : : kan; and after 2 h, Sakai was reduced to 11 c.f.u. ml−1, whereas counts for SakairpoN : : kan averaged 3.8×105 c.f.u. ml−1. Together these results suggest that rpoN inactivation leads to increased GDAR and survival in a model stomach system.
DISCUSSION
There are 21 confirmed RpoN promoters in E. coli K-12 (Pallen, 1999; Reitzer & Schneider, 2001; Skibinski et al., 2002; Vogel et al., 2003), which specify the transcription of over 60 genes. This study identified 103 genes to be differentially expressed in the rpoN-inactivated E. coli O157 : H7 strain SakairpoN : : kan. The large majority of these are likely to be indirectly modulated through secondary regulators, as differential expression was observed in only 7/21 RpoN promoters, all seven of which specified the transcription of operons/regulators involved in the nitrogen regulatory (Ntr) response. This observation emphasizes the conditional nature of transcription from RpoN promoters, which are predisposed to idiosyncrasies in enhancer-binding protein (EBP) availability/activity. Interestingly, regulation by RpoN was growth phase dependent, with decreasing numbers of differentially expressed genes during transition into slower growth. As the abundance of RpoN in E. coli has been reported to be static (Jishage et al., 1996; Jishage & Ishihama, 1997), this may reflect variability in the cellular level of various EBPs, or in the availability of ATP, which energizes EBP activity.
The LEE, encoding type III secretion system structural genes and regulators, was positively regulated by RpoN. This finding is particularly salient in light of recent studies demonstrating a role for RpoN in the regulation of a P. aeruginosa type III secretion system (Matz et al., 2008). Type III secretion is essential to the A/E pathology of E. coli O157 : H7 and its expression is affected by numerous distally encoded factors, whose influence converges on the LEE-encoded activators Ler and GrlA (Spears et al., 2006). In this study, ler was not observed to be altered in SakairpoN : : kan; however, grlA was downregulated. Thus, RpoN may affect LEE expression through indirect stimulation of grlA, the product of which can activate LEE transcription independent of ler (Russell et al., 2007). RpoN may also regulate LEE through rpoS. We have already determined that rpoS is essential to increased GDAR in SakairpoN : : kan (see below), and RpoS is a known transcriptional regulator of the LEE. However, its control of LEE expression has been reported to be through increased transcription of ler, not grlA (Iyoda & Watanabe, 2005; Laaberki et al., 2006). RpoN could also regulate LEE through GadE. This protein has been reported to repress LEE expression independent of ler (Tatsuno et al., 2003), and gadE expression was upregulated in SakairpoN : : kan in this study.
Inactivation of rpoN was observed to affect the expression of stress resistance genes; most notably, gad genes of the GDAR system were shown to be upregulated in SakairpoN : : kan during exponential growth. Moreover, upregulation of gad genes in SakairpoN : : kan correlated with increased GDAR. This phenotype was validated in three distinct E. coli O157 : H7 strains representing major clonal lineages (Manning et al., 2008), suggesting that regulation of GDAR by RpoN is conserved among E. coli O157 : H7.
SakairpoN : : kan was dependent on the GDAR system for acid resistance. Both the exclusion of exogenous glutamate and deletion of the central regulator gadE in SakairpoN : : kan abrogated GDAR. gadE and gadX were shown to be repressed by RpoN in microarray analysis; however, there are no RpoN-binding sites in the core promoters of these genes. RpoN may therefore alter gad gene expression through a secondary trans-/cis-acting factor(s). H-NS and RpoS are two important regulators of the GDAR system. H-NS represses gad transcriptionally through gadA and gadX (Giangrossi et al., 2005; Tramonti et al., 2006). Since gad transcription increases substantially in an hns-inactivated background (De Biase et al., 1999), we initially hypothesized that RpoN may act indirectly to repress gad expression through hns activation. However, upregulation of gad genes in SakairpoN : : kan was not associated with a decrease in hns transcript levels, and exponential cultures of an hns-inactivated strain of Sakai could not be recovered after 0.5 h of acid challenge (data not shown). Alternatively, RpoS activates gad transcription through the GadX/W circuit (Ma et al., 2003). The results of this study reveal that the inactivation of rpoS in SakairpoN : : kan abolishes exponential-phase GDAR, despite there being no difference in rpoS expression between SakairpoN : : kan and Sakai (Fig. 2e). One hypothesis to explain this outcome is that reduced growth rates in rpoN-inactivated backgrounds (Supplementary Fig. S1) stimulate RpoS accumulation, which then leads to increased gad transcription. This argument is not well supported, however, as growth-rate attrition has only been shown to induce rpoS expression under conditions that increase generation times (g) to >140 min, or when cultures are grown under conditions of starvation (Gentry et al., 1993; Lange & Hengge-Aronis, 1991). In this study, GDAR was observed in exponentially growing cultures of SakairpoN : : kan (g=54 min) grown in nutrient excess. A more plausible hypothesis is that RpoN is controlling rpoS expression post-transcriptionally through an auxiliary regulator(s), of which at least ten have been described for rpoS (Hengge-Aronis, 2000). As RpoS is known to regulate several mechanisms of acid resistance in E. coli (Foster, 2004), interplay between RpoN and RpoS could have a broad impact on the acid resistance phenotype and on stress resistance in general. Interestingly, an RpoN-RpoS regulatory pathway has been recently described in the enteric pathogen S. enterica, and in the aetiological agent of Lyme disease, Borrelia burgdorferi. In S. enterica, RpoN-RpoS have been determined to co-regulate O antigen LPS production through rfaH (Bittner et al., 2002, 2004), and in B. burgdorferi, RpoN-RpoS regulation has been shown to be essential to transmission and pathogenesis through control of membrane lipoproteins OspC and DspA (Boardman et al., 2008; Hubner et al., 2001; Yang et al., 2005). Collectively, these findings testify to the importance of RpoN-RpoS-directed regulation in bacterial stress resistance and pathogenesis.
Conclusions
In this study, rpoN inactivation in E. coli O157 : H7 resulted in the differential expression of 103 genes by microarray analysis, including acid resistance and LEE genes, which are essential to transmission and pathogenesis. GDAR genes were upregulated in SakairpoN : : kan during exponential growth, which correlated strongly with increased acid resistance by the GDAR mechanism, and in a model stomach system. Acid resistance by the GDAR mechanism in SakairpoN : : kan during exponential growth required intact rpoS and gadE genes. Genes encoded throughout the LEE were downregulated in SakairpoN : : kan during exponential growth. This included the LEE regulator grlA. Together these results suggest that RpoN is an important growth-phase-dependent regulator of acid resistance and LEE expression. Future research will explore further the mechanism by which RpoN directs GDAR expression, its regulatory dependence on rpoS, and the full contribution of rpoN to stress resistance and virulence in E. coli O157 : H7.
Acknowledgments
This work was supported by grants from the US Department of Agriculture (USDA) Food Safety NRI (2005-35201-16362), the National Institutes of Health (NIH) and the National Institutes of Allergies and Infectious Diseases (NIAID) to T. S. W. through the Food and Waterborne Diseases Integrated Research Network (N01-AI-30058). The authors would like to dedicate this manuscript to the memory of Thomas S. Whittam.
Abbreviations
A/E, attaching and effacing
GDAR, glutamate-dependent acid resistance
GSEA, gene set enrichment analysis
LEE, locus of enterocyte effacement
SAM, significance analysis of microarrays
Footnotes
Normalized microarray intensity data files have been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE17467.
Two supplementary tables and a supplementary figure are available with the online version of this paper.
References
- Bergholz, T. M. & Whittam, T. S. (2007). Variation in acid resistance among enterohaemorrhagic Escherichia coli in a simulated gastric environment. J Appl Microbiol 102, 352–362. [DOI] [PubMed] [Google Scholar]
- Bergholz, T. M., Tarr, C. L., Christensen, L. M., Betting, D. J. & Whittam, T. S. (2007a). Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. Mol Biol Evol 24, 2323–2333. [DOI] [PubMed] [Google Scholar]
- Bergholz, T. M., Wick, L. M., Qi, W., Riordan, J. T., Ouellette, L. M. & Whittam, T. S. (2007b). Global transcriptional response of Escherichia coli O157 : H7 to growth transitions in glucose minimal medium. BMC Microbiol 7, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, R. E., Leskiw, B. K., Hodges, R. S., Kay, C. M. & Weiner, J. H. (1998). The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol 280, 583–596. [DOI] [PubMed] [Google Scholar]
- Bittner, M., Saldias, S., Estevez, C., Zaldivar, M., Marolda, C. L., Valvano, M. A. & Contreras, I. (2002). O-antigen expression in Salmonella enterica serovar Typhi is regulated by nitrogen availability through RpoN-mediated transcriptional control of the rfaH gene. Microbiology 148, 3789–3799. [DOI] [PubMed] [Google Scholar]
- Bittner, M., Saldias, S., Altamirano, F., Valvano, M. A. & Contreras, I. (2004). RpoS and RpoN are involved in the growth-dependent regulation of rfaH transcription and O antigen expression in Salmonella enterica serovar Typhi. Microb Pathog 36, 19–24. [DOI] [PubMed] [Google Scholar]
- Boardman, B. K., He, M., Ouyang, Z., Xu, H., Pang, X. & Yang, X. F. (2008). Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun 76, 3844–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannon, D. E., Connell, N., Keener, J., Tormo, A., Espinosa-Urgel, M., Zambrano, M. M. & Kolter, R. (1991). Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J Bacteriol 173, 4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales, R. D., Luo, Y., Willey, J. C., Austermiller, B., Barbacioru, C. C., Boysen, C., Hunkapiller, K., Jensen, R. V., Knight, C. R. & other authors (2006). Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 24, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet, M. P., Penfound, T. A., Smith, D., Elliott, J. F. & Foster, J. W. (1999). Control of acid resistance in Escherichia coli. J Bacteriol 181, 3525–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chart, H. (2000). VTEC enteropathogenicity. Symp Ser Soc Appl Microbiol 29, 12S–23S. [DOI] [PubMed] [Google Scholar]
- Crawford, J. A., Blank, T. E. & Kaper, J. B. (2002). The LEE-encoded type III secretion system in EPEC and EHEC: assembly, function and regulation. In Escherichia coli, Virulence Mechanisms of a Versatile Pathogen, pp. 337–359. Edited by M. S. Donnenberg. San Diego, CA: Academic Press.
- Dalet, K., Briand, C., Cenatiempo, Y. & Hechard, Y. (2000). The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr Microbiol 41, 441–443. [DOI] [PubMed] [Google Scholar]
- Darwin, A. J. (2005). The phage-shock-protein response. Mol Microbiol 57, 621–628. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase, D., Tramonti, A., Bossa, F. & Visca, P. (1999). The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32, 1198–1211. [DOI] [PubMed] [Google Scholar]
- Dong, T., Kirchhof, M. G. & Schellhorn, H. E. (2007). RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Molecular Genetics and Genomics. Berlin: Springer [DOI] [PubMed]
- Donnenberg, M. S. & Whittam, T. S. (2001). Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest 107, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2, 898–907. [DOI] [PubMed] [Google Scholar]
- Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B. & Cashel, M. (1993). Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J Bacteriol 175, 7982–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, H. M., Styrvold, O. B., Kaasen, I. & Strom, A. R. (1988). Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol 170, 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrossi, M., Zattoni, S., Tramonti, A., De Biase, D. & Falconi, M. (2005). Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J Biol Chem 280, 21498–21505. [DOI] [PubMed] [Google Scholar]
- Gorden, J. & Small, P. L. (1993). Acid resistance in enteric bacteria. Infect Immun 61, 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis, R. (2000). The general stress response in Escherichia coli. In Bacterial Stress Responses, pp. 161–178. Edited by G. Storz & R. Hengge-Aronis. Washington, DC: American Society for Microbiology.
- Hengge-Aronis, R. & Fischer, D. (1992). Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol 6, 1877–1886. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis, R., Klein, W., Lange, R., Rimmele, M. & Boos, W. (1991). Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol 173, 7918–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner, A., Yang, X., Nolen, D. M., Popova, T. G., Cabello, F. C. & Norgard, M. V. (2001). Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda, S. & Watanabe, H. (2005). ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J Bacteriol 187, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage, M. & Ishihama, A. (1997). Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J Bacteriol 179, 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage, M., Iwata, A., Ueda, S. & Ishihama, A. (1996). Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol 178, 5447–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic, M., Lilic, M., Janjusevic, R., Jovanovic, G. & Savic, D. J. (1999). tRNA synthetase mutants of Escherichia coli K-12 are resistant to the gyrase inhibitor novobiocin. J Bacteriol 181, 2979–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailasan Vanaja, S., Bergholz, T. M. & Whittam, T. S. (2009). Characterization of the Escherichia coli O157 : H7 Sakai GadE regulon. J Bacteriol 191, 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, T., Takahashi, K. & Inoue, H. (2006). Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J Biochem 140, 703–711. [DOI] [PubMed] [Google Scholar]
- Laaberki, M. H., Janabi, N., Oswald, E. & Repoila, F. (2006). Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157 : H7: interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol 296, 197–210. [DOI] [PubMed] [Google Scholar]
- Lange, R. & Hengge-Aronis, R. (1991). Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol 5, 49–59. [DOI] [PubMed] [Google Scholar]
- Large, T. M., Walk, S. T. & Whittam, T. S. (2005). Variation in acid resistance among shiga toxin-producing clones of pathogenic Escherichia coli. Appl Environ Microbiol 71, 2493–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Livak, K. J. & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative
PCR and the
 method. Methods
25, 402–408. [DOI] [PubMed] [Google Scholar]
method. Methods
25, 402–408. [DOI] [PubMed] [Google Scholar] - Ma, Z., Richard, H., Tucker, D. L., Conway, T. & Foster, J. W. (2002). Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J Bacteriol 184, 7001–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z., Richard, H. & Foster, J. W. (2003). pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J Bacteriol 185, 6852–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, S. D., Motiwala, A. S., Springman, A. C., Qi, W., Lacher, D. W., Ouellette, L. M., Mladonicky, J. M., Somsel, P., Rudrik, J. T. & other authors (2008). Variation in virulence among clades of Escherichia coli O157 : H7 associated with disease outbreaks. Proc Natl Acad Sci U S A 105, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, N. & Church, G. M. (2003). Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol 48, 699–712. [DOI] [PubMed] [Google Scholar]
- Matz, C., Moreno, A. M., Alhede, M., Manefield, M., Hauser, A. R., Givskov, M. & Kjelleberg, S. (2008). Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J 2, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead, P. S. & Griffin, P. M. (1998). Escherichia coli O157 : H7. Lancet 352, 1207–1212. [DOI] [PubMed] [Google Scholar]
- Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., Griffin, P. M. & Tauxe, R. V. (1999). Food-related illness and death in the United States. Emerg Infect Dis 5, 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michino, H., Araki, K., Minami, S., Takaya, S., Sakai, N., Miyazaki, M., Ono, A. & Yanagawa, H. (1999). Massive outbreak of Escherichia coli O157 : H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol 150, 787–796. [DOI] [PubMed] [Google Scholar]
- Model, P., Jovanovic, G. & Dworkin, J. (1997). The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol 24, 255–261. [DOI] [PubMed] [Google Scholar]
- Murphy, K. C. & Campellone, K. G. (2003). Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, Y., Okada, N., Makino, S., Asakura, H., Yamamoto, S. & Igimi, S. (2006). The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett 263, 54–60. [DOI] [PubMed] [Google Scholar]
- Pallen, M. (1999). RpoN-dependent transcription of rpoH? Mol Microbiol 31, 393. [DOI] [PubMed] [Google Scholar]
- Perna, N. T., Mayhew, G. F., Posfai, G., Elliott, S., Donnenberg, M. S., Kaper, J. B. & Blattner, F. R. (1998). Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157 : H7. Infect Immun 66, 3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer, L. & Schneider, B. L. (2001). Metabolic context and possible physiological themes of sigma54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65, 422–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer, L. J., Bueno, R., Cheng, W. D., Abrams, S. A., Rothstein, D. M., Hunt, T. P., Tyler, B. & Magasanik, B. (1987). Mutations that create new promoters suppress the sigma54 dependence of glnA transcription in Escherichia coli. J Bacteriol 169, 4279–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichon, D., Gouin, E., Debarbouille, M., Cossart, P., Cenatiempo, Y. & Hechard, Y. (1997). The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J Bacteriol 179, 7591–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R. M., Sharp, F. C., Rasko, D. A. & Sperandio, V. (2007). QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J Bacteriol 189, 5387–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski, D. A., Golby, P., Chang, Y. S., Sargent, F., Hoffman, R., Harper, R., Guest, J. R., Attwood, M. M., Berks, B. C. & Andrews, S. C. (2002). Regulation of the hydrogenase-4 operon of Escherichia coli by the sigma54-dependent transcriptional activators FhlA and HyfR. J Bacteriol 184, 6642–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears, K. J., Roe, A. J. & Gally, D. L. (2006). A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol Lett 255, 187–202. [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R. & other authors (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuno, I., Nagano, K., Taguchi, K., Rong, L., Mori, H. & Sasakawa, C. (2003). Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157 : H7. Infect Immun 71, 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis, P., Takumi, K. & Shinagawa, K. (2004). Dose response for infection by Escherichia coli O157 : H7 from outbreak data. Risk Anal 24, 401–407. [DOI] [PubMed] [Google Scholar]
- Tramonti, A., Visca, P., De Canio, M., Falconi, M. & De Biase, D. (2002). Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184, 2603–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti, A., De Canio, M., Delany, I., Scarlato, V. & De Biase, D. (2006). Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J Bacteriol 188, 8118–8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, D. L., Tucker, N. & Conway, T. (2002). Gene expression profiling of the pH response in Escherichia coli. J Bacteriol 184, 6551–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., Tibshirani, R. & Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Axmann, I. M., Herzel, H. & Hess, W. R. (2003). Experimental and computational analysis of transcriptional start sites in the cyanobacterium Prochlorococcus MED4. Nucleic Acids Res 31, 2890–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadington, M. C., Ladner, J. E., Stourman, N. V., Harp, J. M. & Armstrong, R. N. (2009). Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochemistry 48, 6559–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H., Polen, T., Heuveling, J., Wendisch, V. F. & Hengge, R. (2005). Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, L., Brissette, J. L. & Model, P. (1991). Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma54 and modulated by positive and negative feedback mechanisms. Genes Dev 5, 1912–1923. [DOI] [PubMed] [Google Scholar]
- Yang, X. F., Lybecker, M. C., Pal, U., Alani, S. M., Blevins, J., Revel, A. T., Samuels, D. S. & Norgard, M. V. (2005). Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol 187, 4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]