Abstract
Background & Aims
We followed persons with ongoing hepatitis C virus (HCV) exposure following control of an initial HCV infection to determine whether primary control conferred protection against future persistent infections.
Methods
Twenty-two active injection drug users (IDU) who had cleared a primary hepatitis C viremia for at least 60 days were monitored monthly. Reinfection was defined as the detection of a new hepatitis C virus infection. Protection was assessed based on the magnitude and duration of viremia following reinfection and generation of T-cell and neutralizing antibody (nAb) responses
Results
Reinfection occurred in 11 IDUs (50%) who previously spontaneously controlled primary HCV infection. Although viral clearance occurs in approximately 25% of patients with primary infections, spontaneous viral clearance was observed in 83% of reinfected patients. The duration and maximum level of viremia during subsequent episodes of reinfection were significantly decreased, compared with those of the primary infection in the same subjects. In contrast to chronic infection, reinfection was associated with a significant increase in the breadth of T-cell responses. During acute infection, nAbs against heterologous viral pseudoparticles were detected in 60% of reinfected subjects; cross-reactive nAbs are rarely detected in patients who progress to chronic infection.
Conclusions
HCV reinfection is associated with a reduction in the magnitude and duration of viremia (compared with the initial infection), broadened cellular immune responses, and the generation of cross-reactive humoral responses. These findings are consistent with the development of adaptive immunity that is not sterilizing but protects against chronic disease.
Intravenous drug use is the most significant risk factor for infection with hepatitis C virus (HCV) in the United States, as most new infections occur in intravenous drug users (IDU)1 and most IDUs are infected with HCV2. The incidence rate of HCV infection among IDUs in metropolitan areas of the United States, Europe, and Australia has been shown to be 10–30% per year3–5 and a recent meta-analysis by Hagan et al revealed that by the tenth year of drug use 75–80% of IDUs have been infected with HCV, at least once6. At the same time, spontaneous HCV clearance typically occurs in less than 30% of infected IDUs7.
The occurrence and outcome of HCV reinfection has implications regarding vaccine development. Currently, no HCV vaccine has been licensed despite the public health need. However, work in the chimpanzee model suggests that immunity against HCV can be generated by initial infection8–10 and vaccination11. Clearance of multiple infections with homologous and heterologous virus has been observed in chimpanzees9, 10, 12. Moreover, Lanford, et al. reported that clearance of both homologous and heterologous viral rechallenges was associated with decreased duration and magnitude of viremia12. However, the existence of protective immunity in humans remains controversial.
Similar evidence for protective immunity has been reported in two studies in humans. Mehti, et al13 and Grebely, et al14 demonstrated a decreased risk of the development of viremia in previously infected IDUs compared to naïve IDUs even after accounting for risk behavior. Conversely, a, recent study by Aitken, et al15 reported a higher rate of infection in previously cleared young IDUs compared to naïve young IDUs. There are major technical limitations in these human studies that would cause protective immunity to be underestimated. Instances of reinfection with rapid clearance of viremia would often be missed by infrequent sampling and viral sequencing is necessary to distinguish new infection from recrudescence of preexisting viremia. However, persons who recovered once might recover more often a second time even without generation of protective immunity due to host factors. More rapid and effective control of subsequent infections than the first infection would suggest a role for adaptive protective immunity rather than fixed genetic factors. Thus, it is also important to assess the magnitude and duration of viremia in the same person with sequential infections.
In this study, we have longitudinally followed young IDUs with ongoing exposure following control of their initial infection to assess the kinetics and immunological parameters of HCV reinfection. Using a strict definition of reinfection as viremia with heterologous virus with at least 60 days of aviremia following control of the primary infection, we were able to document a new infection in half of individuals who had previously cleared infection. Previous clearance of a primary HCV infection was associated with high reinfection clearance rates, and, compared to initial infection, with a decreased duration and magnitude of viremia during subsequent infections. Reinfection with a heterologous virus also resulted in the generation of new T cell responses not seen with the initial infection and cross-reactive anti-HCV neutralizing antibodies. Taken together, these data demonstrate that previous clearance of HCV alters the outcome and kinetics of secondary infections, providing further evidence that generation of protective immunity against HCV is possible.
Materials and Methods
Participants
In a prospective study of young IDUs in Baltimore, MD, the incidence, immunology, and virology of HCV infection are examined. Participants are invited to enroll in a prospective study of acute hepatitis C if they are at risk for HCV infection because of drug use but are anti-HCV antibody and HCV RNA negative. Those who consented are provided counseling to reduce the risks of drug use and monthly blood samples were collected, as previously described1. All participants with acute HCV infection were referred for evaluation for possible treatment. The study protocol was approved by the institutional review board of the Johns Hopkins School of Medicine.
We identified people who developed antibodies to HCV between 1997 and 2007 with sufficient follow-up to evaluate outcome of infection. HCV RNA testing was performed on serum or plasma separated from blood using a quantitative HCV RNA assay, described below. HCV RNA testing was performed on samples collected before seroconversion until a negative result was obtained to determine the time of initial viremia and after seroconversion to evaluate the outcome of infection. PBMC (~108 – 1010) were collected from infected persons who were eligible for and consented to unit blood donation or apheresis.
HCV RNA Assays
Determination of viral load
Total RNA was extracted from serum using a Qiagen MinElute Virus column (Qiagen, Valencia, CA) according to the manufacturer’s instructions. To determine the concentration of HCV RNA in blood samples, we used a quantitative RT-PCR assay (TaqMan® HCV analyte-specific reagent, Roche Molecular Diagnostics, USA) with the generation of DNA amplification products monitored on a Cobas TaqMan® Analyzer (Roche Molecular Diagnostics, USA). This assay has a lower limit of detection of 50 IU/ml.
Phylogenetic analysis
HCV Core-E1 sequences were obtained from the first and last viremic specimen of the initial period of viremia and from the first and last viremic specimen of any subsequent period of viremia. E2 sequences were obtained from viremic specimens within the first 100 days of an initial infection, following the detection of new genotype during initial infection, and during reinfections. Total RNA was extracted from serum using a QIAamp viral RNA mini column (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Direct sequencing of RT-PCR products from the Core-E1 region was performed as previously described16. Sequences were aligned using ClustalX17 and trimmed to equal length using BioEdit. The E2 region was reverse transcribed using genotype-specific primers and amplified using a nested PCR strategy with genotype-specific primers (Supplementary Table 1). Purified E2 PCR products were cloned into the entry vector, pDONR221, using Gateway technology (Invitrogen, USA). Sequences were assembled into contigs using Aligner (CodonCode Corporation, Dedham, MA). Genetic divergence between Core-E1 sequences and E2 protein divergence from H77 was determined using the DNADist DNA matrix or ProtDist Phylip programs, resepectively, included in the BioEdit software package. Viral sequences were identified as unique when the Core-E1 divergence between two sequences was ≥ 0.05. Genotypes were determined by comparing Core-E1 sequences to HCV genotype reference sequences using the Los Alamos National Laboratories HCV Phylogenetic Placement Service with pairwise distance analysis18. All E2 sequences have been deposited into Genbank/EMBL/DDBJ with accession numbers (will deposit upon acceptance).
Viral recovery and reinfection
HCV controllers were defined as individuals with anti-HCV antibodies and HCV RNA undetectable by the COBAS Taqman RT-PCR quantitative assay for a period of at least 60 days. Chronic infection was defined as continuous viremia or recurrent viremia with the same virus as determined by Core-E1 phylogenetic analysis. Reinfection was defined as the presence of new viremia, defined as a genetically unique hepatitis C virus by Core-E1 phylogenetic analysis, in HCV controllers. The outcome of reinfection was analyzed in subjects who had follow-up for at least 120 days post-reinfection. Control of reinfection was defined as undetectable HCV RNA for at least 60 days. Persistent reinfection was defined as continuous viremia or recurrent viremia with the same virus following the detection of reinfection. The midpoint between sample collection dates was used to determine duration. Subjects were excluded from analysis of HCV reinfection if they met any of the following criteria: 1) had gaps in follow-up greater than 365 days 2) had detectable HBsAg at time of enrollment or 3) could not successfully undergo Core-E1 sequencing of the initial HCV infection.
Alanine aminotransferase (ALT) assay
The ALT level in of plasma samples was determined by the Johns Hopkins Hospital clinical laboratory.
IFN-γ ELISpot assay
HCV-specific CD8+ T-cell responses were quantified by interferon-gamma (IFN-γ) ELISpot assay as previously described7. Briefly, PBMC were screened for recognition of HCV-specific antigens using overlapping peptides and previously described optimal HCV epitopes. Pooled cytomegalovirus, Epstein-Barr virus, and influenza antigens and phytohemmaglutinin (PHA) were used as positive controls. A new T cell response was defined as recognition of a peptide by PBMCs collected greater than 200 days following the initial detection of viremia that was not recognized by PBMCs collected at earlier time points.
H77 pseudoparticle production and neutralization assays
HIV-H77 (HCV genotype 1a) and HIV-murine leukemia virus (MLV) pseudoparticles (pp) containing the luciferase reporter gene were produced as described elsewhere19, 20. Neutralization assays were performed as previously described21. Pseudoparticle infection, indicated by luciferase activity, was measured in terms of relative light units (RLUs) in the presence of subject serum (RLUtest) versus average infection in the presence of two HCV-negative serum specimens (RLUcontrol). Percent neutralization was calculated as 100 × [1−(RLUtest/RLUcontrol)]. Results are reported as 50% inhibitory dilution (ID50) values. As a control, all subject serum neutralized MLVpp < 50%.
Statistical analysis
Wilcoxon rank-sum test, paired t-test, Kruskal-Wallis One Way ANOVA by Ranks followed by Dunn’s multiple comparisons, and Fisher Exact test were used to evaluate statistically significant differences between groups. Differences were considered statistically significant when P < 0.05.
Results
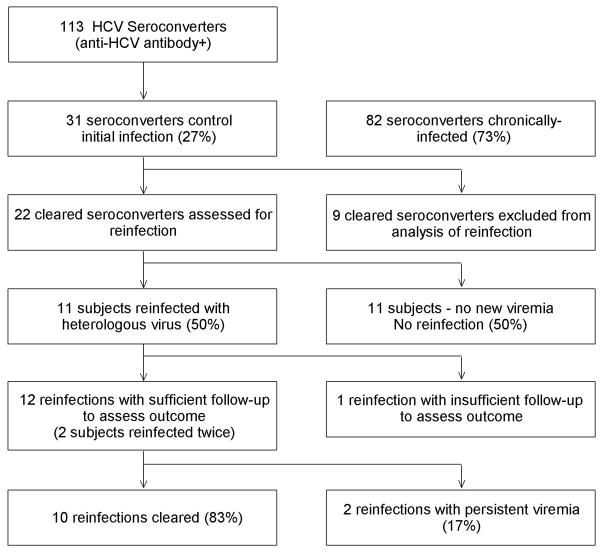
From 1997 to 2007, we detected anti-HCV antibody seroconversion in 113 individuals (Figure 1). No period of aviremia greater than 60 days was observed in 82 of these seroconverters, indicative of chronic infection. Of the 31 persons who controlled viremia, nine were excluded from subsequent analysis for reinfection, permitting analysis of reinfection in 22 subjects.
Figure 1.
Flow of participants in the study. The ratio of cleared to persistent subjects during reinfection was significantly greater than during primary infection (P=0.001).
Analysis of reinfection
Reinfection with a heterologous virus was documented in 11 cleared subjects while no new viremia was detected in 11 subjects. Overall, the incident reinfection rate was 30.1 reinfections per 100 person-years. The median interval between HCV testing, total follow-up from seroconversion, follow-up post-clearance, subject age at the time of seroconversion, and subject gender composition did not differ between the Reinfection and No reinfection groups, respectively (Table 1). All subjects were Caucasian.
Table 1.
Follow-up data and demographics of Control - Reinfection and Control - No reinfection subjects.
| Control - Reinfection | Control – No reinfection | P | |
|---|---|---|---|
| Subjects | 11 | 11 | |
| Follow-up from serovconversion1 | 1269 (981–1856) | 984 (741–1791) | 0.39 |
| Follow-up post-clearance1 | 702 (505–1397) | 815 (344–966) | 0.47 |
| Testing interval1 | 32 (31–33) | 33 (32–35) | 0.19 |
| % female | 55 | 55 | 1.00 |
| Age2 | 25 (24–27) | 25 (24–26) | 0.62 |
median (IQR), days
median (IQR), yr at time of seroconversion
Phylogenetic analysis of viral sequences during initial infection and subsequent infections was performed to confirm reinfection. Seven subjects were reinfected with genotype 1a, two with genotype 1b and two with genotype 3a (Table 2). Reinfection with a new virus of the same subtype occurred in six subjects, while reinfection with a new virus of a different subtype or genotype occurred in three and five subjects, respectively.
Table 2.
Phylogenetic analysis of viral sequences and plasma ALT levels during primary infection and reinfection within the same subject.
| Subject ID | ALT1 | Primary Infection | Reinfection A | Reinfection B | Divergence3 | Outcome4 | |||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | ALT2 | Genotype | ALT2 | Genotype | ALT2 | ||||
| 18 | 13 | 1a | 74 | 1a | 55 | 0.109 | P | ||
| 19 | 16 | 1a | 411 | 1a | 577 | 0.071 | P | ||
| 27 | 30 | 3a | 559 | 1a | 553 | 0.562 | C | ||
| 48 | 16 | 3a/1a | 258 | 1a | 63 | 3a | 29 | 0.592, 0.595 | C, C |
| 57 | 10 | 1a | 211 | 1a | 20 | 0.103 | C | ||
| 133 | 17 | 1a | 351 | 1a | 42 | 0.116 | C | ||
| 152 | 17 | 1a | 535 | 1b | 347 | 1a | N/A5 | 0.375, 0.383 | C, C |
| 170 | 6 | 1a | 38 | 3a | 61 | 0.628 | N/A | ||
| 172 | 16 | 1b | 387 | 1b | 64 | 0.163 | C | ||
| 176 | 22 | 1a | 785 | 3a | 62 | 0.625 | C | ||
| 180 | 18 | 3a/1a | 475 | 1a | 84 | 0.102 | C | ||
baseline ALT value prior to infection; all subjects returned to baseline prior to reinfection.
maximum ALT value during infection.
First value represents divergence between primary infection and reinfection A. Second value represents divergence between reinfection A and reinfection B.
C, cleared; P, persistent; N/A, excluded from analysis of outcome. The first and second letters indicate the outcome of the first and second reinfections, respectively.
N/A, subject never returned to baseline following reinfection A.
To examine the frequency of virus replacement in chronically-infected subjects, we performed phylogenetic analysis of viral sequences obtained during various time points of chronic infection. This analysis revealed the appearance of a genetically-distinct virus in 19 of 82 chronically-infected subjects. Ten of these unique viruses were a different genotype while 9 were the same genotype as the initial infecting virus (data not shown).
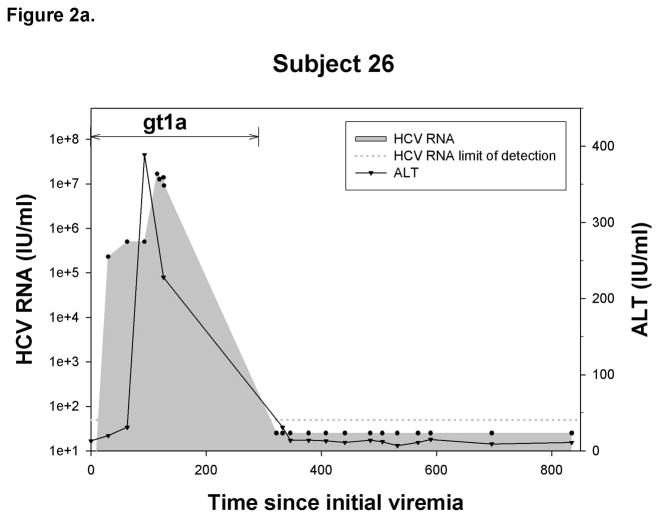
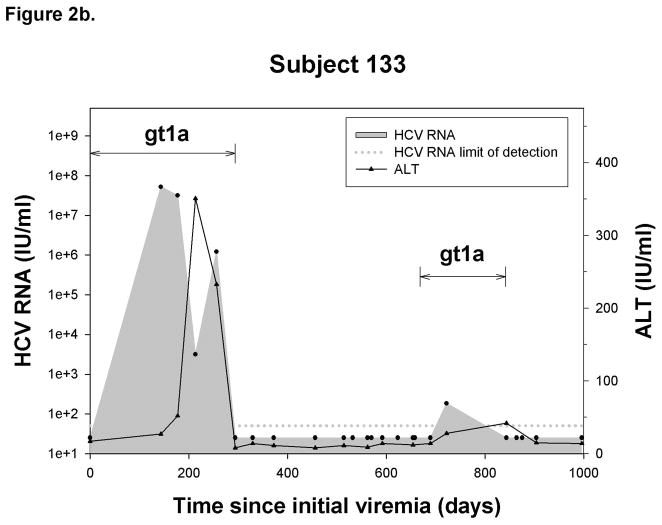
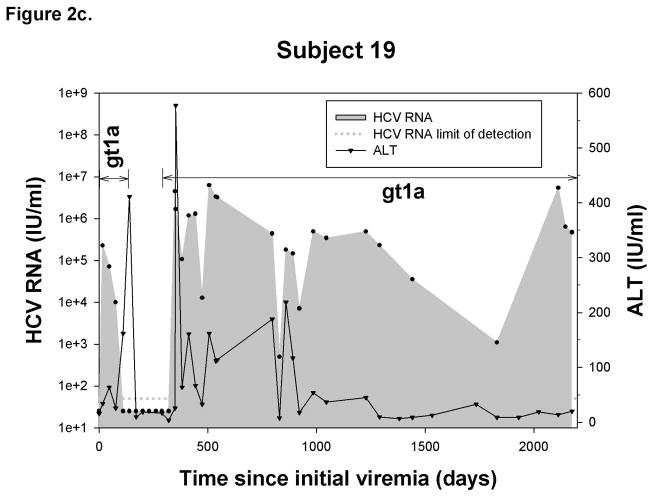
Outcome of reinfection
The eleven reinfected subjects were monitored to assess the outcome of reinfection. One reinfected subject was excluded from analysis due to inadequate follow-up after initial detection of reinfection to determine outcome, while two subjects cleared two reinfections each. Therefore, we were able to analyze outcomes of a total of 12 reinfections in 10 subjects (Figure 1). Representative graphs are shown in Figure 2, illustrating HCV RNA and ALT levels throughout the history of infection. Various outcomes following control of initial infection are shown; no reinfection (2a), cleared reinfection (2b), and persistent reinfection (2c). Additional graphs are included in Supplementary Figure 1. Interestingly, increases in blood ALT levels of at least 2.5-fold over baseline were observed during 10 of 12 reinfections (Table 2). In total, 10 reinfections were controlled and two reinfections resulted in persistent viremia (follow-up during persistent reinfection: 1435 and 2268 days). The ratio of clearance to persistence in reinfection was nearly reversed and significantly different from that of primary infection (P = 0.001). The median total follow-up time following the first detection of viremia and median testing interval in subjects with a primary infection and subjects with a reinfection were not different (P = 0.217 and P = 0.663, respectively; data not shown). These data suggest that spontaneous clearance of a reinfection is significantly more likely than spontaneous clearance of a primary infection.
Figure 2.
Representative graphs demonstrating the history of viremia and plasma ALT levels in A) No reinfection, B) Reinfection-cleared and C) Reinfection-persistent groups. HCV genotype of primary infection and reinfection is indicated and the time over which a unique virus is detected is denoted by a horizontal black line. Black circles represent HCV RNA concentrations (IU/ml) detected in serum or plasma samples obtained at given time points from date of first detection of viremia. Dotted-line denotes the HCV RNA limit of detection. Samples below the HCV RNA limit of detection were assigned a value of 25 IU/ml. Black triangles denote ALT activity (IU/ml) detected in plasma samples obtained at given time points from date of first detection of viremia.
Infection kinetics during primary infection and reinfection
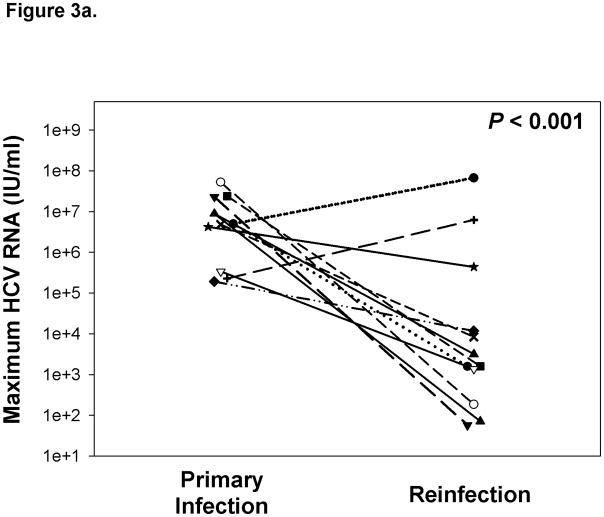
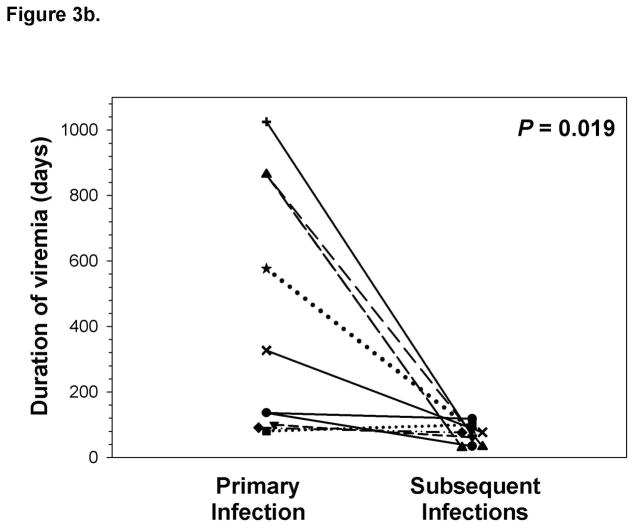
Frequent monitoring of HCV infection status allowed for assessment of the kinetics of viremia during initial and subsequent infections within the same subjects, as has previously been done with experimental infection of chimpanzees. The maximum concentration of HCV RNA detected in blood during reinfection was approximately three logs lower compared to initial infection even when persistent reinfections were included in the analysis (Figure 3a). Similarly, the duration of viremia during primary infection was nearly four-fold longer than in subsequent infections in subjects who cleared reinfections (Figure 3b). These results suggest that immunologic events associated with clearance of an initial infection alter the kinetics of subsequent infections.
Figure 3.
Clearance of a primary infection attenuates the infection kinetics of subsequent infections. A) Maximum HCV RNA concentrations (IU/ml) detected in serum samples obtained during primary and subsequent infections in subjects with sufficient follow-up following the detection of a reinfection. Triangles represent maximum viremia detected in Reinfection – persistent subjects. The maximum viremia in each subject during initial infection and reinfection is connected by a line. Median maximum HCV RNA concentration of reinfections was significantly lower than primary infections (P < 0.001). B) Duration of viremia (days) during primary infections and subsequent infections in Reinfection – cleared subjects. The duration of initial and reinfection viremia in each subject is connected by a line. The duration of viremia during reinfection was significantly lower than in primary infection (P = 0.019).
Cellular immune responses following exposure to a new virus
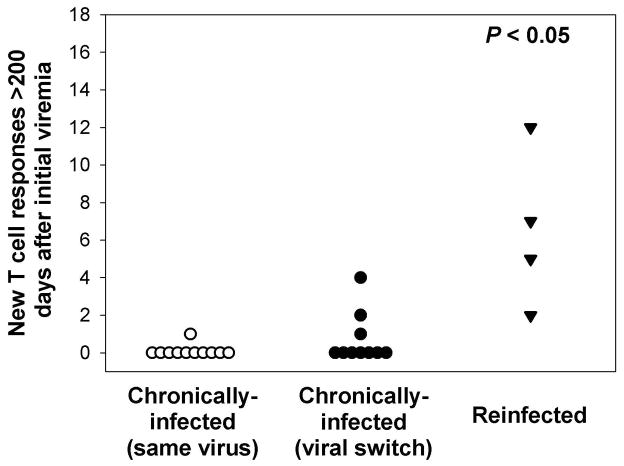
The loss of breadth and specificity of T cells generated in the acute phase of HCV infection with a resultant paucity of cellular immune responses is one of the hallmarks of chronic HCV infection7, 22, 23. Because reinfection is associated with altered infection kinetics, we investigated whether reinfection altered cellular immune responses to HCV, and specifically whether reinfection was associated with the generation of new T cell responses. Exposure to a genetically distinct virus following a period of aviremia was associated with acquisition of a significantly greater number of new T cell responses than in persistent viremia (Figure 4) whether chronic infection was with the same virus or genetically distinct viruses (viral switch). In reinfected subjects, exposure to a new virus elicited new T cell responses in all four subjects from whom sufficient PBMC were obtained to evaluate cellular immune responses. Three out of four subjects with new responses were infected with only a genotype 1a virus during both initial and subsequent infections. These new T cell responses were directed against epitopes evenly distributed across the HCV polyprotein (Supplementary Figure 3a and Supplementary Table 2). There was no significant correlation between the number of new T cell responses and decreases in maximum HCV RNA concentrations during reinfection (R2 = 0.36; P = 0.40; Supplementary Figure 3b). While the number of new T cell responses appeared to be negatively correlated with increased ALT during reinfection (R2 = 0.89), this correlation was not statistical significant due to small sample size (P = 0.087; Supplementary Figure 3c). In contrast, evolution of the same viral sequence in subjects with chronic infection resulted in new cellular responses in only one of 11 subjects tested and new cellular responses were detected in only three out of 10 subjects in whom multiple dominant viral sequences were detected during initial infection. The number of new T cell responses in these two groups was significantly less than in reinfected subjects. These results suggest that exposure to a new virus is not sufficient to elicit new cellular immune responses and that previous complete control of viremia is associated with enhanced generation of new cellular immune responses in response to a new HCV infection. However, one reinfected subject with new T cell responses developed a persistent reinfection, suggesting that the development of new T cell responses does not provide absolute protection against persistence. This led us to investigate the humoral response to reinfection.
Figure 4.
Significantly greater numbers of new T cell responses are detected during reinfection than during chronic infection. Values represent new T cell responses in chronically-infected subjects in whom the same virus is detected during chronic infection, chronically-infected subjects in whom more than one genetically-distinct virus is detected during chronic infection, and reinfected subjects. A new T cell response was defined as a peptide that was recognized, in an IFN-γ ELISpot, by PBMCs obtained greater than 200 days following the first detection of viremia but not by PBMCs collected at earlier time points in reinfected subjects and chronically-infected individuals. P > 0.05 compared to both chronically-infected groups.
Humoral immune responses during reinfection
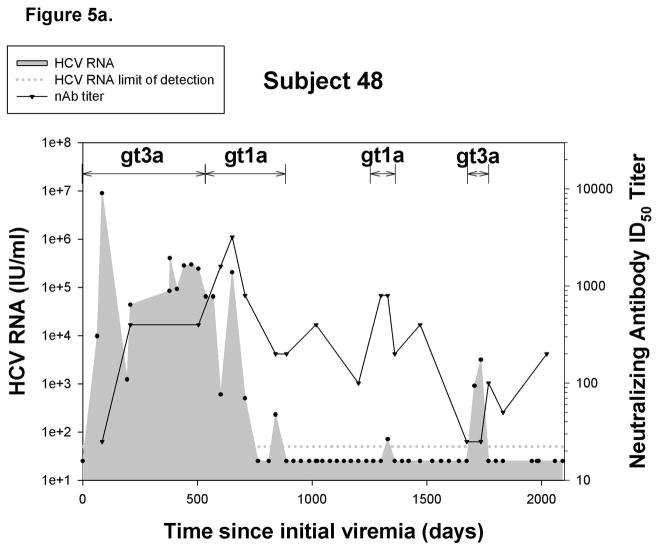
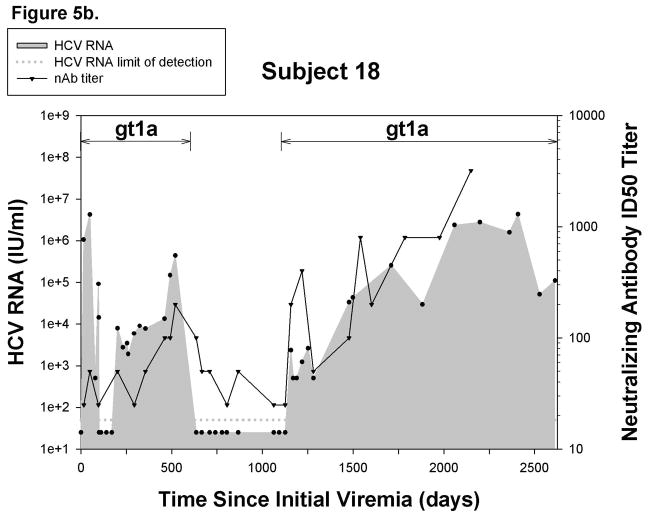
Since the appearance of neutralizing antibodies (nAb) corresponds with clearance of initial infections in some individuals24, we investigated the role of nAb in control of reinfection. nAbs against a genotype 1a pp were detected in 6 out of 10 reinfected subjects and in 8 out of 12 reinfections. Six of the reinfections with detectable nAbs were controlled, but nAbs were also detected in the two persistent reinfections. Representative graphs are shown in Figure 5, illustrating nAb titers in a cleared reinfection (5a) and a persistent reinfection (5b). Additional graphs are included in Supplementary Figure 2. Surprisingly, nAbs against a genotype 1a pseudoparticle were detected during reinfections in which the stimulating virus was not genotype 1a.
Figure 5.
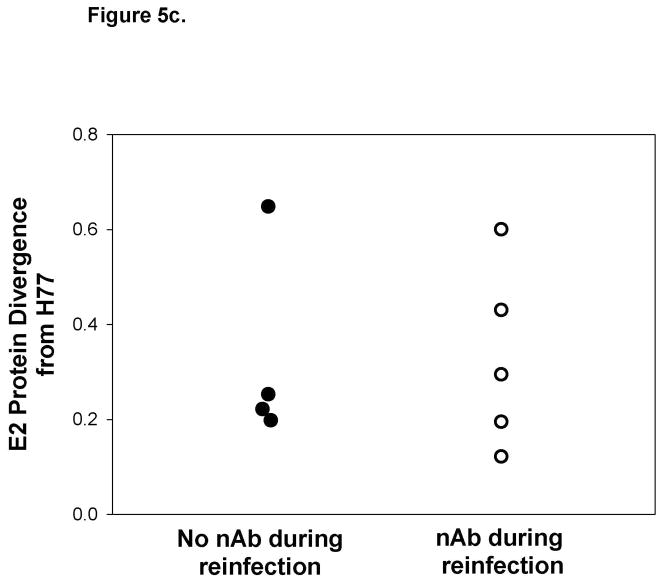
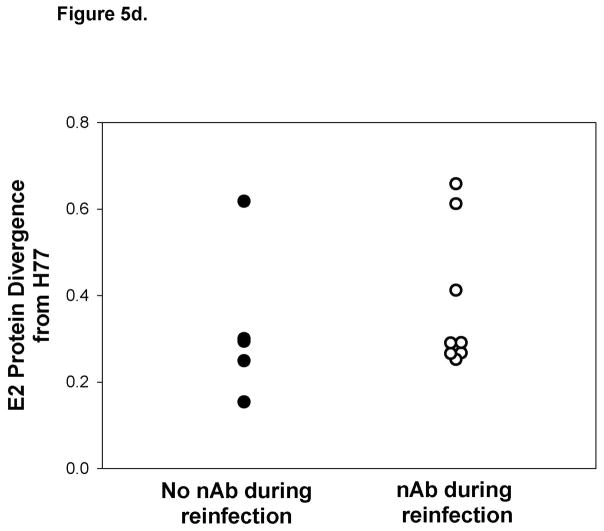
Reinfection is associated with generation of cross-reactive nAb. Representative graphs demonstrating nAb titers in A) a subject who controls reinfection and B) a persistently-reinfected subject. Solid line represents reciprocal ID50 nAb titer against a genotype 1a HCVpp (H77). Undetectable nAb titers were assigned a value of 25. Black circles represent HCV RNA concentrations (IU/ml) detected in serum or plasma samples obtained at given time points from date of first detection of viremia. Dotted-line denotes the HCV RNA limit of detection. Samples below the HCV RNA limit of detection were assigned a value of 25 IU/ml. Viral E2 protein sequence divergence from H77 E2 protein sequences of viruses obtained during C)reinfection and D) initial infection from subjects in whom nAb were and were not detected during reinfection. No significant difference in the E2 protein sequence divergence between groups was observed during reinfection and initial infection.
In order to rule out the possibility that detection of nAb is associated with the similarity of the infecting virus to H77, we compared HCV E2 protein sequences of viruses obtained during reinfection in subjects with and without nAb responses to the H77 E2 protein sequence. Sequences were not obtainable from three reinfection viruses due to low HCV RNA levels, but E2 divergence from H77 is shown for the remaining 9 viruses in Figure 5c. E2 sequence analysis revealed similar amino acid divergence from H77 of viruses obtained during reinfections with and without detectable nAbs. Interestingly, some viral sequences were highly divergent from H77 during reinfections in which nAb responses were detectable. Likewise, the sequence divergence of the initial infection stimulating virus from H77 within these groups was not different (Figure 5d). Taken together, these results suggest that, in some subjects, clearance of reinfection is associated with the generation of cross-reactive nAb independent of sequence similarity to stimulating viruses during both initial infection and reinfection.
Discussion
The results of our longitudinal analysis of IDUs demonstrate that control of an initial HCV infection is associated with abbreviated subsequent viremia, reduced magnitude of viremia, generation of new T cell responses, the appearance of cross-reactive anti-HCV nAbs, and a higher rate of clearance. Frequent monitoring of HCV infection status in IDUs who clear a primary infection demonstrated that reinfection of IDUs following control of a primary infection is common, with detection of one or more subsequent infections in 50% of cleared subjects. The incident reinfection rate in our study was 30.1 reinfections/100 person-years. This reinfection rate is very similar to the incident primary infection rate previously reported in the same cohort (27.2 infections/100 person-years)1; strongly suggesting that prior clearance of HCV infection does not provide sterilizing immunity against reinfection.
Some previous studies demonstrated lower rates of reinfection13, 14, but these studies had long intervals between testing. Given the brief duration of viremia with reinfection, these longer testing intervals and therefore less frequent HCV RNA testing likely missed multiple episodes of recurrent viremia. Consistent with this, our reinfection rate is similar to that shown in a study by Aitken, et al in which shorter intervals between HCV RNA testing were employed and in which reinfection was documented in 46% of previously cleared IDUs15. Since approximately half of IDUs are demonstrated to have been reinfected, studies with limited longitudinal analysis of HCV infection status might incorrectly identify viremia as persistent infection following initial exposure when, in fact, it is a reinfection following control of the primary infection. This would lead to underestimation of the actual clearance rate of a single infection.
Studies in older, seropositive IDUs have shown that prior clearance of HCV decreases the rate of persistent reinfection compared to initial infection in 13,14. In contrast, a study in younger, seropositive IDUs recently demonstrated that reinfection of previously cleared IDUs occurred at a higher rate than primary infections after accounting for risk factors15. These conflicting results may in part be explained by the limitations of these previous studies. In these studies, the cohorts of reinfected subjects consisted primarily of individuals in whom seroconversion had occurred at an unknown time prior to assessment of reinfection. It is possible that the cohorts of older IDU’s represent a selection of subjects who have controlled multiple infections. The duration of reinfection in subjects who have cleared multiple infections may be progressively shorter with each reinfection, thereby decreasing the likelihood of detecting reinfection in these individuals.
In addition, the inclusion of seropositive, aviremic subjects precludes phylogenetic confirmation that any newly observed viremia is genetically distinct from previous infections. Longitudinal analysis of HCV infection has demonstrated that recurrent, short periods of aviremia followed by viral resurgence are common during acute primary infections1, 7. Therefore, reinfection criteria based solely on sequential HCV RNA negative blood samples prior to an HCV RNA positive sample, without sequencing, allow for remergence of the same virus during early primary infection to be misclassified as reinfection. In the current study, phylogenetic analysis of viral sequences coupled with the requirement of a temporal association between viral sequence divergence and aviremia providing complementary methods of confirming reinfection, thereby increasing confidence that reinfections were accurately classified.
While a reduced incidence of reinfection would indicate the presence of sterilizing immunity, protective immunity need not be sterilizing. In addition to a higher rate of control of subsequent infections, the duration and magnitude of secondary HCV viremia were significantly decreased relative to primary infection. The lower duration and magnitude of viremia we observed in subsequent infections suggest that prior exposure to HCV provides protective immunity against persistent reinfection since fixed genetic factors associated with control of infection would not be expected to adapt and become more efficient at viral control like the immune response. An effective vaccine must be able to evoke protective immunity against multiple HCV subtypes and genotypes due to the broad genetic diversity of HCV. Even though reinfection was common in our study, prior clearance was associated with protective immunity against subsequent persistent infection with viruses of similar and differing genotypes.
Adaptive immunity has been shown to play a vital role in clearance of primary HCV infection as well as reinfection. In humans, sustained and broad CD4+ and CD8+ T cell responses are associated with viral control7, 25. Additionally, increased levels of serum ALT, which have been associated with T cell activity in some studies, have been observed during acute infection25. However, the magnitude and breadth of CD8+ T cell responses declines with increasing duration of viremia in chronically-infected individuals, suggesting that the effectiveness of cellular immunity in controlling infection declines with time7. The role of adaptive immune cells in controlling reinfection has been investigated in chimpanzees, the only animal model for HCV infection. Control of secondary infections, in chimpanzees, was associated with robust CD8+ T cell activity resulting in rapid clearance26, 27. Likewise, depletion of CD8+ T cells during reinfection resulted in viral persistence while restoration of CD8+ T cells led to rapid clearance27. However, serum ALT levels in chimpanzees remain stable during rechallenge, perhaps indicative of the differences in HCV infection parameters in chimpanzees compared to humans12, 26.
Increased blood ALT levels were observed during reinfection and new T cell responses were detected in reinfected individuals at a higher frequency than during chronic infection, suggesting cellular immune responses may contribute to the protective immunity afforded by previous clearance. Moreover, all reinfected subjects with detectable new T cell responses had lower maximum viremia during reinfection. However, new T cell responses and increased levels of ALT during reinfection were observed in a subject who developed persistent viremia, indicating that enhanced cellular immune responses provide incomplete protection against persistent reinfection. Our cellular immune response studies were limited by the fact that not all reinfected subjects were eligible for or consented to large volume blood draws thereby precluding analysis of T cell responses in these subjects. Also, T cell responses were screened using peptides derived from a genotype 1a virus, possibly underestimating T cell responses during reinfection with non-genotype 1a virus. More detailed characterization of these responses in reinfected individuals may provide further insight into the role of cellular immunity in controlling reinfection and may illustrate immune mechanisms to target with a vaccine.
While the appearance of neutralizing antibodies has been shown to drive E2 sequence evolution and correspond with control of viremia during initial infection24, the role of neutralizing antibodies in clearance of reinfection in humans is unknown. In our study, 60% of reinfected subjects generated nAb against a heterologous HCVpp that was highly divergent from the stimulating virus detected during both the initial infection and reinfection in these subjects. Previous experiments have demonstrated that during chronic infection nAbs fail to or weakly neutralize heterologous HCVpp during the acute phase of infection (< 33 weeks), while neutralization titers against autologous HCVpp are significantly higher20, 24. These findings suggested that the reactivity of nAbs during initial infection is dependent upon the stimulating virus. In contrast, nAb with high titers against heterologous virus were detected in reinfected individuals independent of the sequence of the stimulating virus during initial infection and reinfection, suggesting the presence of cross-reactive nAbs able to neutralize heterologous HCVpp in these subjects. Detection of cross-reactive nAbs during reinfections of short duration suggests that clearance of an initial infection alters subsequent humoral responses to repeated HCV infection, thereby resulting in rapid generation of broadly-neutralizing antibodies. Conversely, the ability to generate cross-reactive nAbs may be a marker of the ability to control infection rather than the result of repeated infection. The appearance of broadly neutralizing nAb before detectable reinfection supports this. However, nAb responses were detected in both individuals with persistent reinfection, demonstrating that, nAbs, like enhanced T cell responses, provide incomplete protection against persistent reinfection.
Moreover, the high prevalence of cross-reactive nAbs observed in our study versus the absence of cross-reactive nAbs during chronic infection suggests that nAb responses generated during reinfection may be qualitatively different than in initial infection. Cross-reactive human monoclonal antibodies against conserved epitopes important in CD81-mediated viral entry have been described28, 29. It is possible that prior clearance of an HCV infection results in selection of antibodies targeting conserved epitopes, thereby allowing for neutralization of heterologous HCVpp. Further investigation of the epitopes targeted by nAb responses during reinfection may lead to identification of relevant immunogenic targets for inclusion in future vaccine development.
One limitation of our study is that a period of 60 days of aviremia may be unnecessarily conservative. We detected a genetically-distinct virus in 19 subjects with constant viremia or aviremic periods of less than 60 days prior to detection of the new virus, precluding classification of these subjects as reinfected. It is possible that these individuals controlled a primary infection and were subsequently reinfected between clinical visits, resulting in the absence of sufficient aviremia to meet our stringent criteria. This would lead to an underestimate of both the true rate of primary infection clearance and the frequency of reinfection. However, defining the appearance of a new virus as a reinfection without sufficient aviremia might potentially overestimate the incidence of reinfection. Sequence-specific PCR analysis of viral sequences in one transfusion recipient infected with multiple HCV strains by exposure to blood from different donors revealed a newly dominant viral sequence late in infection that was detected in the inoculum but not observed in earlier samples from the infected subject30. In individuals with new dominant viruses without a period of aviremia, distinguishing new infection from infection with multiple strains with alternating dominance will require the development of highly sensitive assays for tracking specific viral sequences.
Collectively, the results of our study provide clear evidence supporting protective immunity associated with clearance of primary HCV infection. The fact that protective immunity following HCV infection is possible highlights the plausibility of a preventive vaccine. In the absence of sterilizing immunity, it may still be possible to target immune mechanisms with immunization that favor clearance in naïve individuals following infection. Careful study of subjects who clear a primary infection and are subsequently reinfected will be important in determining the immunological correlates critical in the development of such vaccination strategy.
Supplementary Material
Acknowledgments
This work was supported by: NIH grants U19AI040035, R01DA024565 and T32AI07247, as well as grants from The Damon Runyon Cancer Research Foundation and The Dana Foundation.
Abbreviations used
- IFN-γ
interferon-gamma
- IDU
intravenous drug user
- PHA
phytohemmaglutinin
- HCVpp
HCV pseudoparticle
Footnotes
There are no conflicts of interest to disclose.
The authors contributed the following to the preparation of this manuscript:
W. O. – Study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript
B. F. – Acquisition of data
K. D. – Acquisition of data
G. U. – Acquisition of data
L. L. – Acquisition of data
S. R. – Critical revision of the manuscript for important intellectual content; obtained funding
D. T. - Critical revision of the manuscript for important intellectual content; study concept and design
A. C. – Obtained funding; study concept and design; critical revision of the manuscript for important intellectual content
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox A, Netski D, Mosbruger T, et al. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin Infect Dis. 2005;40:951–8. doi: 10.1086/428578. [DOI] [PubMed] [Google Scholar]
- 2.Prevention CfDCa. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. MMWR. 1998;47:1–39. [PubMed] [Google Scholar]
- 3.Garfein RS, Doherty MC, Monterroso ER, et al. Prevalence and incidence of hepatitis C virus infection among young adult injection drug users. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18 (Suppl 1):S11–9. doi: 10.1097/00042560-199802001-00004. [DOI] [PubMed] [Google Scholar]
- 4.Villano SA, Vlahov D, Nelson KE, et al. Incidence and risk factors for hepatitis C among injection drug users in Baltimore, Maryland. J Clin Microbiol. 1997;35:3274–7. doi: 10.1128/jcm.35.12.3274-3277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagan H, Thiede H, Weiss NS, et al. Sharing of drug preparation equipment as a risk factor for hepatitis C. Am J Public Health. 2001;91:42–6. doi: 10.2105/ajph.91.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagan H, Pouget ER, Des Jarlais DC, et al. Meta-regression of hepatitis C virus infection in relation to time since onset of illicit drug injection: the influence of time and place. Am J Epidemiol. 2008;168:1099–109. doi: 10.1093/aje/kwn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox AL, Mosbruger T, Lauer GM, et al. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–12. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanford RE, Bigger C, Bassett S, et al. The chimpanzee model of hepatitis C virus infections. Ilar J. 2001;42:117–26. doi: 10.1093/ilar.42.2.117. [DOI] [PubMed] [Google Scholar]
- 9.Bassett SE, Guerra B, Brasky K, et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology. 2001;33:1479–87. doi: 10.1053/jhep.2001.24371. [DOI] [PubMed] [Google Scholar]
- 10.Prince AM, Brotman B, Lee DH, et al. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J Infect Dis. 2005;192:1701–9. doi: 10.1086/496889. [DOI] [PubMed] [Google Scholar]
- 11.Folgori A, Capone S, Ruggeri L, et al. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat Med. 2006;12:190–7. doi: 10.1038/nm1353. [DOI] [PubMed] [Google Scholar]
- 12.Lanford RE, Guerra B, Chavez D, et al. Cross-genotype immunity to hepatitis C virus. J Virol. 2004;78:1575–81. doi: 10.1128/JVI.78.3.1575-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta SH, Cox A, Hoover DR, et al. Protection against persistence of hepatitis C. Lancet. 2002;359:1478–83. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 14.Grebely J, Conway B, Raffa J, et al. Hepatitis C virus reinfection in injection drug users. Hepatology. 2006;44:1139–45. doi: 10.1002/hep.21376. [DOI] [PubMed] [Google Scholar]
- 15.Aitken CK, Lewis J, Tracy SL, et al. High incidence of hepatitis C virus reinfection in a cohort of injecting drug users. Hepatology. 2008 doi: 10.1002/hep.22534. [DOI] [PubMed] [Google Scholar]
- 16.Ray SC, Arthur RR, Carella A, et al. Genetic epidemiology of hepatitis C virus throughout egypt. J Infect Dis. 2000;182:698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 17.Jeanmougin F, Thompson JD, Gouy M, et al. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 18.Los Alamos National Laboratory. 2008 retrieved from http://hcv.lanl.gov/content/sequence/phyloplace/PhyloPlace.html.
- 19.Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A. 2003;100:7271–6. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logvinoff C, Major ME, Oldach D, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A. 2004;101:10149–54. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowd KA, Hershow RC, Yawetz S, et al. Maternal Neutralizing Antibody and Transmission of Hepatitis C Virus to Infants. J Infect Dis. 2008;198:1651–1655. doi: 10.1086/593067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner F, Gruener NH, Urbani S, et al. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000;30:2479–87. doi: 10.1002/1521-4141(200009)30:9<2479::AID-IMMU2479>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 23.Cooper S, Erickson AL, Adams EJ, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–49. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 24.Dowd KA, Netski DM, Wang XH, et al. Selection Pressure from Neutralizing Antibodies Drives Sequence Evolution during Acute Infection with Hepatitis C Virus. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thimme R, Oldach D, Chang KM, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nascimbeni M, Mizukoshi E, Bosmann M, et al. Kinetics of CD4+ and CD8+ memory T-cell responses during hepatitis C virus rechallenge of previously recovered chimpanzees. J Virol. 2003;77:4781–93. doi: 10.1128/JVI.77.8.4781-4793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoukry NH, Grakoui A, Houghton M, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med. 2003;197:1645–55. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keck ZY, Li TK, Xia J, et al. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol. 2008;82:6061–6. doi: 10.1128/JVI.02475-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owsianka AM, Tarr AW, Keck ZY, et al. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J Gen Virol. 2008;89:653–9. doi: 10.1099/vir.0.83386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskus T, Wang LF, Radkowski M, et al. Exposure of hepatitis C virus-negative recipients to > or =2 infected blood donors. J Infect Dis. 2001;183:666–9. doi: 10.1086/318531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.