Abstract
Copy number variation in the human genome is prevalent but relatively little is known about the dynamics of DNA rearrangement. We therefore used the duplicated γ-globin genes as a simple system to explore de novo copy number changes. Rearrangements that changed gene number were seen in both germline and somatic DNA, and mainly arose by unequal sister chromatid exchange between homologous sequences, with evidence from recurrent mosaic rearrangements that many, if not all, of these events in sperm arise before meiosis. Unequal exchange frequencies are apparently controlled primarily by the degree of sequence identity shared by the duplicate genes, leading to substantial variation between haplotypes in copy number instability. Additional, more complex rearrangements generated by mechanisms not involving homologous recombination, and in some cases showing DNA transfer between chromosomes, were also detected but were rare. Sequence changes were also seen in γ-globin DNA molecules, with strong evidence that some were genuine de novo base substitutions. They were present in sperm at a frequency far higher than predicted from current estimates of germline mutation rates, raising interesting questions about base mutation dynamics in the male germline.
Keywords: base mutation, blood, ectopic, recombination, sperm
Copy number variation in the human genome is extensive (1–4) and has a substantial impact on health (5, 6). Analysis of DNA rearrangements seen in populations and patients has revealed a diversity of mechanisms that create this variation, including unequal crossover between homologous sequences, transposition, nonhomologous end-joining of DNA breaks, and frequently complex events probably arising from a fork-stalling/template-switching replication process (5–8). However, few studies have directly addressed the dynamics and processes involved. Single DNA molecule analysis of megabase-scale rearrangements underlying genomic disorders, such as Charcot-Marie-Tooth disease type 1A, has revealed a meiosis-specific process involving ectopic recombination (nonallelic homologous recombination) between dispersed repeats (9). In contrast, kilobase-scale rearrangements in the α-globin gene cluster are not restricted to the germline, with evidence that both meiotic and mitotic recombination pathways play an important role in copy number instability (10, 11).
To explore these mechanistic issues further and to investigate the relative roles of meiotic recombination, mitotic exchanges, and other processes in driving rearrangements, we developed single DNA-molecule methods to recover de novo rearrangements in the fetal Gγ- and Aγ-globin genes. These duplicate genes arose as part of a tandem 5-kb duplication ∼34 Myr ago (12–14) and subsequently diverged to create a patchwork of low- and high-sequence divergence (Fig. 1A), with regions of low divergence maintained by recombination between the duplicates (12, 15). Ongoing interactions are evidenced by the existence of chromosomes carrying one, three, and four γ-globin genes in human populations (16). The patchwork sequence divergence and a high density of SNPs make the γ-globin genes particularly informative for copy number analysis.
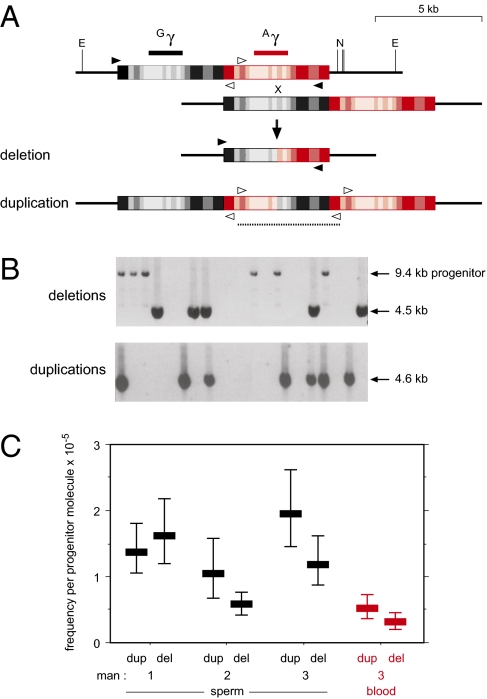
Fig. 1.
Detecting copy number instability in human γ-globin genes. (A) Location of Gγ- and Aγ-globin genes in a 5-kb tandem duplication (black, red boxes), with sequence divergence between these homology blocks indicated by shading from <5% (light) to >20% (dark). Unequal crossover can generate –γ and γγγ rearrangements that were purified from genomic DNA by digestion with EcoRV (E) and NdeI (N), followed by electrophoretic fractionation to separate –γ and γγγ molecules away from progenitor γγ molecules. Deletion molecules were recovered by PCR amplification with the black primers; duplications were amplified with the divergent white primers that converge following duplication to allow specific amplification of the exchange interval (dotted line). (B) Examples of molecules amplified from multiple aliquots of DNA size-enriched for deletions or duplications, each containing DNA derived from 1.8 × 105 haploid genomes. PCR products were visualized after agarose gel electrophoresis by staining with ethidium bromide. Some remaining progenitor molecules were also detected in the deletion-enriched DNA. (C) Frequencies of rearrangements, with confidence intervals (Materials and Methods), in sperm and blood DNA, as determined from 69 to 398 mutants of each class detected in 1.2 to 3.3 × 107 haploid genomes surveyed per man.
Results
Detecting de Novo γ-Globin Gene Rearrangements.
Genotype analysis of the γ-globin genes revealed intense linkage disequilibrium across the entire region with just four common haplotype classes, A to D, present in northern Europeans that accounted for 93% of all haplotypes (Table S1). We selected three men of northern European origin, aged 55 to 59, for mutation analysis, who between them contained all four haplotypes (man 1 A/B, man 2 C/D, man 3 D/D), to allow the haplotype of origin of rearrangements to be determined and to investigate any influence of haplotype on copy number instability. We purified genomic DNA molecules carrying de novo rearrangements of γ-globin genes by electrophoretic size enrichment (10, 11). DNA fractions containing –γ deletions were heavily depleted in progenitor γγ molecules (<0.003% remaining), allowing deletion molecules to be amplified in their entirety by PCR. In contrast, inverse PCR (11) was used to amplify the exchange junction in γγγ duplication molecules, given the significant levels of progenitor (0.2–1%) remaining in duplication-enriched DNA (Fig. 1 A and B).
Substantial instability was seen in sperm DNA from all three men analyzed. Deletion and duplication molecules were correctly distributed across size fractions, providing strong evidence that these rearrangements are not PCR artifacts derived from contaminating progenitor molecules (Fig. S1). Duplications and deletions were detected at a similar mean frequency of 1.5 × 10−5 and 1.1 × 10−5 per progenitor molecule, respectively, although there was some variation between men and between duplication and deletion frequencies in a given man (Fig. 1C). Instability was also seen in somatic (blood) DNA tested for man 3, at 27% of the level seen in sperm.
Most Rearrangements Arise by Ectopic Recombination.
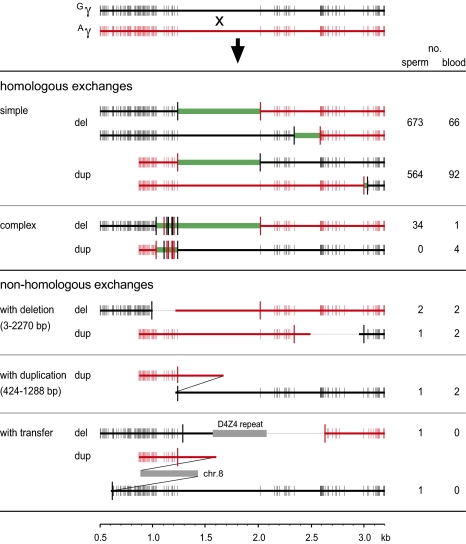
Sequence analysis of mutant DNA molecules (4.3 kb per molecule) revealed a wide variety of rearrangements (Fig. 2 and Fig. S2). Almost all (99.2%) arose by unequal crossover between homologous sequences to produce –γ and γγγ rearrangements. In 2.6% of cases, the rearranged molecule showed a patchwork of paralogous sequence variants (PSVs) derived from one or another homology block that presumably arose through patchwork repair of heteroduplex DNA generated during recombination (10). Using multiple SNP heterozygosities in men 1 and 2 to determine haplotype of origin (10) showed that only 0.6% of sperm rearrangements had recombinant haplotypes (Fig. S2). Exchange between homologous chromosomes is therefore rare, pointing to unequal sister chromatid exchange as the dominant process.
Fig. 2.
Representative examples of γ-globin gene duplications and deletions detected in sperm and blood DNA. Sequences derived from Gγ and Aγ homology blocks shown at top are indicated in black and red, respectively, with positions indicated from the start of the 5-kb homology blocks. PSVs are indicated by vertical lines, in bold for differences around the exchange interval (green rectangle). The number of mutants seen in each class of rearrangement is given at right, with sperm data pooled across all three men analyzed. The size ranges of local deletions and duplications seen in nonhomologous exchanges are indicated. Deleted sequences are shown as dashed lines, and sequences acquired from remote locations as gray rectangles. The 503-bp D4Z4 insertion is identical to a member of the D4Z4 repeat family (37) of unknown chromosomal location. The 542-bp chromosome 8 insertion is a perfect match to chromosome 8 bases 91,554,957 to 91,555,498 (assembly GRCh37, release 56). Structures of all rearrangements seen are provided in Fig. S2.
These exchanges are not exclusively meiotic, as shown by instability in blood and by mutational mosaicism in sperm, indicating premeiotic exchanges that had spread to multiple descendants of a germ cell following exchange. Specifically, 30 different types of rearrangement were detected in sperm that were unlikely to occur recurrently, namely complex patchwork exchanges plus exchanges that mapped to a very short (<21 bp) interval of perfect sequence identity (IPSI) shared by the Gγ- and Aγ-globin genes (Fig. S2). Nine of these rearrangements were seen repeatedly, with replicates observed 2 to 10 times in a given individual and with mosaic mutants being shared on average by 1 sperm in 5,000,000. Including recurrences, 65 of these rare rearrangements were detected, of which 44 were recurrent and therefore premeiotic. This finding indicates that at least 68% (44/65) of rare rearrangements seen in sperm must arise by mitotic recombination, an estimate that presumably also applies to more common classes of rearrangement where mosaicism arising from a single event cannot be distinguished from the repeated generation of the same rearrangement.
Influence of Sequence Homology on Ectopic Exchange Frequencies.
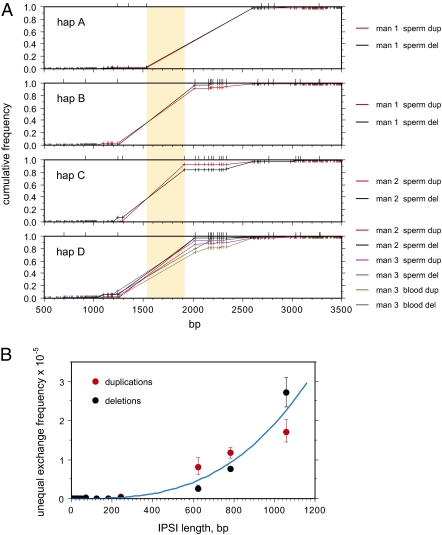
Unequal exchanges strongly clustered into the longest IPSI shared by homology blocks (Fig. 3A) and appeared to signal the existence of an unequal crossover hotspot. However, SNPs within homology blocks altered the length of the longest IPSI in different haplotypes and defined a minimum active region of 376 bp, far narrower than the 1- to 2-kb meiotic recombination hotspots characterized to date (9, 17–21). Instead, IPSI length l appears to be the main, and very strong, determinant of unequal crossover frequency r (Fig. 3B), with r ≈ 1.9 × 10−14l3 in sperm. This simple relationship successfully predicted overall exchange frequencies, typically within 30% of the observed value, and faithfully recapitulated the exchange distributions seen on different haplotypes. Exchange frequencies should therefore vary substantially between haplotypes depending on IPSI length. Major haplotype skews were indeed seen in men 1 and 2, who each carry different haplotypes (Fig. S2); for example, haplotypes A and B in man 1 have a longest IPSI of 1,058 and 783 bp, respectively, and as expected, most deletions and duplications were derived from haplotype A (338 from A, 115 from B).
Fig. 3.
Distribution of ectopic exchanges across the γ-globin homology blocks. (A) Cumulative frequencies of exchanges in duplications and deletions on each of the four haplotypes (A–D) present in the men tested (man 1 A/B, man 2 C/D, man 3 D/D), with positions along the homology blocks indicated as in Fig. 2. Haplotype-specific PSVs created by SNPs are shown by ticks above each plot. The minimum region of overlap of the longest IPSIs, within which most recombination activity would have to occur under a hotspot model, is shaded in yellow. (B) Relationship between IPSI length and ectopic recombination activity. Sperm duplication and deletion data were pooled over all haplotypes and the mean ectopic exchange frequency r was determined for each IPSI length l following binning of data across shorter IPSIs, together with 95% confidence intervals estimated from the number of exchanges scored at each length. Least-squares analysis of power functions produced the best-fit curve for combined deletion plus duplication data shown in blue, where r = 1.9 × 10−14l3.
Evidence for Nonreciprocal Exchanges.
Duplications and deletions should arise as reciprocal products of unequal sister chromatid exchange. However, complex patchwork exchanges in sperm were only seen in deletions (Fig. 2 and Fig. S2); even after elimination of mosaic duplicates, this duplication/deletion disparity remained significant (P = 0.004). Furthermore, the dependence of exchange frequencies on IPSI length is weaker for duplications than deletions (Fig. 3B); for example, over the longest IPSI man 1 shows 66% (133/202) of duplications derived from haplotype A versus 84% (182/217) for deletions (P = 0.00002) (Fig. S2). Such nonreciprocities might point to two distinct ectopic-exchange pathways, one fully reciprocal plus a second, highly dependent on perfect sequence identity, that only generates deletions [perhaps by intrachromatid exchange (9)] that often show patchwork exchanges. However, there is no consistent excess of deletions over duplications, as predicted by this model (Fig. 1C) and as seen in meiotic rearrangements in genomic disorders (9), although such an excess could be masked by chance high-level mosaicism of duplications (10, 11).
Complex Rearrangements.
Only a few γ-globin gene rearrangements in sperm and blood did not arise by ectopic recombination, as shown by minimal (<5 bp) microhomologies at breakpoints (Fig. 2 and Fig. S2). These rearrangements were complex, showing additional deletions or duplications plus, in two instances, transfer of a short segment of DNA from a remote genomic location, perhaps via a fork-stalling/template-switching replication process (8).
Base Changes in γ-Globin Gene Rearrangements.
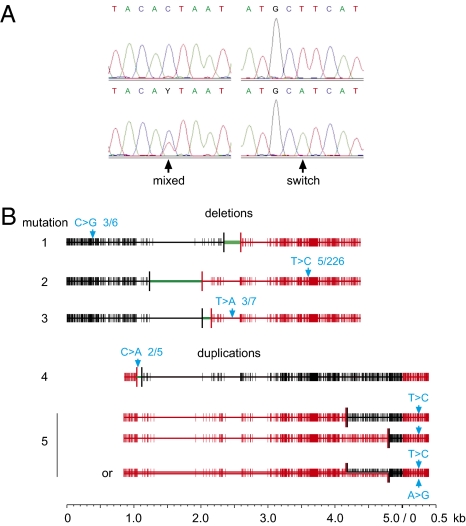
Sequence traces from amplified molecules revealed numerous base changes (Fig. 4A and Fig. S3), including sites showing a mixture of the correct base and an incorrect base as expected for base misincorporations during the early stages of PCR, plus apparently complete switches of DNA sequence with the correct base undetectable in sequence traces (Fig. 4A and Fig. S3). These base switches were seen at similar frequencies in rearranged sperm and blood molecules and in control unrearranged progenitor molecules in sperm. Their high incidence (83 different switches seen in 4.6 Mb sequenced DNA, frequency 1.8 × 10−5/bp) is wholly incompatible with estimates of the frequency of de novo base substitution in the male germline [∼3.9 × 10−8/bp, or ∼10−7/bp in the men analyzed allowing for their age (22–25)]. PCR must therefore be capable of generating these switches as artifacts, for example by once-only misincorporation at a damaged base in one template strand coupled with total failure of the other strand to amplify; this distinct process is consistent with the different spectrum of base changes seen at switched versus mixed sites (Fig. S4).
Fig. 4.
Sequence changes in γ-globin gene rearrangements. (A) Sequence traces from amplified single molecules showing a mixed site or a base switch. (B) Recurrent base switches 1 to 5 seen in rearranged molecules, with the base change plus the number of recurrences versus the total number of rearrangements of each class shown in blue. Mutation 3 was seen in blood DNA; all other mutations were detected in sperm. Mutation 5 showed mixed Gγ plus Aγ sequences over a 609-bp interval, signaling either two different rearrangements or an unrepaired heteroduplex molecule with one strand showing a T→C switch and the other strand the complementary A→G change. See Table S2 for further details of these candidate mutations and Fig. S3 for sequence traces.
However, we could not dismiss all of these switched bases as PCR artifacts because four proved to be recurrent (Fig. 4B, Fig. S3, and Table S2). For example, we found three molecules of a sperm deletion carrying exactly the same C→G switch (mutation 1). It is unlikely that this switch would have occurred three times independently, and only on rearranged molecules of this uncommon class derived from the same haplotype in the same man (P < 10−9). A fifth instance of recurrence (mutation 5) (Fig. 4B) was found in two different duplications detected in the same PCR that might instead derive from a single molecule of meiotic origin containing an unrepaired heteroduplex tract; if so, then both DNA strands must carry an identical (complementary) base switch.
It is extremely difficult to explain away these recurrent base changes as PCR artifacts because PCR reactions showing these changes were correctly distributed across size fractions and randomly distributed across PCR plates, and showed strong PCR signals fully consistent with molecules present in the starting fractionated DNA (Materials and Methods). Contaminating genomic DNA also seems highly implausible, as this would require repeated trace contamination with DNA from multiple individuals, each carrying one of these base changes as a SNP on a rearranged haplotype. We have never seen a rearrangement carrier and none of the recurrent base switches matches a known SNP. Furthermore, such a hypothetical contaminant would be easily detected as such, unless it fully matched one of the haplotypes present in the man being analyzed. We therefore conclude that these recurrent base switches are strong candidates for genuine mosaic de novo base mutations that, in sperm, must have arisen before meiosis and spread to multiple descendant cells.
Discussion
The high level of copy number instability seen at the γ-globin genes makes this an informative system for analyzing instability processes. The frequency of rearrangements seen in sperm (1.3 × 10−5) is comparable with that seen at the similarly organized α-globin gene pair (4 × 10−5) and, as with the α-globin genes, contrasts with the scarcity of –γ and γγγ chromosomes in most populations, suggesting that stable copy number is being actively maintained by purifying selection against rearrangements (10, 11).
This γ-globin gene survey revealed diverse mechanisms that contribute to de novo copy number variation, with ectopic recombination being by far the dominant process, as expected from the organization of this duplicated gene. These γ-globin rearrangements arise almost exclusively by unequal sister chromatid exchange, with little evidence for meiotic interactions between homologous chromosomes. Instability in blood and the presence of rearrangement mosaicism in sperm point to a major role for mitotic recombination in driving copy number instability, although it is not possible to exclude a minor role for meiotic sister chromatid exchange in the germline. Similar mosaic mutants have been seen in α-globin gene rearrangements, and as noted previously, can contribute to interindividual variation in instability levels and to disparities in the frequency of duplications and deletions (10, 11). Instability processes at the γ-globin genes contrast with large-scale rearrangements seen in genomic disorders [such as Charcot-Marie-Tooth disease type 1A (9)] that are largely, if not exclusively, meiotic in origin, and also with the α-globin genes, where ∼30% of sperm rearrangements arise by unequal exchanges between homologous chromosomes, most likely at meiosis (10, 11). Therefore, the γ-globin genes provide a system for analyzing copy number instability driven almost exclusively by sister chromatid exchange.
Instability in γ-globin genes appears to be mainly controlled by levels of sequence identity shared by homology blocks. Haplotype differences in IPSI length will also contribute to variation in rearrangement frequencies between men and the effect can be substantial, with haplotype A and D homozygotes (Fig. 3A) predicted to show a difference in instability of ∼5-fold. This major impact of sequence mismatches has also been seen in allelic and particularly ectopic recombination in yeast (26, 27), and suggests that new base substitutions within homology blocks could substantially reduce recombinational interactions between repeat sequences, facilitating their evolutionary divergence. Curiously, the human α-globin genes did not show this IPSI length dependence, with unequal exchanges being more randomly distributed along homology blocks and with disruption of IPSIs by SNPs having little, if any, effect on unequal exchange frequencies (10, 11). The reason for this major difference is unclear, although one possibility is that mismatched heteroduplexes in unequal exchange intermediates are more readily detected by mismatch repair systems in γ-globin than α-globin genes, and result in exchange events spanning PSVs being preferentially aborted in the former, thereby focusing the remaining exchanges into long IPSIs.
The scale of this γ-globin gene survey was sufficient to capture additional rearrangements that did not arise through homologous recombination, and showed that such de novo changes, including deletions, duplications, and interchromosome transfer of DNA, can be amenable to direct detection in genomic DNA. These nonhomologous exchanges were rare, arising at a frequency of 5 × 10−8 per sperm (95% CI 2–11 × 10−8), 240-fold lower than the frequency of unequal crossover and similar to the frequency of nonrecombinational deletions seen in sperm in the β-globin gene cluster (4 × 10−8) (28).
The detection of what appear to be genuine de novo base mutations in γ-globin genes was wholly unexpected. With the exception of specific mutations underlying inherited disorders, such as Apert syndrome, which are driven to high levels by germ-cell selection (29–32), this result would be a unique direct detection of base mutations in human sperm. We saw 11 to 12 of these candidate mutations in 3.85 Mb of sequenced sperm DNA. All would have to be present in different sperm, suggesting a de novo base-mutation frequency of (11 − 12)/(3.85 × 106) = 3.0 × 10−6 per base pair per sperm (95% CI 1.7–5.4 × 10−6). This result is wholly incompatible with the conventional mutation frequency estimate of ∼10−7/bp in the men analyzed (P = 5 × 10−13). Only one instance of a recurrent mutation was seen in blood, preventing meaningful estimation of somatic mutation frequency. The sperm frequency will be underestimated because of singleton mutations that cannot be validated by recurrence; however most of the rare γ-globin rearrangements showed recurrence, suggesting that this underestimation might not be substantial.
These recurrent base mutations were not caused by hypermutation at CpG doublets, because none changed CpG to CpA/TpG, nor obviously by gene conversion given the absence of likely donor sequences (Materials and Methods). Therefore, how can their remarkably high incidence be explained, particularly in sperm? One possibility is that conventional but indirect estimates of germline base-mutation frequencies (22–25), in particular for older men as analyzed in the present survey, are seriously underestimated. A second explanation is that these changes are associated specifically with rearranged γ-globin DNA molecules, perhaps being generated by mutagenic DNA repair during ectopic recombination. However, there was no obvious association between the location of the base switch and the site of ectopic exchange, and mutation 5 on two different molecules or a single heteroduplex molecule would most likely have been present prior to the recombination event (Fig. 4B). Instead, perhaps a subset of cells are particularly prone to accumulating γ-globin gene rearrangements and de novo base mutations. If this arises through loss of systems that maintain genome integrity, then copy number instability and base mutations could be strongly enhanced across the entire genome. Further analysis of this intriguing possibility will require the identification of other copy number variable loci at which instability is driven by mitotic exchanges. We note that base switches were detected in previous α-globin instability surveys (10, 11) but none were recurrent, not surprisingly given the small number of switches detected (six in total) and the limited sequence survey (0.2 Mb). Whatever the explanation for these strong candidates for de novo mutants, whether through highly enhanced mutation in older men or through “mutator” cell lineages producing heavily mutated sperm, there are important implications for genetic risk assessment and the generation of population diversity.
Materials and Methods
Identification of Informative Men.
Semen and blood samples were collected with approval from the Leicestershire Health Authority Research Ethics Committee and with informed consent. Nine semen donors of northern European descent were resequenced over a 10.5-kb interval spanning the Gγ-/Aγ-globin gene region, yielding 62 SNPs, of which 52 were present in dbSNP. All SNPs were genotyped on an additional 83 United Kingdom semen donors, as described previously (33). Details of common haplotypes are provided in Table S1. None of these men contained –γ, γγγ, or γγγγ chromosomes.
DNA Fractionation to Recover γ-Globin Gene Rearrangements.
DNAs were extracted and manipulated under conditions designed to minimize the risk of contamination (34), with sperm being subjected to prelysis with SDS to eliminate any somatic DNA (35). For each blood and sperm sample, 230 μg genomic DNA were digested with EcoRV-HF (New England BioLabs) plus NdeI (Fermentas) according to the manufacturers’ recommendations, then electrophoresed in a 0.8% SeaKem HGT agarose gel (Cambrex Bio Science Rockland), as described previously (10). Twenty-two size fractions ranging from 5.5 to >30 kb were recovered by electroelution, spanning deletion molecules (6.94 kb), duplications (16.78 kb), and progenitors (11.86 kb). Progenitor molecules were monitored in each fraction by long PCR with the γ-globin primers G18.4F (CGT ACT TTA GGC TTG TAA TGT G) and G23.2R (GTT AGA GAG AAG GGC GCA G) to amplify a 4.76-kb region spanning the Aγ-globin gene. Fractions containing duplications and deletions were identified by assaying for control genomic restriction fragments of the same size as duplication and deletion molecules (Fig. S1), as described elsewhere (10, 11).
Recovery of Deletions and Duplication Junctions by Single Molecule PCR Amplification.
Deletion molecules were amplified in their entirety by long PCR for 25 cycles using Taq and Pfu polymerases (36) plus primers G12.7F (TGC CTC TGT TCG AAT ACT TTC) and G23.3R (TAC AAG GCT AGA GTA AAG CAT G), then reamplified for 26 cycles using the nested primers G13.7F (TCT ATA CAC ACC CAT CTC AC) and G23.2R. This nesting strategy minimized the risk of carry-over contamination. Limited cycle numbers were chosen to ensure that only DNA molecules present at the outset would generate enough secondary PCR products for detection, thus eliminating artifact rearrangements generated late during amplification. Pilot assays on deletion fractions were used to estimate mutant frequencies, and subsequent full-scale mutant recovery was conducted with fraction inputs adjusted to less than one mutant molecule per PCR (10, 11). Duplication exchange junctions were recovered from duplication-enriched fractions using inverse PCR amplification (11) with primers G19.3R (GAC ATA AGA TGT GCG TGT AC) and G19.6F (CAT ACA TAC CTG AAT ATG GAA TCA), followed by reamplification with the nested primers G19.3aR (CTC TAA AGA GGC AAG GGT TG) and G19.6aF (GAT ATT GAG GTA AGC ATT AGG). These primers are located near the beginning of the Aγ homology block in a region diverged between Gγ and Aγ. Each duplication fraction (40 μL) was digested with 20 units restriction endonuclease Bst1107I (Fermentas) at 37 °C for 1 h before PCR amplification. This enzyme cleaves between the inverse primer sites and minimizes the risk of artifacts arising by template switching between homologous sequences on the same DNA template molecule during the first cycles of PCR (11), reducing their observed incidence per progenitor molecule from ∼10−4 to < 5 × 10−6. Pilot and full-scale recovery of duplication junctions proceeded as for deletions.
Size Validation of Rearrangements.
The number of deletions or duplications in each fraction was Poisson-adjusted for instances of a positive PCR containing more than one mutant molecule (these corrections were usually negligible and increased the estimated number of mutants by, at most, 32% in any fraction). Cumulative frequencies of deletion and duplication molecules across fractions were then compared with the cumulative frequency of progenitor molecules and of deletion and duplication control molecules (Fig. S1).
Estimation of Rearrangement Frequencies.
Gel-electrophoretic analysis of size fractions (10, 11) recovered from 230 μg digested genomic DNA indicated DNA yields of ∼65%. Aliquots of all fractions were pooled and the numbers of amplifiable progenitor molecules, plus deletion and duplication control molecules (Fig. S1), were estimated by Poisson analysis of limiting dilutions of DNA. Estimated counts of progenitor, deletion control, and duplication control molecules were indistinguishable and data from all three were combined to estimate the number of haploid genomes present in each enrichment experiment. Typically, 4.1 × 107 amplifiable molecules were recovered from fractionated DNA containing 5.0 × 107 molecules (equivalent to 150 μg input genomic DNA after fractionation losses). Thus, 82% of DNA molecules were amplifiable.
Deletion and duplication frequencies were estimated from the Poisson-corrected number of rearrangements, summed over all fractions, and compared with the total number of amplifiable progenitor molecules. Confidence intervals were determined using Poisson analysis to estimate the likelihood of obtaining the rearrangement-detection PCR data for each fraction as a function of the number of rearranged molecules present. Corresponding likelihood tables were determined for the numbers of progenitor/deletion control/duplication control molecules. Likelihoods over all fractions were then combined with each other and with the input molecule data. Confidence intervals were set as the lowest and highest numbers of rearrangements per progenitor molecule that gave a composite likelihood 20-fold less than the maximum likelihood.
Sequence Analysis of Rearrangements.
Up to 188 deletions and duplications from each man were reamplified and fully sequenced using BigDye Terminator v3.1 Cycle Sequencing (Applied Biosystems) on a 3730 DNA Analyzer (Applied Biosystems). If more rearrangements were available, we avoided those from fractions showing the highest contamination with progenitor molecules to minimize the risk of sequencing any possible PCR artifacts. In total, 1,150 rearrangement-positive PCRs were sequenced. On average, 7% of sequences showed mixed traces and were thus derived from two or more different mutant molecules present in the same PCR. In most cases these could be fully explained by admixture of two common classes of mutant. In remaining cases, we separated the constituent mutants using PCR primers specific for SNPs or PSVs before sequencing (10). A full inventory of each mutant type over all DNA size fractions was then used to Poisson-correct for PCRs containing two or more mutant molecules of the same type. This correction only affected the most common classes of mutant, with modest increases in mutant frequency of at most 28% per fraction. The overall number of mutants of a given class was then estimated by summing Poisson-corrected data over all fractions. The final corrected number of sequenced mutant molecules was estimated at 1,448.
Analyzing Mixed and Switched Bases in Sequence Traces.
Base changes were analyzed in deletion and duplication sequences, and also in control progenitor molecules (Fig. S4). Mixed base positions were analyzed as described in Fig. S4. All complete base switches were inspected by eye, and any showing evidence for the presence of the correct base in the sequence trace was rejected as mixed. The frequency of base switches in sequenced DNA was estimated from the number of switches seen and from the total length of DNA sequenced in PCR products derived from single input DNA molecules, as estimated by Poisson analysis of the numbers of PCR reactions positive for amplifiable DNA molecules.
Screening for Potential Gene-Conversion Donor Sequences.
There are no matching sequences in the Gγ and Aγ homology blocks that could serve as gene-conversion donors for any of the five candidate base mutations seen in sperm and blood. We extended the search for possible donors to a 200-kb interval centered on the Gγ/Aγ-globin genes using 21-nt probes centered on the mutant sequence and, as a control, equivalent probes corresponding to the progenitor sequence. The best perfect match that included the switched base was 12 bp, with similar match lengths seen equally frequently with the control probes. Equivalent whole-genome searches using BLAST again showed no significant difference in match frequencies between mutant and controls, with both showing longest perfect matches of only 18 bp. There is therefore no evidence for a likely conversion donor for any of the five validated switches, although we cannot exclude base switching by microconversion with extremely short donor sequences as a source of these base substitutions, a conclusion that would apply to almost any base substitution in humans.
Supplementary Material
Acknowledgments
We thank the volunteers for semen and blood samples, J. Sutherland for assistance with sequencing, and colleagues and reviewers for helpful discussions. This work was supported in part by the Medical Research Council, the Biotechnology and Biological Sciences Research Council, the Wellcome Trust (ref. 081227/Z/06/Z), the Royal Society, and the Louis-Jeantet Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003634107/-/DCSupplemental.
References
- 1.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 2.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad DF, et al. The Wellcome Trust Case Control Consortium. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 7.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam KWG, Jeffreys AJ. Processes of copy-number change in human DNA: The dynamics of α-globin gene deletion. Proc Natl Acad Sci USA. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam KWG, Jeffreys AJ. Processes of de novo duplication of human α-globin genes. Proc Natl Acad Sci USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slightom JL, Blechl AE, Smithies O. Human fetal G γ- and A γ-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980;21:627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- 13.Barrie PA, Jeffreys AJ, Scott AF. Evolution of the β-globin gene cluster in man and the primates. J Mol Biol. 1981;149:319–336. doi: 10.1016/0022-2836(81)90476-9. [DOI] [PubMed] [Google Scholar]
- 14.Shen SH, Slightom JL, Smithies O. A history of the human fetal globin gene duplication. Cell. 1981;26:191–203. doi: 10.1016/0092-8674(81)90302-0. [DOI] [PubMed] [Google Scholar]
- 15.Smithies O, Powers PA. Gene conversions and their relation to homologous chromosome pairing. Philos Trans R Soc Lond B Biol Sci. 1986;312:291–302. doi: 10.1098/rstb.1986.0008. [DOI] [PubMed] [Google Scholar]
- 16.Hill AVS, Bowden DK, Weatherall DJ, Clegg JB. Chromosomes with one, two, three, and four fetal globin genes: Molecular and hematologic analysis. Blood. 1986;67:1611–1618. [PubMed] [Google Scholar]
- 17.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 18.Webb AJ, Berg IL, Jeffreys AJ. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc Natl Acad Sci USA. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiter LT, et al. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- 20.Reiter LT, et al. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet. 1998;62:1023–1033. doi: 10.1086/301827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes J, et al. Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: A study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet. 1999;8:2285–2292. doi: 10.1093/hmg/8.12.2285. [DOI] [PubMed] [Google Scholar]
- 22.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 23.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 25.Xue Y, et al. Asan. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borts RH, Haber JE. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237:1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holloway K, Lawson VE, Jeffreys AJ. Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum Mol Genet. 2006;15:1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- 29.Tiemann-Boege I, et al. The observed human sperm mutation frequency cannot explain the achondroplasia paternal age effect. Proc Natl Acad Sci USA. 2002;99:14952–14957. doi: 10.1073/pnas.232568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goriely A, McVean GAT, Röjmyr M, Ingemarsson B, Wilkie AOM. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science. 2003;301:643–646. doi: 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- 31.Goriely A, et al. Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc Natl Acad Sci USA. 2005;102:6051–6056. doi: 10.1073/pnas.0500267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin J, et al. The molecular anatomy of spontaneous germline mutations in human testes. PLoS Biol. 2007;5:e224. doi: 10.1371/journal.pbio.0050224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffreys AJ, Ritchie A, Neumann R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet. 2000;9:725–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 34.Jeffreys AJ, Murray J, Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 35.May CA, et al. Minisatellite mutation frequency in human sperm following radiotherapy. Mutat Res. 2000;453:67–75. doi: 10.1016/s0027-5107(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 36.Jeffreys AJ, Neil DL, Neumann R. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 1998;17:4147–4157. doi: 10.1093/emboj/17.14.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.