Abstract
Studying the cell cycle process is crucial for understanding cell growth, proliferation, development, and death. We uncovered some key factors in determining the global robustness and function of the budding yeast cell cycle by exploring the underlying landscape and flux of this nonequilibrium network. The dynamics of the system is determined by both the landscape which attracts the system down to the oscillation orbit and the curl flux which drives the periodic motion on the ring. This global structure of landscape is crucial for the coherent cell cycle dynamics and function. The topography of the underlying landscape, specifically the barrier height separating basins of attractions, characterizes the capability of changing from one part of the system to another. This quantifies the stability and robustness of the system. We studied how barrier height is influenced by environmental fluctuations and perturbations on specific wirings of the cell cycle network. When the fluctuations increase, the barrier height decreases and the period and amplitude of cell cycle oscillation is more dispersed and less coherent. The corresponding dissipation of the system quantitatively measured by the entropy production rate increases. This implies that the system is less stable under fluctuations. We identified some key structural elements for wirings of the cell cycle network responsible for the change of the barrier height and therefore the global stability of the system through the sensitivity analysis. The results are in agreement with recent experiments and also provide new predictions.
Keywords: oscillation, barrier height, entropy production and dissipation, sensitivity, global stability
Understanding the global stability and function of cellular networks is a grand challenge to the current systems biology. The conventional way of describing the networks in terms of either deterministic or stochastic [due to the intrinsic fluctuations from a finite number of the molecules in the cell and external fluctuations (1, 2)] chemical kinetics often probes only the local properties of the network (3, 4). The global nature is hard to see. When the state space of the cellular network is huge, then a related challenge is to understand globally how the seemingly infinite number of genotypes can result in a finite of number of functional phenotypes. The probabilistic landscape description may provide an answer because the importance of each state can be discriminated from its associated weight. Functional states may correspond to higher probability of appearing and occupy low potential valleys (5–15). For example, a one-dimensional nonequilibrium potential under a periodic boundary condition (6) can be quantified (6). The multidimensional nonequilibrium potential under natural boundary conditions can also be quantified and explored (detailed discussions are in SI Appendix). The global stability of the functional states can be quantified through the topography of the underlying probabilistic landscape. The barrier heights between the functional basins may provide such a measure of the degree of difficulty to change from one functional state to another. The corresponding stabilities under different fluctuations, perturbations of wirings, and mutations of the network quantify robustness of the system (11–14).
The probability evolution of the cellular network can be mathematically determined by the probabilistic diffusion equation for environmental fluctuations and master equation for intrinsic statistical fluctuations due to the finite number of molecules in the cell. It is impossible to solve the equation exactly due to the large number of states involved. In this paper, we will use the self-consistent mean field approximation to approach a specific cellular network-budding yeast cell cycle. It can reduce the computational task from exponential to polynomial.
The cell cycle is the series of events that takes place in a cell leading to its division and duplication (replication) crucial for growth, proliferation, development, and death (16–23). The cell cycle consists of several distinct phases: G1 phase, S phase (synthesis), G2 phase (collectively known as interphase), and M phase (mitosis). Activation of each phase is dependent on the proper progression and completion of the previous one. In a budding yeast, the cell cycle can often be thought of as alternative “states” (G1 and S-G2-M) separated by two transitions (start and finish) (16).
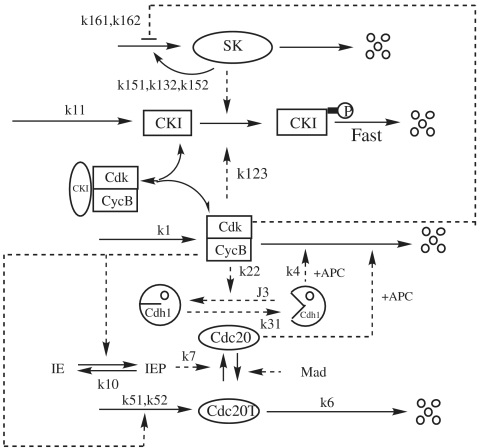
Regulation of the cell cycle involves processes crucial to the survival of a cell. The molecular events that control the cell cycle are ordered and directional. Two key classes of regulatory molecules, cyclins and cyclin-dependent kinases (CDKs), determine a cell’s progress through the cell cycle. There are four kinds of main proteins in the cell cycle process: CDKs including cyclin B (clb2, clb5); enemies including CKIs (Sic1, Cdc6), Cdh1/APC (anaphase-promoting complex); start kinase (SK) including cln1,cln2, cln3; and exit proteins (EP) including Cdc20, Cdc14 (17, 23). At the core of the cell cycle is a hysteresis loop from the fundamental antagonism between CDKs and APC: the APC represses CDK activity by destroying its cyclin partners, whereas cyclin/CDK dimers deactivate APC activity by phosphorylating Cdh1 (Fig. 1 is a highly simplified illustration of the complicated process (16–20). This mutual repression creates bistable states of the control system: a G1 state, with high Cdh1/APC activity and low cyclin/CDK activity, and an S-G2-M state, with high cyclin/CDK activity and low Cdh1/APC activity. For a newborn cell in the G1 phase, as the cell grows, the CycB (cyclin B) starts to increase and drives the cell into the S phase and mitosis (start). This turns on synthesis of Cdc20. As Cdc20 accumulates, Cdh1/APC is activated by Cdc20, and the cell exits mitosis (finish), with CycB destroyed. At cell division, the cell mass is reduced to two parts. The system is captured by the stable G1 steady state until the cell size once more increases to the critical size (17) with the continuation of nutrition supply.
Fig. 1.
The basic cell cycle engine in eukaryotic cells.
Fig. 1 introduces additional levels of control. Cdc20 is synthesized only during S-G2-M phase of the cell cycle. A Cdc20-activatory signal is illustrated by the mitotic process through intermediate enzyme (IE), and a Cdc20-inhibitory signal is generated by mitotic spindle abnormalities (through Mad pathways). Furthermore, CycB/CDK is inhibited by a binding partner (CKI) that is prevalent in the G1 phase of the cell cycle. The abundance of this inhibitor is controlled by phosphorylation reactions, which label CKI for rapid proteolysis. CKI can be phosphorylated by CycB/CDK, but there is little or no CycB/CDK activity in G1 cells. In order to leave G1 and enter the S phase, the cell must produce a “starter kinase,” of which its whole job is to phosphorylate and thereby remove CKI. SK is a dimer of CDK and a different type of cyclin (called Cln2 in budding yeast). The starter kinase is not inhibited by CKI and not destroyed by Cdh1, so when this alternative cyclin protein is produced in late G1, SK can help CycB/CDK to overcome its repressions.
We will study the budding yeast cell cycle network to explore the nature and topography of the underlying potential landscape for stable cell cycle oscillations. We will also study the robustness of the system (in terms of barrier height separating the basins of attractions of oscillations) under stochastic fluctuations, perturbations of the wirings, and mutations. We will identify some key structural elements critical for maintaining the function and stability of the network.
Results and Discussions
Cell Cycle Landscape.
We first start with the reaction scheme in Fig. 1 and write down explicitly the associated chemical reaction equations (16–23). We can then establish the corresponding probabilistic diffusion equation and use a self-consistent mean field method to solve the steady state probability distributions P in terms of the protein concentrations of the cell cycle network for a given diffusion coefficient D characterizing the environmental fluctuation level (5, 7–14). From the nonequilibrium steady state distribution, we can identify the underlying potential as U(x) = - ln P(x,t → ∞) = - ln Pss (when ). In this way, we map out the potential landscape (5, 7–14).
The potential landscape is multidimensional in concentration configuration space x and it is difficult to visualize U(x), so we select only two important protein concentration variables x1(CycB) and x3(Cdc20) for the process from eight in the network to calculate the landscape by integrating out the other six variables. This makes it possible for visualization.
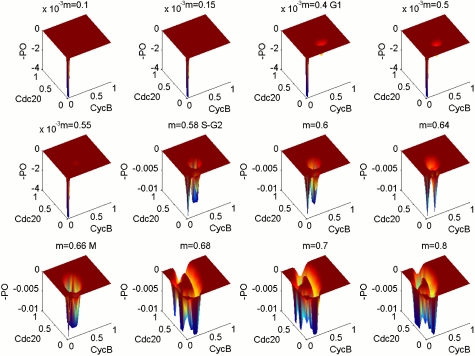
We monitor the cell mass to explore different stages of the cell growth, and we obtain different landscapes when cell mass (often represented as Mass) is increased from 0.1 to 0.8. Fig. 2 shows three-dimensional landscapes for different Mass at small environmental fluctuation strength D = 0.0005. We can see from the figure that at the initial location when the Mass is small (< 0.4), the system is monostable around G1. As mass starts to grow (beyond mass = 0.4), two basins of attraction appear for the underlying potential landscape, the system is in a weakly bistable state, and the bistable state is not symmetric and is still heavily biased towards G1. This still corresponds to a stable G1 steady state. As the mass increases further (beyond mass = 0.58), the second basin of attraction becomes deeper and more stable and G1 loses its stability. The system begins to enter into the S-G2 phase. The network dynamics starts to shift from bistable states towards oscillations (G1-S transition at cell mass = 0.58) (21, 22). Furthermore, from m = 0.6 to 0.8, the landscape changes from two basins of attraction to strong oscillation with a Mexican hat shape. This process is just like digging along a ring, first two holes, then three holes, etc., and at the last becoming a groove. From the shape of landscape, m = 0.66 should be the beginning of the M stage, and when m = 0.8, it is the anaphase of mitosis, with very stable oscillation. This gives a global picture and structure of the underlying landscape for the cell cycle network.
Fig. 2.
Figures show the three-dimensional landscape picture from monostable, bistable, to oscillation state; m = 0.4–0.55 are G1, m = 0.58–0.64 are S-G2, and m = 0.68–0.8 are M stage.
Flux Flow.
The probabilistic evolution of diffusion equation, , represents a conservation law of probability (local change is due to net flux in or out). And the probability flux vector J of the system in concentration space x is defined as .
In general, the dynamic driving force F can be decomposed into a gradient of a potential and a curl flow flux (14) (. Pss represents steady state probability distribution and potential and U is defined as U = - ln Pss. With detailed balance, the gradient of potential controls the underlying dynamics as the driving force. For nonequilibrium systems, the gradient of potential landscape and flux of probability determine the dynamics and global properties together. The dynamics of a nonequilibrium network can be described as a spiral, along the gradient direction, not like the case of the equilibrium state only following the gradient. It is similar to electrons moving in both electric and magnetic fields (14).
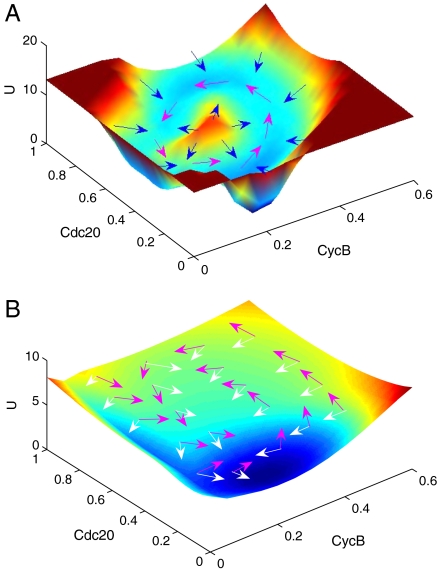
The detailed potential landscape for stable cell cycle oscillation at large mass (mass = 0.8) in the M phase has a closed ring or Mexican hat structure in the projection concentration space of x1 (CycB) and x3(Cdc20) as shown in Fig. 3A.
Fig. 3.
The potential energy landscape with m = 0.8 for diffusion coefficient D = 0.001 (A) with Mexican hat-like closed ring valley shape and D = 0.01 (B) with shallow shape. The magenta arrows represent the flux and white and blue arrows represent the force from negative gradient of the energy landscape.
We can see that the closed ring is around the deterministic oscillation trajectories. This means that the oscillation path or the closed ring have lower potential (and higher probability). Inside and outside the closed ring the potentials are both higher, which form a Mexican hat shape. The system is therefore attracted to the closed ring instead of a specific stable basin.
In Fig. 3A, the magenta arrows represent the steady state probability flux and white or blue arrows represent the force from the negative gradient of the potential landscape. We can see the direction of the flux near the ring is parallel to the oscillation path and circulates along the closed ring. The forces from the negative gradient of the potential landscape are insignificant along the closed ring and significant inside and outside the ring (see also SI Appendix for details). The direction of the negative gradient of the potential is almost vertical to the ring. Therefore, the landscape attracts the system towards the closed ring and is driven by the flux flow for oscillation along the closed ring valley. The nonequilibrium system is an open one with constant exchange of energy and information from outside environments. The nonzero flux is from the energy input from the nutrition supply.
As the diffusion coefficient D measuring the degrees of fluctuations increases, the potential landscape becomes flatter. Fig. 3B shows that the landscape transforms from a distinct ring valley into a shallower structure. This implies that there is more freedom to go from one state to another. Therefore, coherent oscillation is hard to maintain and the system becomes less stable. On the contrary, the weaker the fluctuation is, the more robust the oscillation is.
Notice that the mass acts as a control parameter to measure the progression of the cell cycle. We can think of a picture of the cell cycle as follows: Cell mass itself is also a measure of the energy intake from outside. When cell mass is small, the energy pump is insufficient to drive the state out of the basin of attraction. As the mass grows larger, energy intake is enough to drive the system to the next basins of attraction (digging another hole on the landscape). As the cell mass grows even larger, the energy intake is enough to push the system around through the flux flow and create a cycle of oscillations.
Robustness, Barrier Height, and Entropy Production Rate.
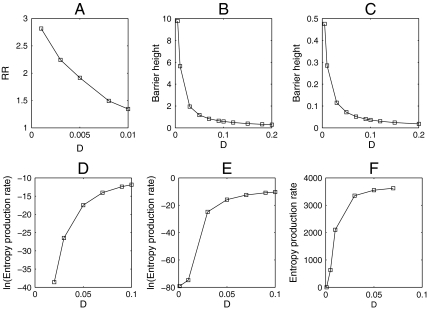
Having the underlying potential landscape, we can furthermore study the robustness of the cell cycle network by exploring the underlying landscape topography. For the monostable state, the slope of the basin attraction against local trapping termed as robustness ratio RR measures the degree of stability. For bistability, the barrier height between the basins of attraction and for oscillation, the barrier height from the minimum of the ring valley to the top of the barrier at the oscillation center becomes the quantitative measure of the degree of global robustness. We compute RR and barrier heights separately for the monostable state, bistable state, and oscillation.
For monostability with small cell mass in the initial stage of the cell cycle, we define RR (robustness ratio) for the network as RR = δU/ΔU. Here the δU is the difference between the global minimum Uglobal-minimum, and the average of U, 〈U〉, and ΔU is the half-width of the distribution of U. The δU measures the bias or the slope toward the global minimum (G1) of the potential landscape, whereas ΔU is a measure of the averaged roughness or the local trapping of the potential landscape. Fig. 4A shows the robustness measured by RR decreases with external noise increased. This means under fluctuations the system will become less stable.
Fig. 4.
(A–C) Robustness or barrier height versus external noise D for monostable, bistable, and oscillation separately. (D–F) Entropy production rate versus external noise D for monostable, bistable, and oscillation separately.
For a bistable condition in the middle range of cell mass, we define ΔUbarrier(barrier height) as U max - U min. Here, U max is the potential at the saddle point between the two stable basins of attraction (G1 and SG2M) and U min is the potential at one of the basins of attraction. Then, ΔUbarrier represents the potential energy barrier height between two stable basins of attraction. In Fig. 4B, we can see that as the fluctuation characterized by diffusion coefficient D decreases, the ΔUbarrier increases. The two basins of attraction become more stable since it is harder to go from one well to another. Therefore, large barrier heights and small fluctuations serve as the robustness and stability conditions for bistability (12).
For oscillation with a large cell mass, we define the barrier height in the same way, as U max - U min. U min is the potential minimum along the limit cycle attractor. U max is the potential at the local maximum point inside the limit cycle circle. In Fig. 4C, when the diffusion coefficient is small, the barrier height associated with escaping from the limit cycle attractor is higher. The resulting limit cycle attractor becomes more stable because it is more difficult to go from the ring to the inside or outside. Thus, small fluctuations and a large barrier height leads to robustness and stability in the oscillatory protein network (14). As we can see, the barrier height becomes a quantitative measure of global stability and degree of robustness of the cell cycle network.
In addition, we can compute the entropy production rate for different fluctuations (24). Because the entropy production is a feature of the global properties of the network combining both the effects of landscape and flux just like the heat generated from the voltage and current of an electrical circuit, we can use it to analyze global features of the network. In Fig. 4 D, E, and F, we plotted the entropy production (per unit time) or the dissipation cost of the network in steady state for monostable state, bistable state, and oscillation regime versus diffusion coefficient D. We can see that the entropy production rate decreases as the diffusion coefficient D decreases, which may reflect the fact that the fewer perturbations make the system more robust and cost less energy in the mean time. This might provide a design criterion, which will optimize the connections for the network (13).
Period and Amplitude and Coherence for Oscillation.
We also show the distributions of the period and amplitude of oscillations for x1(CycB) for different fluctuations (SI Appendix). We can see that, when the fluctuations increase, the distribution of period and amplitude becomes more scattered, and the standard deviation σ of period and amplitude from the mean increases, which means more possible values of the period and amplitude of oscillations can appear. This implies that less fluctuations corresponding to a more stable network make more coherent oscillations with asingle period and amplitude rather than spread periods and amplitudes.
The robustness of the oscillation can be quantified further by the phase coherence ξ, which is a measure of the degree of periodicity of the time evolution of a given variable (SI Appendix) (25). The larger of ξ value represents the more periodic evolution, in the presence of fluctuations. ξ decreases when the diffusion coefficient increases. This means larger fluctuations tend to destroy the coherence of the oscillations and further decrease the stability of the system.
Analysis of Sensitivity.
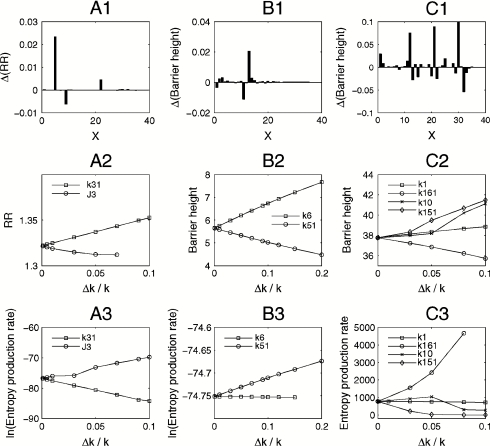
We did an analysis of sensitivity to uncover the local key connections or wirings of the cell cycle network responsible for stability of monostability, bistability, and oscillation. This is through changing the chemical reaction rate constants to see their effects on robustness ratio RR (monostable state), and barrier heights (bistable state and oscillation). Fig. 5 A1, B1, and C1 show effects of rate constants on the robustness for monostability, bistability, and oscillation separately. We chose some top important rate constants and then studied the effects of these parameters on robustness and entropy production rate of the system, which are shown in Fig. 5 A2–A3, B2–B3, and C2–C3.
Fig. 5.
(A1), (B1), (C1) Effects of rate constants on the robustness for monostable, bistable, and oscillation separately. (A2, A3) Effects of parameter k31 and J3 to RR and entropy production rate for monostable. (B2, B3) Effects of parameter k51 and k6 to barrier height and entropy production rate for bistable. (C2, C3) Effects of parameter k1, k16, k10, and k151 to barrier height and entropy production rate for oscillation. (A1), (B1), (C1) x axis: 1:k1, 2:k21, 3:k22, 4:k23, 5:k31, 6: k32, 7:k41, 8:k4, 9:J3, 10:J4, 11:k51, 12:k52, 13:k6, 14:J5, 15:k7, 16: k8, 17:J7, 18:J8, 19:Mad, 20:k9, 21:k10, 22:k11, 23:k121, 24:k122, 25:k123, 26:Keq, 27:k131, 28:k132, 29:k14, 30:k151, 31:k152, 32:k161, 33:k162, 34:J15, 35:J16. (A2, A3), (B2, B3), (C2, C3) x axis: Δk/k represents the percent of parameters increased.
For monostability with small cell mass, we can see that the increase of k31 and decrease of J3 makes the network more stable. By checking the network, we found that the increase of k31 and decrease of J3 at monostability will activate Cdh1 (Enemies). Cdh1 represses CycB/CDK, which must increase the concentration in order to leave G1 and enter the S phase as seen in previous studies (17). So the activation of Cdh1 makes the network inclined to stay at the monostable (G1 state) state. Therefore, increasing k31 makes monostability (G1 state) more stable, and it is opposite for J3.
For bistability with a middle range of cell mass, we found two important rate constants, k6 and k51, and analyzed their effects on barrier height and the entropy production rate. These are shown in Fig. 5 B2–B3. From these figures we can see that the increase of k6 makes the system more stable, and the increase of k51 makes the system less stable. Checking the network, we found that at bistability k6 and k51 repress and activate Cdc20, respectively. From formal discussions and previous studies (26–29), the presence of Cdc20 makes Cdh1 (Enemies) more active and represses CycB/CDK and cell exits miosis (G1 state again). This means that the system is inclined to be in the G1 state (monostable state) when Cdc20 increases, making the bistable state less stable. Therefore, the increase of k51 makes the bistable state less robust, and the increase of k6 strengthens the bistable state.
For oscillation with a large cell mass, we explored the effects of k1, k10, k151, and k161 on the stability and dissipations of the system (Fig. 5 C2–C3). From the figures we can see that increasing k1, k10, and k151 augments the robustness and causes less dissipations of the system, while it is the opposite for parameter k161. From the wiring diagram, k1 for oscillation increases CycB/CDK and k161 inactivates SK (cln2). SK weakens Enemies (cdh1,sic1), allowing CycB/CDK activity to rise. So inactivation of SK means CDK is deactivated and the oscillation system is inclined to be unstable. Therefore, the increase of k161 makes oscillation less stable (30, 31). For the same reason, since k1 increases CycB, the increase of k1 makes oscillation more stable (17, 23). Some experimental explorations have found the importance of CycB in both G1 and mitosis stages (31). As far as k10 is concerned, it represses production of IEP(x5) and further deactivates Cdc20(x3) since IEP activates Cdc20. Cdc20 increases Cdh1 (Enemies) and then decreases CycB/CDK. So increasing k10 activates CycB/CDK and makes the oscillation system more stable (32). For parameter k151, it is related to a positive feedback loop of SK (cln2), so increasing k151 makes oscillation more stable (23, 26–29, 33, 34).
Furthermore, we studied the effect of a positive feedback loop in the network on the stability of the system. The rate constants related with a positive feedback loop include k152, k132, k22, k4, and k123. See SI Appendix for the changes of the barrier height with respect to the changes of these parameters. We can see that when these rate parameters related to a positive feedback loop increase, the corresponding barrier height of the landscape increases. This means that the network becomes more stable. It demonstrates that positive feedback loops provide the network system with greater robustness and reliability. This is consistent with some recent studies (23, 26–29, 33, 34). In addition, k152 and k132 are positive feedback parameters related with Cln2 (SK). This result is in agreement with recent experimental studies, which showed that positive-feedback-amplified expression of Cln1, Cln2 drives stable budding and rapid, coherent regulon expression (26).
Methods
There have been intensive explorations on the cell cycle regulatory system for budding yeast (Saccharomyces cerevisiae) (17, 19, 20). See details in SI Appendix.
Our method is to uncover the potential landscape. The statistical nature of the chemical reactions can be captured by the corresponding diffusion equation, which describes the evolution of the networks probabilistically. It is hard to solve a diffusion equation due to its inherent huge dimensions. We therefore used the self-consistent mean field approximation to reduce the dimensionality (7, 12, 13). In this way, we could follow the time evolution and steady state probability of the protein concentrations and finally map out the potential landscape, which is closely associated with steady state probability distribution.
Self-Consistent Mean Field Approximation.
The P follows probabilistic diffusion equation: P(X1,X2, ...,Xn,t) where X1,X2,… is the concentration of proteins. We expected to have a N-dimensional partial differential equation, which is not feasible to solve since if every variable can have M values, then the dimensionality of the system becomes MN. Following a mean field approach (7, 12, 13), we split the probability into the products of individual ones: and solve the probability self-consistently. Now the degrees of freedom are reduced to M × N. Therefore the problem is computationally tractable from exponential to polynomials.
Although self-consistent mean field approximation reduce the dimensionality of system, it is still hard to solve diffusion probability directly. However the moment equations are relatively easy to obtain. In principle, once we know all moments, then we can construct probability distribution, but in many cases, we cannot get all moments. We may start from moment equations and then we may simply assume specific probability distribution based on physical argument, which means we give some specific relation between moments (12, 13). Here we use gaussian distribution as an approximation (5, 35), and then we need two moments, mean and variance.
When diffusion coefficient D is small, the moment equations could be approximated to (5, 35)
| [1] |
| [2] |
Here, x, σ(t), and A(t) are vectors and tensors, and AT(t) is the transpose of A(t). The matrix elements of A are . According to this equation, we can solve x(t) and σ(t). We consider here only the diagonal element of σ(t) from the mean field splitting approximation. Therefore, the evolution of distribution for one variable could be obtained using the mean and variance by Gaussian approximation:
| [3] |
The total probability is the product of probability for each individual variable from the mean field splitting approximation. Finally, once we have the steady state probability distribution, we can construct the potential landscape by the relationship U(x) = - ln P(x).
Conclusions
We explored the global nature of a budding yeast cell cycle network in terms of the potential landscape. To reduce the degrees of freedom of the system from exponential to polynomial, a self-consistent mean field approximation method was developed. We used the experimentally inferred rate parameters to study the network by computing RR (robustness ratio), barrier height, coherence, entropy production rate, etc. for both a simplified 8-variable model (19) and more sophisticated 38-variable cell cycle model (20) (details in SI Appendix).
The global shape of the underlying landscape of the oscillation cell cycle has a closed ring valley shape attracting the system down, and the curl flux (originated from energy input through nutrition supply) along the ring is the driving force for oscillation. We can see the cell cycle network can be globally characterized by the landscape topography. The barrier height between basins of attraction provides a quantitative measure of the stability. The network is more stable and oscillation is more coherent when there is less environmental fluctuations. The corresponding dissipation of the network is also reduced. This might provide a way of selecting the suitable parameter subspace of the cellular network, guarantee the robustness, cost less dissipations, and perform specific biological functions, which are useful for the network design.
To explore the different stages of cell growth, we uncovered different landscapes when cell mass increases from 0.1 to 0.8 for an 8-variable model. For small mass, the network is in monostable state G1 (energy intake is insufficient to drive the system out). When the mass increases (energy intake is sufficient), the network starts to become bistable (m≥0.4). For m = 0.6 to 0.8 (energy intake is sufficient), landscape changed significantly from bistable to oscillation. This process is like digging along a ring, first two holes, then three holes, etc., at last becoming a groove driven by the energy intake through nutrition supply. This provides an evolution of landscapes along the cell cycle process. Exploring the landscape of a cell cycle network may provide insights and methods to study what happens from G1 to S-G2-M in the cell cycle process.
Analysis of sensitivity provides one approach to study the influences of specific rate constants or wirings or mutations to robustness of the network. This helps us to identify the key structural elements or wirings that are responsible for the stability and function of the whole network. Some results are in agreement with experiments and others are unique insights and predictions for future experiments to verify.
We also found that increasing the perturbation levels of rates or wirings (details in SI Appendix) does not necessarily weaken the cell cycle stability from wild-type. Nature might be at the balance where the barrier height is just enough to maintain the stable oscillation while not so high to adapt external changes.
The methods in this paper can be applied to other complicated networks and also more realistic systems to explore the underlying global potential landscape and flux of biological networks.
Supplementary Material
Acknowledgments.
J.W. acknowledges support from the National Science Foundation Career Award. C.H.L. and E.K.W. are supported by the National Natural Science Foundation of China (Grants 90713022 and 20735003) and the 973 project 2009CB930100.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910331107/DCSupplemental.
References
- 1.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thattai M, Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 4.Huang CY, Ferrell JE. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1996;93:10078–10082. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu G. In: Stochastic Forces and Nonlinear Systems. Hao BL, editor. Shanghai: Shanghai Scientific and Technological Education Press; 1994. pp. 68–74. [Google Scholar]
- 6.Hatano T, Sasa S. Steady state themodynamics of langevin systems. Phys Rev Lett. 2001;86:3463–3466. doi: 10.1103/PhysRevLett.86.3463. [DOI] [PubMed] [Google Scholar]
- 7.Sasai M, Wolynes PG. Stochastic gene expression as a many-body problem. Proc Natl Acad Sci USA. 2003;100:2374–2379. doi: 10.1073/pnas.2627987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ao P. Potential in stochastic differential equations: Novel construction. J Phys A Math Gen. 2004;37:L25–L30. [Google Scholar]
- 9.Hornos JEM, et al. Self-regulating gene: An exact solution. Phys Rev E. 2005;72:051907. doi: 10.1103/PhysRevE.72.051907. [DOI] [PubMed] [Google Scholar]
- 10.Qian H, Reluga TC. Nonequilibrium thermodynamics and nonlinear kinetics in a cellular signaling switch. Phys Rev Lett. 2005;94:028101. doi: 10.1103/PhysRevLett.94.028101. [DOI] [PubMed] [Google Scholar]
- 11.Han B, Wang J. Quantifying robustness and dissipation cost of yeast cell cycle network: The funneled energy landscape perspectives. Biophys J. 2007;92:3755–3763. doi: 10.1529/biophysj.106.094821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K, Wang J. Potential energy landscape and robustness of a gene regulatory network: Toggle switch. PLOS Comput Biol. 2007;3:0565–0577. doi: 10.1371/journal.pcbi.0030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapidus S, Han B, Wang J. Intrinsic noise, dissipation cost, and robustness of cellular networks: The underlying energy landscape of MAPK signal transduction. Proc Natl Acad Sci USA. 2008;105:6039–6044. doi: 10.1073/pnas.0708708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Xu L, Wang EK. Potential landscape and flux framework of non-equilibrium networks: Robustness, dissipation and coherence of biochemical oscillations. Proc Natl Acad Sci USA. 2008;105:12271–12276. doi: 10.1073/pnas.0800579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prost J, Joanny JF, Parrondo JMR. Generalized fluctuation-dissipation theorem for steady-state systems. Phys Rev Lett. 2009;103:090601. doi: 10.1103/PhysRevLett.103.090601. [DOI] [PubMed] [Google Scholar]
- 16.Novak B, et al. Mathematical model of the fission yeast cell cycle with checkpoint controls at the G1/S, G2/M and metaphase/anaphase transitions. Biophys Chem. 1998;72:185–200. doi: 10.1016/s0301-4622(98)00133-1. [DOI] [PubMed] [Google Scholar]
- 17.Tyson JJ, Novak B. Regulation of the eukaryotic cell cycle: Molecular antagonism, hysteresis, and irreversible transitions. J Theor Biol. 2001;210:249–263. doi: 10.1006/jtbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 18.Kar S, Baumann WT, Paul MR, Tyson JJ. Exploring the roles of noise in the eukaryotic cell cycle. Proc Natl Acad Sci USA. 2009;106:6471–6476. doi: 10.1073/pnas.0810034106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen KC, et al. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen KC, et al. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battogtokh D, Tyson JJ. Bifurcation analysis of a model of the budding yeast cell cycle. Chaos. 2004;14:653–661. doi: 10.1063/1.1780011. [DOI] [PubMed] [Google Scholar]
- 22.Csikasz-Nagy A, et al. Analysis of a generic model of eukaryotic cell cycle regulation. Biophys J. 2006;90:4361–4379. doi: 10.1529/biophysj.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyson JJ, Novak B. Temporal organization of the cell cycle. Curr Biol. 2008;18:R759–R768. doi: 10.1016/j.cub.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian H. Mesoscopic nonequilibrium thermodynamics of single macromolecules and dynamic entropy-energy compensation. Phys Rev E. 2001;65:016102. doi: 10.1103/PhysRevE.65.016102. [DOI] [PubMed] [Google Scholar]
- 25.Yoda M, Ushikubo T, Inoue W, Sasai M. Roles of noise in single and coupled multiple genetic oscillators. J Chem Phys. 2007;126:115101–115111. doi: 10.1063/1.2539037. [DOI] [PubMed] [Google Scholar]
- 26.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 28.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijnen H, Landman A, Futcher B. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol Cell Biol. 2002;22:4402–4418. doi: 10.1128/MCB.22.12.4402-4418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln/Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surana U, et al. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 32.Lim HH, Goh PY, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 33.Tsai TY, et al. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos SD, Ferrell JE., Jr On the cell cycle and its switches. Nature. 2008;454:288–289. doi: 10.1038/454288a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Kampen NG. Stochastic Processes in Physics and Chemistry. Amsterdam: North-Holland; 1992. pp. 120–127. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.