Abstract
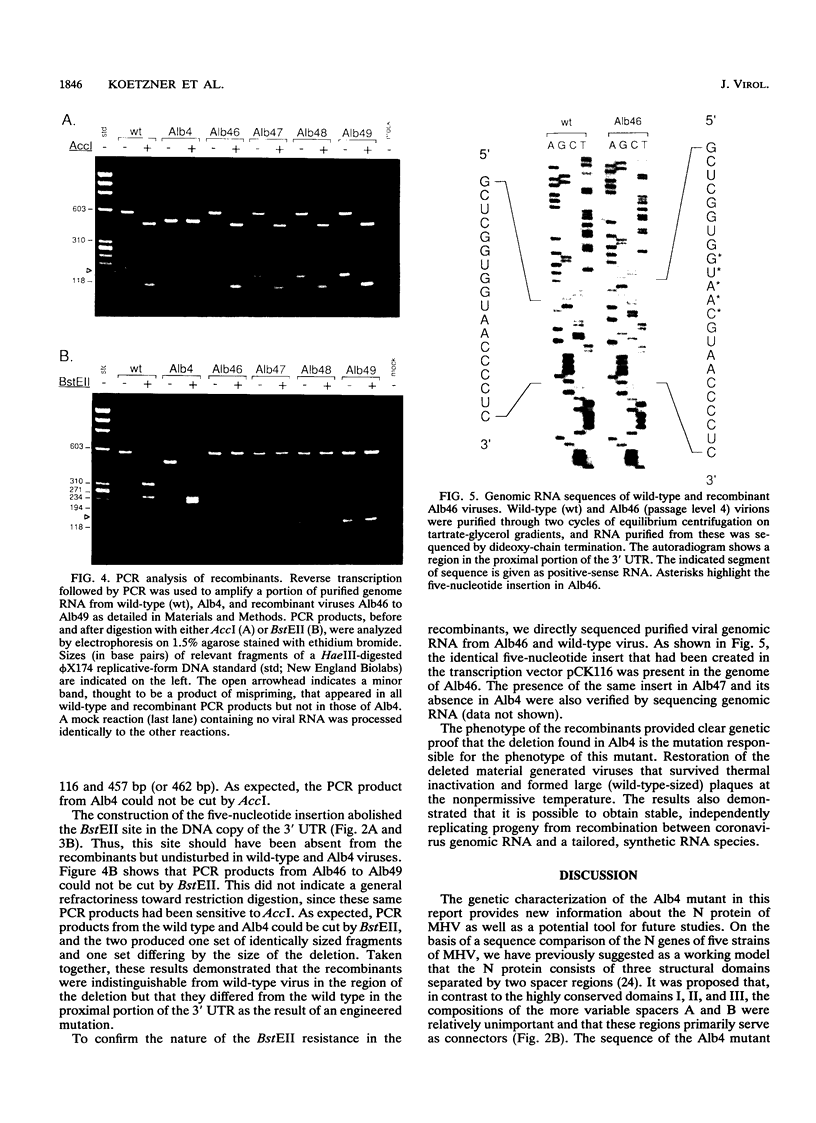
The genetic characterization of a nucleocapsid (N) protein mutant of the coronavirus mouse hepatitis virus (MHV) is described. The mutant, Albany 4 (Alb4), is both temperature sensitive and thermolabile. Analysis of the progeny of a mixed infection showed that the defective Alb4 allele is recessive to wild type, and its gene product is diffusible. The N protein of Alb4 was found to be smaller than its wild-type counterpart, and sequence analysis of the Alb4 N gene revealed that it contains an internal deletion of 87 nucleotides, producing an in-frame deletion of 29 amino acids. All of these properties of Alb4 made it ideal for use as a recipient in a targeted RNA recombination experiment in which the deletion in Alb4 was repaired by recombination with synthetic RNA7, the smallest MHV subgenomic mRNA. Progeny from a cotransfection of Alb4 genomic RNA and synthetic RNA7 were selected for thermal stability. Polymerase chain reaction analysis of candidate recombinants showed that they had regained the material that is deleted in the Alb4 mutant. They also had acquired a five-nucleotide insertion in the 3' untranslated region, which had been incorporated into the synthetic RNA7 as a molecular tag. The presence of the tag was directly verified, as well, by sequencing the genomic RNA of purified recombinant viruses. This provided a clear genetic proof that the Alb4 phenotype was due to the observed deletion in the N gene. In addition, these results demonstrated that it is possible to obtain stable, independently replicating progeny from recombination between coronavirus genomic RNA and a tailored, synthetic RNA species.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison R., Thompson C., Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner L. R., Keck J. G., Lai M. M. A clustering of RNA recombination sites adjacent to a hypervariable region of the peplomer gene of murine coronavirus. Virology. 1990 Apr;175(2):548–555. doi: 10.1016/0042-6822(90)90439-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner L. R., Lai M. M. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology. 1991 Nov;185(1):441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J. J., Kaesberg P. Genetic recombination between RNA components of a multipartite plant virus. 1986 May 29-Jun 4Nature. 321(6069):528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer C. W., Stenzler L., Fay N., Howell S. H. Nonmutant forms of the avirulent satellite D of turnip crinkle virus are produced following inoculation of plants with mutant forms synthesized in vitro. Virology. 1991 Jul;183(1):251–259. doi: 10.1016/0042-6822(91)90137-z. [DOI] [PubMed] [Google Scholar]
- Golub E. I., Kim H., Volsky D. J. Transfection of DNA into adherent cells by DEAE-dextran/DMSO method increases drastically if the cells are removed from surface and treated in suspension. Nucleic Acids Res. 1989 Jun 26;17(12):4902–4902. doi: 10.1093/nar/17.12.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Keck J. G., Matsushima G. K., Makino S., Fleming J. O., Vannier D. M., Stohlman S. A., Lai M. M. In vivo RNA-RNA recombination of coronavirus in mouse brain. J Virol. 1988 May;62(5):1810–1813. doi: 10.1128/jvi.62.5.1810-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J. G., Soe L. H., Makino S., Stohlman S. A., Lai M. M. RNA recombination of murine coronaviruses: recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J Virol. 1988 Jun;62(6):1989–1998. doi: 10.1128/jvi.62.6.1989-1998.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J. G., Jager E. J., Niesters H. G., van der Zeijst B. A. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine. 1990 Dec;8(6):605–608. doi: 10.1016/0264-410X(90)90018-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani S., Colonno R. J. In vitro synthesis of an infectious RNA from cDNA clones of human rhinovirus type 14. J Virol. 1985 Nov;56(2):628–632. doi: 10.1128/jvi.56.2.628-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obijeski J. F., Marchenko A. T., Bishop D. H., Cann B. W., Murphy F. A. Comparative electrophoretic analysis of the virus proteins of four rhabdoviruses. J Gen Virol. 1974 Jan;22(1):21–33. doi: 10.1099/0022-1317-22-1-21. [DOI] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. M., Masters P. S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990 Nov;179(1):463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981 Nov 20;214(4523):916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Sturman L. S., Eastwood C., Frana M. F., Duchala C., Baker F., Ricard C. S., Sawicki S. G., Holmes K. V. Temperature-sensitive mutants of MHV-A59. Adv Exp Med Biol. 1987;218:159–168. doi: 10.1007/978-1-4684-1280-2_20. [DOI] [PubMed] [Google Scholar]
- Sturman L. S., Holmes K. V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980 Jan;33(1):449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L. S., Holmes K. V. The molecular biology of coronaviruses. Adv Virus Res. 1983;28:35–112. doi: 10.1016/S0065-3527(08)60721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weiss B. G., Schlesinger S. Recombination between Sindbis virus RNAs. J Virol. 1991 Aug;65(8):4017–4025. doi: 10.1128/jvi.65.8.4017-4025.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]