Abstract
Background and Aims
Expected increases in world population will continue to make demands on agricultural productivity and food supply. These challenges will only be met by increasing the land under cultivation and by improving the yields obtained on existing farms. Genetic engineering can target key traits to improve crop yields and to increase production on marginal soils. Soil acidity is a major abiotic stress that limits plant production worldwide. The goal of this study was to enhance the acid soil tolerance of wheat by increasing its resistance to Al3+ toxicity.
Methods
Particle bombardment was used to transform wheat with TaALMT1, the Al3+ resistance gene from wheat, using the maize ubiquitin promoter to drive expression. TaALMT1 expression, malate efflux and Al3+ resistance were measured in the T1 and T2 lines and compared with the parental line and an Al3+-resistant reference genotype, ET8.
Key Results
Nine T2 lines showed increased TaALMT1 expression, malate efflux and Al3+ resistance when compared with untransformed controls and null segregant lines. Some T2 lines displayed greater Al3+ resistance than ET8 in both hydroponic and soil experiments.
Conclusions
The Al3+ resistance of wheat was increased by enhancing TaALMT1 expression with biotechnology. This is the first report of a major food crop being stably transformed for greater Al3+ resistance. Transgenic strategies provide options for increasing food supply on acid soils.
Keywords: Acid soil, aluminium resistance, TaALMT1 gene, transgenic wheat, Triticum aestivum, aluminum
INTRODUCTION
The world population is predicted to increase by >2 billion by 2050. Most of this increase will occur in developing countries such as those in sub-Saharan Africa where the population is expected to double. To support this growth, cereal production for food and animal feed will need to reach 3 billion tonnes per annum, an increase of 50 % on current production levels (FAO, 2009). The majority of these gains are expected to come from improving yields and increasing cropping intensity. However, an estimated 120 million ha of extra land are also likely to come into production, mostly in Africa and Latin America. Much of the land currently not in use suffers from chemical, physical or biological constraints, making it suitable for the production of a limited range of low value crops. While conventional plant breeding and improved practices have kept up with the demands of population increases in the past, all options, including transgenic approaches, should be considered to meet the challenges of the future (Hoisington, 2002; Bhalla, 2006).
Soil acidity is one of the important abiotic stresses limiting plant production. Almost 60 % of the soil in tropical and sub-tropical regions is acidic (von Uexküll and Mutert, 1995), making it a major limitation to food production in many developing countries. Acid soils present many stresses to plants, but it is their high concentration of soluble aluminium cations, especially Al3+, which is largely responsible for reducing root elongation, disrupting nutrient and water uptake, and increasing the susceptibility of plants to drought and heat stress. Many species have evolved mechanisms to resist Al3+ stress and these are generally divided into those that exclude Al3+ from the root and shoot tissues (resistance) and those that can safely accommodate Al3+ once it is taken up by the plant (tolerance). Genotypes within many species show a significant variation in their ability to cope with Al3+ toxicity (Taylor, 1988, 1991; Kochian et al., 2004), and this variation has been used by breeders to develop more Al3+-resistant lines (Garvin and Caver, 2003).
Soil acidity can be neutralized with the regular application of lime, but it can often take years to ameliorate sub-soil acidity, and these amendments are prohibitively expensive for farmers in many countries. Therefore, the combination of liming and the use of Al3+-resistant germplasm is a common management strategy for cropping acid soils. Among the common cereals, rye (Secale cereale) and triticale (×Triticosecale) are generally more tolerant of acid soils than common wheat (Triticum aestivum), whereas durum wheat (Triticum turgidum) and barley (Hordeum vulgare) perform very poorly on acid soils. Farmers with acid soils are often left with little choice but to grow feed crops such as rye and triticale even though wheat, durum and barley are more profitable.
Although Al3+ resistance is a multigenic trait in wheat (Garvin and Carver, 2003; Cai et al., 2008; Ryan et al., 2009), most of the phenotypic variation can be attributed to a single locus on chromosome 4DL which controls the Al3+-activated efflux of malate from the root apices (Delhaize et al., 1993a, b; Ryan et al., 1995). The malate anions protect the sensitive root apices by chelating the Al3+ and rendering it non-toxic. The TaALMT1 gene controlling this response encodes an Al3+-activated anion channel that is permeable to malate (Sasaki et al., 2004; Raman et al., 2005; Zhang et al., 2008).
It is now clear that other members of the ALMT family perform similar functions in other species including arabidopsis (Hoekenga et al., 2006), Brassica napus (Ligaba et al., 2006) and rye (Fontecha et al., 2007). Members of a second family of genes named the multidrug and toxic compound extrusion (MATE) family also contribute to the Al resistance in plants via organic anion efflux (Furukawa et al., 2007; Magalhaes et al., 2007; Wang et al., 2007; Liu et al., 2009). The evidence to date indicates that ALMT proteins facilitate malate efflux while the MATE proteins facilitate citrate efflux. In some species, such as arabidopsis and wheat, genes from both these families contribute to resistance (Liu et al., 2009; Ryan et al., 2009).
Enhanced Al3+ resistance is generally correlated with higher expression of these resistance genes, regardless of whether they are from the ALMT family or the MATE family (Raman et al., 2005; Hoekenga et al., 2006; Sasaki et al., 2006; Magalhaes et al., 2007; Fujii et al., 2009). In wheat, high TaALMT1 expression is associated with large tandem repeats of sequence in the promoter region, and it has been proposed that these repeats contain enhancer elements that increase expression of the gene (Sasaki et al., 2006). The strong correlation between gene expression and Al3+ resistance validates transgenic strategies to increase Al3+ resistance in plants by increasing TaALMT1 expression. When expressed in barley (Delhaize et al., 2004) and tobacco suspension cells (Sasaki et al., 2004; Zhang et al., 2008) TaALMT1 conferred an Al3+-activated efflux of malate which was associated with improved resistance to Al3+ stress. Transgenic barley plants showed a >2-fold increase in grain production on acid soil compared with the untransformed controls (Delhaize et al., 2009).
Common or bread wheat is one of world's major crops, providing one-fifth of the calories consumed worldwide (FAO, 2006). Although the variation found in Al3+ resistance of wheat has been successfully exploited by breeders, the level of resistance is still considerably lower than that of species such as rice and triticale. Transgenic approaches provide additional options for enhancing the acid soil tolerance of wheat, and this report describes experiments in which a wheat Al3+ resistance gene TaALMT1 was over-expressed in a sensitive wheat cultivar. We show that even with a relatively small number of successful transgenic events, this strategy is a viable option for increasing the acid soil tolerance of important crop species.
MATERIALS AND METHODS
Genotypes
The wheat gene TaALMT1 was over-expressed in the Triticum aestivum Bob White 26 ‘SH9826’ line (BW26). BW26 is an Al3+-sensitive wheat identified as being highly efficient for generating transgenic plants using the microparticle bombardment technique (Pellegrineschi et al., 2002). The genotype ET8 was included in all experiments as a positive control line for Al3+ resistance (Delhaize et al., 1993a).
Plasmid vectors
The pWUbi vector containing the TaALMT1 coding region [allele TaALMT1-1 (Sasaki et al., 2004)] used in the transformation experiments was the same as described by Delhaize et al. (2004). Plasmid pCMneoSTLS2 (Maas et al., 1997), containing the neomycin phosphotransferase gene (nptII), that confers resistance to geneticin (G418), was used as selectable marker.
Explant preparation and microparticle bombardment
Caryopses from BW26 plants were collected 14–16 d after anthesis and seeds were sterilized (20 % sodium hypochlorite). Immature embryos, 1·0–1·5 mm in width, were excised and placed onto an osmotic medium, MSM for 2–4 h. MSM consisted of MS-based salts and vitamins (Murashige and Skoog, 1962), 0·1 g L−1 myo-inositol, 150 g L−1 maltose, pH adjusted to 5·9 and solidified with 8 g L−1 bactoagar.
Prior to bombardment, 5 µg of pWUbi::TaALMT1 and 3 µg of pCMneoSTLS2 vectors were coated on 50 µL of sub-micron (0·6 µm) gold particles (BioRad). Embryos were bombarded using the helium-driven particle delivery system, PDS-1000/He (BioRad) with 900 psi rupture discs.
Culture conditions and recovery of transformed plants
Twenty-four hours after bombardment, the immature embryos were transferred to MSE medium for callus induction. MSE is comprised of MS salts and vitamins with 30 g L−1 sucrose, 2·5 mg L−1 2,4-D and is solidified with 3 g L−1 phytagel. After 14 d incubation in the dark, the calli were then transferred to regeneration medium, MSW. MSW consisted of MS salts and vitamins only, solidified with 8 g L−1 bactogar and supplemented with 50 mg L−1 geneticin (G418) as the selective agent.
Cultures were transferred to fresh MSW medium with geneticin every 2 weeks over 8–12 weeks. Geneticin-resistant shoots with root formation were then transferred to small soil pots and acclimatized in a high-humidity misting unit in the glasshouse. After 2–3 weeks, plants were transferred to pots of soil and T1 seed were harvested from mature transgenic plants.
A total of 876 wheat embryos were isolated and bombarded over seven separate sessions. Transformation efficiency varied from 0 to 6·31 % (Supplementary Data Table S1, available online) among the sessions, with an average efficiency of 1·5 %.
DNA extraction and PCR tests for transformation
Total DNA was extracted using an SDS-based protocol (Martienssen et al., 1989). After the quantification in agarose gel, 100 ng of DNA were used in PCRs with 1× HotStar Taq Master Mix (Qiagen), 10 µm of each primer and 1× Q solution (Qiagen) (only for primers Neo5R and Neo3F). The primers FgpWUbi-F (TGCAGCATCTATTCATATGC) and TR-2 (GATGGTGCCCACCATCTCG) were used for amplification of a TaALMT1 289 bp fragment originated from the pWUbi::TaALMT1 vector. Since the FgpWubi-F primer targets the maize ubiquitin promoter (intron) region of the pWUbi vector, the endogenous TaALMT1 is not amplified. The primers Neo3F (GGCTATTCGGCTATGACTG) and Neo5R (ATCGGGAGCGGCGATACCGTA) were used for the amplification of a nptII 738 bp fragment originated from the pCMneoSTLS2 vector. The amplification program used for both primer pairs was 95 °C for 15 min followed by 40 cycles of 94 °C for 45 s, 55 °C for 45 s and 72 °C for 1 min.
Malate efflux experiments
Seeds were grown in nutrient solution (500 µm KNO3, 500 µm CaCl2, 500 µm NH4NO3, 150 µm MgSO4, 10 µm KH2PO4, 2 µm FeCl3, 11 µm H3BO3, 2 µm MnCl2, 0·35 µm ZnCl2, 0·2 µm CuCl2, pH 4·3) for 4 d with natural light and temperature. A total of 24–30 root apices (4 mm), collected from 6–10 different germinated seeds, were excised in Petri dishes and divided into three tubes (8–10 apices per tube). Malate efflux from the excised root apices was measured as described by Ryan et al. (1995) with modifications.
Analysis of TaALMT1 expression by real-time qRT–PCR
The TaALMT1 expression analysis was based on the procedure previously described by Delhaize et al. (2004), with modifications. The total RNA from 8–10 root apices stored at –80 °C following prior malate efflux experiments was purified using the RNeasy minikit (Qiagen). The RNA purification included a DNase step to eliminate any genomic DNA contamination. The cDNA was prepared using 1 µg of total RNA, 1× RT buffer, 10 mm of each dNTP, 500 ng of oligo(dT)15 primer, 0·2 m dithiothreitol (DTT) and 1 U of SuperScript II Reverse Transcriptase (Invitrogen). The volume of each reaction was adjusted to 20 µL and incubated at 25 °C for 5 min followed by 42 °C for 60 min. An RNaseH degradation step was performed for 30 min at 37 °C. TaALMT1 expression relative to the endogenous gene [glyceraldehyde phosphate dehydrogenase (GAPDH)] was determined by real-time quantitative RT–PCR on a Rotor-Gene 3000 Real Time Cycler (Corbett Research, Australia). The samples for qPCR were prepared in a 10 µL final volume containing 5 µL of SYBR Green JumpStart Taq Ready Mix (Sigma), 0·5 µL of primer solution (mix 1 : 1 from primers Forward and Reverse at 5 µm each) and 4·5 µL of cDNA diluted 1 : 50. The primers RTFwd3 (CGTGAAAGCAGCGGAAAGCC) and RTRev3 (CCCTCGACTCACGGTACTAACAACG) were used to amplify the TaALMT1 transcript, and the primers RiwhGAPDH-R (TCAGACTCCTCCTTGATAGC) and RiwhGAPDH-F (GTTGAGGGTTTGATGACCAC) were used to amplify the GAPDH transcript, used as the endogenous reference. Thermocycling conditions were 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s, 55 °C for 20 s and 72 °C for 40 s. At the end of the amplification steps, the reactions were incubated at 40 °C for 5 min and 55 °C for 1 min, followed by a melting curve program (increase of 1 °C from 55 ° to 99 °C holding for 5 s at each temperature).
Relative root growth (RRG) experiments in hydroponic culture
Seeds were pre-germinated in the dark for 2 d at 4 °C and 2 d at 28 °C. Once the length of the longest root was measured, the seedlings were grown over an aerated nutrient solution (as above) containing different concentrations of AlCl3 for 4 d with natural light. The seedlings were then removed from the nutrient solution and the length of the longest root was measured again. The RRG was estimated as (net root growth in Al treatment/net root growth in control solution) × 100. To account for the accumulation of errors associated with deriving RRG, the errors were calculated as follows: SERRL = RRG [(SEx /x)2 + (SEy/y)2]1/2 where x and y represent the mean net root length in the control treatment and the mean net root length in the Al treatment, respectively. Statistical analysis was based on the average RRG in all lines, independent of Al3+ concentration and experiment. A cluster analysis was employed and all the lines that were similar to BW26 were discarded. An analysis of variance (ANOVA) with 5 % significance was performed on the remaining lines using the R software.
Scoring homozygous T2 families
Homozygous T2 families were identified by a rapid screen for Al3+ resistance. For each T2 family, 16–25 germinated seeds were placed in 500 mL conical flasks containing 70 mL of nutrient solution (pH 4·3) and 30 µm AlCl3. The flasks were capped with aluminium foil and placed on a platform shaker (100 rpm). Solutions were replaced daily. Aluminium resistance of each seedling in each family was scored after 5 d by assessing root length and by examining the root apices for tissue damage under a dissecting microscope. Families in which all the seedlings showed relatively good root growth with undamaged apices were potentially homozygous for the transgene.
Southern analysis
Genomic DNA (10 µg) was isolated from BW26 and nine T2 lines potentially homozygous for the transgene based on an Al3+ resistance assay. Following an overnight digestion with BamHI, the DNA was run on a 1 % agarose gel and transferred to a PALL Biodyne B membrane using the alkaline blotting procedure. The membrane was probed with a 32P-labelled 946 bp fragment targeting the maize ubiquitin intron included in the binary vector.
Soil experiments
Short-term growth experiments compared the root growth of three T2 lines and control plants on a acidic red ferrosol obtained from the Robertson region of New South Wales in Australia (34°35′S, 150°36′E). The pH of an 0·01 m CaCl2 extract was 4·33 and the exchangeable Al comprised 21 % of total cations. Half of this soil was left unamended and half was limed at a rate of 5 g CaCO3 kg−1 dry soil to raise the pH and reduce Al3+ toxicity. Liming increased the pH to 5·18 and reduced the soluble Al to <1 % of the unamended soil. Field capacity was determined to be 271 mL water kg−1 dry soil. Soil was wetted to 90 % field capacity and 622 g added to pots (8 cm diameter and 14·5 cm high) and arranged randomly on benches in a naturally lit glasshouse. Pre-germinated seeds were planted, two per pot at 2 cm depth, after first measuring the length of their longest root. Pots were maintained at 90 % field capacity, and after 4 d the plants were gently removed from the pots. The length of the longest root on each plant was measured a second time. The RRG of each wheat line was calculated by dividing the root lengths in the acid soil by lengths in the limed soil. The errors accounted for the cumulative errors (see above). Statistical analysis for RRG used a one-factor ANOVA where plant line was the factor.
For the long-term soil experiments three seedlings of BW26, ET8 and T2_4·4 were grown in pots (20 cm diameter and 25 cm high) containing 6 kg of the unamended acidic soil. Pots were arranged randomly in a naturally lit glasshouse and growth continued for 31 d. Roots were washed free of soil and the longest root on each plant was measured with a ruler. The roots were then scanned for total root length and root diameter using the software package WinRHIZO Pro v. 2002c (Regent Instruments Inc.). Dry weight of the roots and shoots was determined after drying at 65 °C for 48 h.
RESULTS
Generation of T0 plants
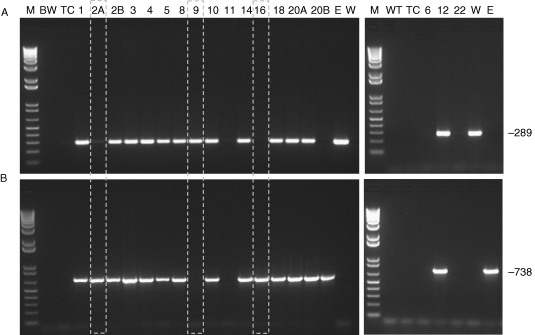
Of the 18 primary transformed (T0) plants recovered from the tissue culture, PCR determined that 13 were positive for the pWUbi::TaALMT1 vector (T0_1, T0_2B, T0_3, T0_4, T0_5, T0_8, T0_9, T0_10, T0_12, T0_14, T0_18, T0_20A and T0_20B). The primers used spanned the ubiquitin intron in the pWUbi vector and the TaALMT1 cDNA which enabled the transgene to be distinguished from the endogenous TaALMT1 gene (Fig. 1A). A second PCR targeting the pCMneoSTLS2 vector (providing the antibiotic resistance) revealed that T0_9 did not contain the selectable marker (Fig. 1B). These PCRs identified two additional lines, T0_2A and T0_16, that possessed the selectable marker but not the pWUbi::TaALMT1 vector.
Fig. 1.
Detection of transgenic lines. (A) Amplification of a 289 bp fragment specific to the inserted TaALMT1 copy. The primer pairs do not amplify the endogenous TaALMT1 as can be seen in the negative controls including BW26 (BW) and a tissue culture control (TC; plants regenerated in tissue culture but not transformed). (B) Amplification of the 738 bp fragment specific to the nptII gene from the selectable marker. M denotes the DNA size ladder while E and W indicate the vectors pCMneoSTLS2 (selectable marker) and pWUbi::TaALMT1, respectively. Numbers indicate the different T0 transformed lines recovered from the tissue culture. Dotted boxes represent lines containing only the selectable marker (T0_2A and T0_16), and a line containing the TaALMT1 transgene but not the selectable marker (T0_9).
Analysis of the T1 plants
T1 seed were harvested from the T0 plants, and the T1 seedlings were analysed for TaALMT1 expression, malate efflux and Al3+ resistance. All experiments included the parental cultivar BW26 and a standard Al3+-resistant line ET8. Relative TaALMT1 expression and Al3+-activated malate efflux from root apices were measured over two experiments (Table 1). The relatively large errors are expected since these T1 plants will be segregating for one or more copies of the transgene. This segregation was confirmed by scoring 16–21 seeds from each of the T1 lines for the presence of pWUbi::TaALMT1 with PCR (data not shown). This also identified plants that lacked the transgene (null segregants). The relationship between TaALMT1 expression and malate efflux among the T1 lines is shown in Supplementary Data Fig. S1A (available online).
Table 1.
TaALMT1 expression and malate efflux in the T1 lines
| Experiment | Wheat lines | TaALMT1 expression (arbitrary units) | Malate efflux (nmol apex−1 h−1) |
|---|---|---|---|
| 1 | BW26 | 0·033 ± 0·001 | 0·3 ± 0·2 |
| ET8 | 0·159 ± 0·005 | 1·1 ± 0·3 | |
| T1_1 | 0·138 ± 0·001 | 1·7 ± 0·1 | |
| T1_4 | 0·029 ± 0·005 | 0·4 ± 0·1 | |
| T1_8 | 0·051 ± 0·010 | 0·4 ± 0·1 | |
| T1_14 | 0·056 ± 0·002 | 1·0 ± 0·2 | |
| T1_18 | 0·077 ± 0·012 | 1·0 ± 0·7 | |
| T1_20B | 0·134 ± 0·018 | 0·9 ± 0·6 | |
| 2 | BW26 | 0·016 ± 0·000 | 0·10 ± 0·10 |
| ET8 | 0·125 ± 0·004 | 0·60 ± 0·10 | |
| T1_2B | 0·083 ± 0·000 | 0·68 ± 0·60 | |
| T1_3 | 0·056 ± 0·003 | 0·36 ± 0·20 | |
| T1_5 | 0·035 ± 0·002 | 0·51 ± 0·40 | |
| T1_9 | 0·022 ± 0·001 | 0·10 ± 0·02 | |
| T1_10 | 0·019 ± 0·002 | 0·10 ± 0·04 | |
| T1_20A | 0·049 ± 0·004 | 0·50 ± 0·30 | |
Expression in the root apices was estimated by qRT–PCR using the GAPDH gene as an internal reference. Data are the mean and standard error of three biological replicates. Malate efflux was measured from excised root apices in the presence of 50 µm AlCl3. Data show the mean and standard error (n = 4).
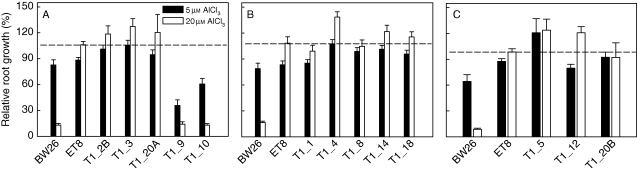
Aluminium resistance of all the T1 lines was estimated over three experiments by measuring net growth in nutrient solution containing 0, 5 or 20 µm AlCl3 and calculating RRG (Fig. 2). Null plants were excluded from the these measurements and so all plants were either hemizygous or homozygous for the transgene. At 5 µm AlCl3 most T1 lines showed similar resistance to BW26, but, at 20 µm, AlCl3 resistance in all T1 lines except T1_9 and T1_10 was significantly greater than that of BW26 and similar to the resistant line ET8.
Fig. 2.
Aluminium resistance of the T1 lines. Al3+ resistance was estimated by measuring relative root growth in nutrient solution containing AlCl3. The 13 T1 transgenic lines were tested over three separate experiments shown in (A–C). Control lines BW26 and ET8 were included in every experiment. Net root growth in 5 and 20 µm AlCl3 (pH 4·3) treatments was calculated relative to the net growth in the zero Al3+ control treatment. The dotted line indicates the relative root growth of the ET8 line in 20 µm AlCl3 in each experiment.
Analysis of T2 plants
Nine T1 transgenic lines were selected to generate families of T2 seeds. Ten plants were grown from each of these nine lines, one of which was a null (previously determined by PCR) and the other nine plants were positive for the transgene (either homozygous or hemizygous). Likely homozygous T2 families from each line were then identified by scoring the individual seeds in each family for Al3+ resistance in a rapid screen (Supplementary Data Table S2). The transgene copy number in these T2 families was then determined by Southern blot (Supplementary Data Fig. S2). The appearance of two bands in the untransformed control plants of BW26 complicated this assessment. Nevertheless, most T2 families had multiple inserts despite the segregation ratios for Al3+ resistance among T2 families in Supplementary Data Table S2 suggesting otherwise. It is possible that the multiple transgenes segregate as a single locus if they become integrated close to another in the wheat genome. Alternatively each of the transgenes might be expressed to different levels so that one drives most of the phenotype. Transgene copy number becomes an important consideration if these transgenic lines are used in crosses to increase the Al3+ resistance of other elite cultivars since multiple inserts makes introgression more difficult.
Malate efflux from the root apices of the nine homozygous lines was 3- to 4-fold greater than that of the parental cultivar BW26 and most were similar to ET8 (Table 2). Quantitative RT–PCR confirmed that TaALMT1 expression in the root apices of these T2 plants remained greater than in those of BW26, whereas the expression level in a null-segregant line included was similar to that of BW26 (Table 2, Supplementary Data Fig. S1B).
Table 2.
TaALMT1 expression and malate efflux from T2 lines
| Wheat lines | TaALMT1 expression (relative to BW26) | Malate efflux (nmol apex−1 h−1) |
|---|---|---|
| Controls | ||
| BW26 | 1·0 | 0·4 ± 0·1 |
| ET8 | 6·7 | 1·7 ± 0·2 |
| T2_1-19 (null) | 0·7 | 0·5 ± 0·1 |
| T2 lines | ||
| T2_1·6 | 5·0 | 2·1 ± 0·2 |
| T2_2B.1 | 2·3 | 2·2 ± 0·2 |
| T2_3·4 | 1·7 | 1·6 ± 0·1 |
| T2_4·4 | 1·8 | 2·0 ± 0·4 |
| T2_5·4 | 3·1 | 1·6 ± 0·1 |
| T2_8·5 | 0·8 | 1·2 ± 0·1 |
| T2_12·7 | 4·5 | 1·7 ± 0·2 |
| T2_18·1 | 5·1 | 2·1 ± 0·1 |
| T2_20A.8 | 5·9 | 1·7 ± 0·2 |
Expression in the root apices was estimated by qRT–PCR using the GAPDH gene as an internal reference. The values shown are expressed relative to the expression in BW26 which was included in each experiment. Data are the mean of three technical replicates. Malate efflux was measured from excised root apices in the presence of 50 µm AlCl3. Data show the mean and standard error (n = 4).
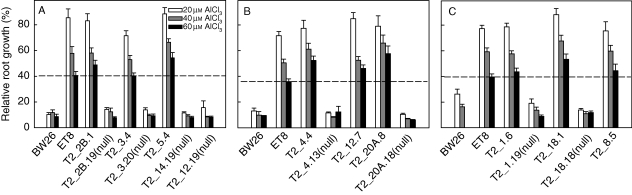
Aluminium resistance in the T2 homozygous lines was estimated from RRG following 4 d growth in nutrient solution containing 0, 20, 40 and 60 µm AlCl3. Relative root growth of the control line BW26 was inhibited to approx. 10 % in all the AlCl3 treatments (Fig. 3). All nine T2 lines were significantly more Al3+ resistant than BW26 across the three Al3+ concentrations while eight null lines performed similarly to BW26. T2_5·4 and T2_18·1 showed statistically greater Al3+ resistance than ET8 at the higher Al3+ concentrations.
Fig. 3.
Aluminium resistance in the T2 transgenic lines. Al3+ resistance was estimated in T2 lines by measuring relative root growth after 4 d in nutrient solution containing 20, 40 and 60 µm AlCl3 (pH 4·3) compared with zero AlCl3 controls. Eight null-segregant lines were also included. The dotted line indicates the relative root growth of the ET8 line in 60 µm Al3+ in each experiment.
Soil experiments
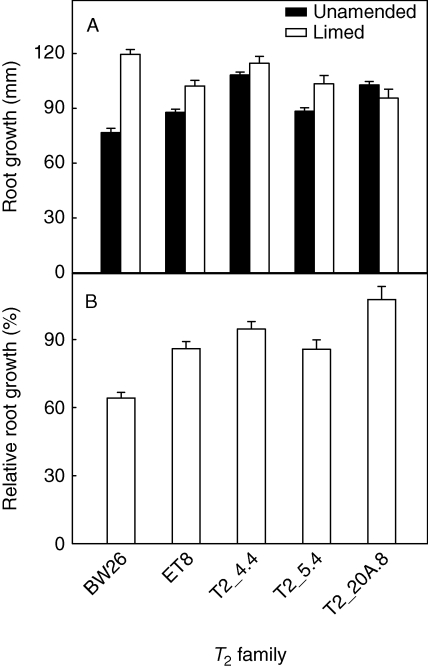
Root growth of BW26, ET8 and three T2 lines, T2_4·4, T2_5·4 and T2_20A.8, was compared in a short-term experiment using an acidic ferrosol soil with and without lime addition. Liming reduced the toxicity of the soil by raising the pH and reducing the concentration of toxic Al3+ cations. Root growth for the lines in each treatment and RRG (growth in acid soil relative to the limed soil) are shown in Fig. 4. All three T2 lines performed better than BW26, and line T2_20A.8 showed significantly greater resistance than ET8.
Fig. 4.
Aluminium resistance of T2 families in a short-term soil experiment. Seedlings of three T2 families homozygous for the transgene as well as BW26 and ET8 were grown on an acid soil with and without lime addition. Seeds were pre-germinated on wetted filter paper and planted in pots. After 4 d the plants were removed and the length of the longest root on each plant measured. (A) Net root growth on unamended acid soil and limed soil, as indicated. (B) Relative root growth of each wheat line was calculated as a percentage of net root growth on acid soil relative to the net root growth in limed soil. Data show the mean and standard error (n = 5) taking into account the cumulative errors.
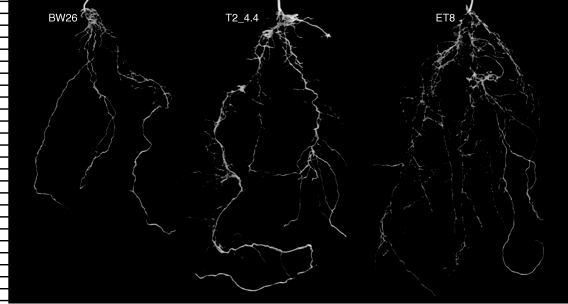
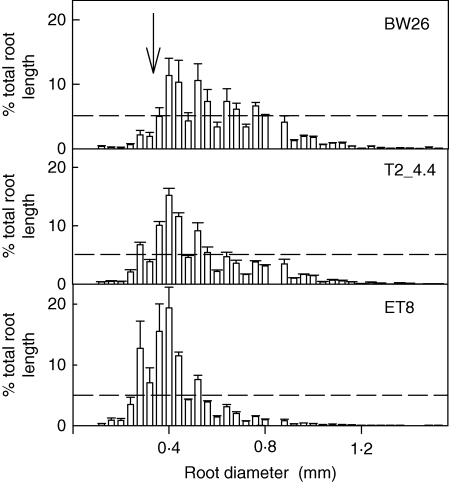
A longer term soil experiment compared the root growth of BW26, ET8 and a single homozygous line (T2_4·4) after 31 d in the unamended and unfertilized acid soil. T2_4·4 and ET8 plants had significantly larger root systems than BW26 based on both total root length and length of the longest root, and T2_4·4 had greater root dry weight than either of the other two lines (Table 3). Representative root systems from each genotype are shown in Fig. 5. Roots of BW26 also appeared to be generally thicker with shorter laterals than the other lines. This observation was confirmed when the distribution of root diameters was analysed on each plant (Fig. 6). As a percentage of the total root system, BW26 had significantly fewer roots with diameters ≤0·28 mm compared with T2_4·4 and ET8. This change in root morphology helps explain why, although the root system of BW26 appears smaller than that of ET8 in Fig. 5, their dry weights are similar. In contrast to the root data, shoot dry weight for T2_4·4 and BW26 plants was significantly greater than for ET8 (Table 3), which is consistent with previous reports suggesting that shoot growth can be relatively unaffected by the early stages of Al3+ toxicity.
Table 3.
Root and shoot analyses from the longer term soil experiment
| Wheat line | Total root length (mm) | Longest root (mm) | % Roots ≤0·28 mm diameter | Root d. wt (mg) | Shoot d. wt (mg) |
|---|---|---|---|---|---|
| BW26 | 300 ± 57a | 258 ± 23a | 10 ± 3a | 56 ± 4a | 95 ± 22a |
| T2_4·4 | 598 ± 21b | 451 ± 57b | 24 ± 1b | 77 ± 5b | 110 ± 9a |
| ET8 | 756 ± 128b | 360 ± 37b | 41 ± 13b | 58 ± 3a | 51 ± 3b |
Plants were grown in the acidic ferrosol soil for 31 d. Provided for each wheat line are the total root length, length of the longest root, the proportion of the total root system with diameters ≤0·28 mm and the total root and shoot dry weights. Data show the mean and standard error (n = 3). Means with the same letter are not significantly different from one another (P > 0·05).
Fig. 5.
Root systems collected from the long-term soil experiment. Plants were grown for 31 d in an acidic ferrosol soil and the soil gently washed from the roots. Photographs were taken of a representative root system from the replicates of each genotype BW26, ET8 and the transgenic T2 line T2_4·4. The intervals on the left-hand scale denote centimetres.
Fig. 6.
Distribution of root diameters in a T2 family and control plants grown in a longer term soil experiment. Plants of BW26, ET8 and the T2_4·4 line were grown for 31 d in the unamended acid soil. Roots were washed from the soil and analysed using the WinRHIZO scanner. The x-axis shows the classes of root diameter increasing by 0·04 mm. Data are the mean frequencies of roots in each diameter class (n = 3). The horizontal line represents the 5 % value and the arrow delineates the 0·28 mm diameter size class below which the data in Table 3 were calculated.
DISCUSSION
Over-expression of TaALMT1 in wheat conferred greater Al3+-activated malate efflux from the roots and improved Al3+ resistance which was maintained in the T1 and T2 generations. This is the first report of stably increasing the Al3+ resistance of a major food crop by genetic engineering. A previous report decribed the transformation of wheat with SbMATE, the Al3+ resistance gene from sorghum which encodes a citrate transporter (Magalhaes et al., 2007). However, the increased Al3+ resistance measured in T1 plants did not appear to be as robust as described for TaALMT1 (Magalhaes et al., 2007) and was not maintained in subsequent generations (L. V. Kochian, USDA-ARS, USA, pers. comm.). Other studies have increased the Al3+ resistance of plants by mutagenesis (Rounds and Larsen, 2008), by over-expressing genes involved in membrane physiology and stress responses (Ezaki et al., 2000; Basu et al., 2001; Stival da Silva et al., 2006; Ryan et al., 2007) or by targeting genes involved in organic anion synthesis and metabolism (de la Fuente et al., 1997; Koyama et al., 1999; Tesfaye et al., 2001; Anoop et al., 2003; Barone et al., 2008; Trejo-Téllez et al., 2010). Although a number of these studies showed enhanced Al3+ resistance in hydroponics, few have demonstrated enhanced tolerance to acid soils. Furthermore, transgenic plants modified for the expression of enzymes involved in malate and citrate synthesis have not consistently increased organic anion concentration in tissues or Al3+ resistance (Delhaize et al., 2001, 2003).
In the present study, nine of the 13 primary trangenic lines generated T2 lines with greater Al3+ resistance than the parental cultivar and some even exceeded the resistance of ET8 in nutrient solution and acid soil (Figs 3 and 4). It is possible that screening of a larger number of transgenic events will identify plants that exceed the Al3+ resistance of even the most resistant wheat cultivars currently available.
We used a constitutive promoter to drive TaALMT1 expression, but future studies could use promoters that restrict expression to the roots. In particular, the use of promoters and coding sequences derived from the species being transformed may enhance public acceptance of genetically modified crops. Furthermore, public acceptance of transgenic crops might also be improved if the selectable marker is absent from the cultivars that are finally commercialized. Including the transgene and selectable marker on different plasmids provides a means of removing the antibiotic resistance in subsequent generations because the two plasmids do not always insert into the same region of the genome.
When grown in acidic soil the T2_4·4 plants generated root systems that were considerably larger than those of the parental line BW26 and more similar to those of the Al3+-resistant genotype ET8 (Fig. 5). The transgenic line had a greater total root length, longer individual roots and a greater percentage of fine roots than BW26, all of which would benefit the plants by improving water and nutrient uptake on acid soil (Table 3).
The finding that the Al3+ resistance of several T2 lines exceeded that of ET8 highlights the potential of genetic engineering to increase the Al3+ resistance of wheat above what is naturally available and opens up the possibility of combining traits. What remains unclear is whether increasing the expression of TaALMT1 in cultivars that are already Al3+ resistant provides an even stronger phenotype or whether other factors become limiting. Interestingly, while the transgenic lines displayed a wide range of TaALMT1 expression levels, none exceeded those measured in ET8 (Table 2). Although the number of transgenic lines is small, this may indicate that there is an upper limit for TaALMT1 expression in wheat. It is unclear what the physiological basis for such a limit would be, but ongoing crosses between the T2 lines and other resistant genotypes will establish whether two loci of the TaALMT1 gene (one transgenic and the other endogenous) generate greater expression than a single locus. Furthermore, the constitutive ubiquitin promoter may extend TaALMT1 expression to additional cells in the root apex or other tissues not normally targeted by the native promoter. Efflux from these new cells might provide relatively greater Al3+ resistance to the plants, perhaps because they are nearer the root surface or because they protect new cells important for growth.
Additional benefits may also be gained by introgressing other Al3+ resistance loci into these transgenic lines. For instance, another mechanism for Al3+ resistance in plants relies on the efflux of citrate (Furukawa et al., 2007; Magalhaes et al., 2007; Wang et al., 2007; Ryan et al., 2009). In wheat, this mechanism confers weaker resistance than the malate efflux controlled by TaALMT1 and appears to be restricted to relatively few genotypes derived from Brazil (Ryan et al., 2009). Nevertheless over-expression of the candidate gene controlling this trait, TaMATE1, could offer other advantages because the stability of the Al3+–citrate complex is substantially stronger than that of the the Al3+–malate complex (Hue et al., 1986). In addition, it is possible to use related genes from barley and sorghum to generate transgenic wheat, although difficulties with this approach are noted above for SbMATE. An additive effect on Al3+ resistance might be achieved by over-expressing two or more genes at the same time, particularly if they control the efflux of different organic anions. If the supply of organic anions becomes limiting for efflux, genes involved in enhancing organic anion biosynthesis could be co-transformed with genes involved in organic anion transport, which might generate transgenics with even greater levels of resistance.
Chief among the challenges facing plant scientists and agronomists are practical options for increasing food production on land presently under cultivation as well as raising the productivity of marginal land which will need to be appropriated for food production in the future. Careful land management is essential for sustainability. Yet the expense of even basic practices such as the application of lime can be prohibitive to many farmers. These expenses are due not only to the costs of the commodities but also to their transportation and distribution. Transgenic strategies, such as the one described here, provide opportunities for sustaining and even increasing grain production on acid soils. We have demonstrated that over-expression of the TaALMT1 gene can increase the Al3+ resistance of wheat, a major food crop. This provides options for improving wheat yields on acid soils as well as helping to expand food production in the future to more marginal lands.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank AOB for supporting the participation of PRR in the “7th International Symposium on Plant-Soil Responses at Low pH” in Guangzhou in 2009. The scholarship of J. F. P. was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – process number 4151-07-0) and Embrapa (Empresa Brasileira de Pesquisa Agropecuária). We thank Muhammad Fahim, Michael Ayliffe and co-workers for supplying primers and advice on the Southern blot analysis.
LITERATURE CITED
- Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ. Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiology. 2003;132:2205–2217. doi: 10.1104/pp.103.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone P, Rosellini D, LaFayette P, Bouton J, Veronesi F, Parrott W. Bacterial citrate synthase expression and soil aluminum tolerance in transgenic alfalfa. Plant Cell Reports. 2008;27:893–901. doi: 10.1007/s00299-008-0517-x. [DOI] [PubMed] [Google Scholar]
- Basu U, Good AG, Taylor GJ. Transgenic Brassica napus plants overexpressing aluminium-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant, Cell and Environment. 2001;24:1278–1269. [Google Scholar]
- Bhalla PL. Genetic engineering of wheat – current challenges and opportunities. Trends in Biotechnology. 2006;24:305–311. doi: 10.1016/j.tibtech.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Cai S, Bai G-H, Zhang D. Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW. Theoretical and Applied Genetics. 2008;117:49–56. doi: 10.1007/s00122-008-0751-1. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Craig S, Beaton CD, et al. Aluminum tolerance in wheat (Triticum aestivum L.) I. Uptake and distribution of aluminum in root apices. Plant Physiology. 1993a;103:685–693. doi: 10.1104/pp.103.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. Aluminum tolerance in wheat (Triticum aestivum L.) II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiology. 1993b;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Hebb D, Ryan PR. Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiology. 2001;125:2059–2067. doi: 10.1104/pp.125.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hocking PJ, Richardson AE. Effects of altered citrate synthase and isocitrate dehydrogenase expression on internal citrate concentrations of tobacco (Nicotiana tabacum L.) Plant and Soil. 2003;248:137–144. [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proceedings of the National Academy of Sciences, USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Taylor P, Hocking P, Simpson R, Ryan PR, Richardson AE. Barley transgenics expressing the wheat aluminium resistance gene (TaALMT1): interactions with phosphorus nutrition and grain yields on an acid soil. Plant Biotechnology. 2009;7:391–400. doi: 10.1111/j.1467-7652.2009.00403.x. [DOI] [PubMed] [Google Scholar]
- Ezaki B, Gardner RC, Ezaki Y, Matsumoto H. Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiology. 2000;122:657–665. doi: 10.1104/pp.122.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecha G, Silva-Navas J, Benito C, et al. Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.) Theoretical and Applied Genetics. 2007;114:249–260. doi: 10.1007/s00122-006-0427-7. [DOI] [PubMed] [Google Scholar]
- Food and Agricultural Organization. Statistics Division. Statistical yearbook 2005–2006. United Nations: Rome; 2006. [Google Scholar]
- Food and Agriculture Organisation of the United Nations. Issue briefs: how to feed the world in 2050. 2009 (http://www.fao.org/wsfs/forum2050/wsfs-background-documents/issues-briefs/en/ ) [Google Scholar]
- de la Fuente JM, Ramírez-Rodríguez V, Cabrera-Ponce JL, Herrera-Estrella L. Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science. 1997;276:1566–1568. doi: 10.1126/science.276.5318.1566. [DOI] [PubMed] [Google Scholar]
- Fujii M, Yamaji N, Sato K, Ma JF. Mechanism regulating HvAACT1 expression in barley. In: Liao H, Yan X, Kochian LV, editors. Plant–soil interactions at low pH: a nutriomic approach – Proceedings of the 7th International Symposium of Plant–Soil Interactions at Low pH. Guangzhou: South China University of Technology Press; 2009. pp. 165–166. [Google Scholar]
- Furukawa J, Yamaji N, Wang H, et al. An aluminum-activated citrate transporter in barley. Plant and Cell Physiology. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- Garvin DF, Carver BF. Role of the genotype in tolerance to acidity and aluminum toxicity. In: Rengel Z, editor. Handbook of soil acidity. New York: Marcel Dekker Inc; 2003. pp. 387–406. [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoisington D. Opportunities for nutritionally enhancing maize and wheat varieties to combat protein and micronutrient malnutrition. Food Nutrition Bulletin. 2002;23:376–377. doi: 10.1177/156482650202300411. [DOI] [PubMed] [Google Scholar]
- Hue NV, Craddock GR, Adams F. Effect of organic acids on aluminum toxicity in subsoils. Soil Science Society of America Journal. 1986;50:28–34. [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorus efficiency. Annual Reviews of Plant Biology. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- Koyama H, Takita E, Kawamura A, Hara T, Shibata D. Over expression of mitochondrial citrate synthase gene improves the growth of carrot cells in Al-phosphate medium. Plant and Cell Physiology. 1999;40:482–488. doi: 10.1093/oxfordjournals.pcp.a029568. [DOI] [PubMed] [Google Scholar]
- Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiology. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Magalhaes JV, Shaff J, Kochian LV. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. The Plant Journal. 2009;57:389–399. doi: 10.1111/j.1365-313X.2008.03696.x. [DOI] [PubMed] [Google Scholar]
- Maas C, Simpson CG, Eckes P, et al. Expression of intron modified NPT II genes in monocotyledonous and dicotyledonous plant cells. Molecular Breeding. 1997;3:15–28. [Google Scholar]
- Magalhaes JV, Liu J, Guimaraes CT, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nature Genetics. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Barkan A, Freeling M, Taylor WC. Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO Journal. 1989;8:1633–1639. doi: 10.1002/j.1460-2075.1989.tb03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revisied medium for rapid growth and bioassys with tobacco tissue culture. Physiologia Plantarum. 1962;15:474–497. [Google Scholar]
- Pellegrineschi A, Noguera LM, Skovmand B, et al. Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome. 2002;45:421–430. doi: 10.1139/g01-154. [DOI] [PubMed] [Google Scholar]
- Raman H, Zhang K, Cakir M, et al. Molecular mapping and characterization of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.) Genome. 2005;48:781–791. doi: 10.1139/g05-054. [DOI] [PubMed] [Google Scholar]
- Rounds MA, Larsen PB. Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Current Biology. 2008;18:1495–1500. doi: 10.1016/j.cub.2008.08.050. [DOI] [PubMed] [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ. Characterisation of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta. 1995;196:103–110. [Google Scholar]
- Ryan PR, Liu Q, Sperling P, Dong B, Franke S, Delhaize E. A higher plant Δ8 sphingolipid desaturase with a preference for (Z)-isomer formation confers aluminum tolerance to yeast and plants. Plant Physiology. 2007;144:1968–1977. doi: 10.1104/pp.107.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiology. 2009;149:340–351. doi: 10.1104/pp.108.129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, et al. A wheat gene encoding an aluminum-activated malate transporter. The Plant Journal. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ryan PR, Delhaize E, et al. Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant and Cell Physiology. 2006;47:1343–1354. doi: 10.1093/pcp/pcl002. [DOI] [PubMed] [Google Scholar]
- Stival da Silva AL, Sperling P, Horst WJ, et al. A possible role of sphingolipids in the aluminium resistance of yeast and maize. Journal of Plant Physiology. 2006;163:26–38. doi: 10.1016/j.jplph.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Taylor GJ. The physiology of aluminum tolerance in higher plants. Communications in Soil Science and Plant Analysis. 1988;19:1179–1194. [Google Scholar]
- Taylor GJ. Current views of the aluminum stress response: the physiological basis of tolerance. Current Topics in Plant Biochemistry and Physiology. 1991;10:57–93. [Google Scholar]
- Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA. Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiology. 2001;127:1836–1844. [PMC free article] [PubMed] [Google Scholar]
- Trejo-Téllez LI, Stenzel R, Gómez-Merino FC, Schmitt JM. Transgenic tobacco plants overexpressing pyruvate phosphate dikinase increase exudation of organic acids and decrease accumulation of aluminum in the roots. Plant and Soil. 2010;326:187–198. [Google Scholar]
- von Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant and Soil. 1995;171:1–15. [Google Scholar]
- Wang JP, Raman H, Zhou MX, et al. High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2007;115:265–276. doi: 10.1007/s00122-007-0562-9. [DOI] [PubMed] [Google Scholar]
- Zhang W-H, Ryan PR, Sasaki T, Yamamoto Y, Sullivan W, Tyerman SD. Electrophysiological characterisation of the TaALMT1 protein in transfected tobacco (Nicotiana tabacum L.) cells. Plant and Cell Physiology. 2008;49:1316–1330. doi: 10.1093/pcp/pcn107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.