Abstract
Background
Photosystem II (PSII) is the light-driven water:plastoquinone oxidoreductase of oxygenic photosynthesis and is found in the thylakoid membrane of chloroplasts and cyanobacteria. Considerable attention is focused on how PSII is assembled in vivo and how it is repaired following irreversible damage by visible light (so-called photoinhibition). Understanding these processes might lead to the development of plants with improved growth characteristics especially under conditions of abiotic stress.
Scope
Here we summarize recent results on the assembly and repair of PSII in cyanobacteria, which are excellent model organisms to study higher plant photosynthesis.
Conclusions
Assembly of PSII is highly co-ordinated and proceeds through a number of distinct assembly intermediates. Associated with these assembly complexes are proteins that are not found in the final functional PSII complex. Structural information and possible functions are beginning to emerge for several of these ‘assembly’ factors, notably Ycf48/Hcf136, Psb27 and Psb28. A number of other auxiliary proteins have been identified that appear to have evolved since the divergence of chloroplasts and cyanobacteria. The repair of PSII involves partial disassembly of the damaged complex, the selective replacement of the damaged sub-unit (predominantly the D1 sub-unit) by a newly synthesized copy, and reassembly. It is likely that chlorophyll released during the repair process is temporarily stored by small CAB-like proteins (SCPs). A model is proposed in which damaged D1 is removed in Synechocystis sp. PCC 6803 by a hetero-oligomeric complex composed of two different types of FtsH sub-unit (FtsH2 and FtsH3), with degradation proceeding from the N-terminus of D1 in a highly processive reaction. It is postulated that a similar mechanism of D1 degradation also operates in chloroplasts. Deg proteases are not required for D1 degradation in Synechocystis 6803 but members of this protease family might play a supplementary role in D1 degradation in chloroplasts under extreme conditions.
Keywords: Photosynthesis, photosystem II, photoinhibition, D1 turnover, assembly factor, membrane protein, FtsH protease, Deg protease
INTRODUCTION
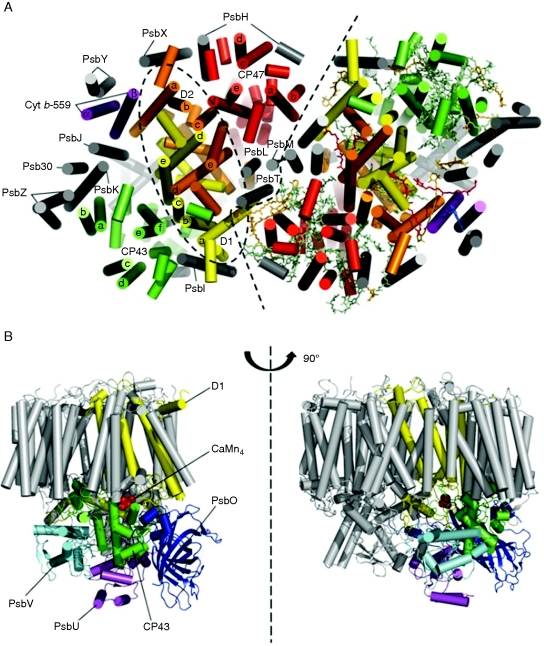
Although the assembly and repair of the photosystem II (PSII) complex has been studied for >30 years (reviewed by Adir et al., 2003), it is only recently, with the advent of detailed structural information for cyanobacterial PSII, that the necessary molecular framework has been established to begin to understand how the complex is assembled from its individual sub-units (Ferreira et al., 2004; Loll et al., 2005; Guskov et al., 2009). Figure 1 presents an overview of the dimeric PSII complex isolated from the thermophilic cyanobacterium Thermosynechococcus elongatus (Guskov et al., 2009; see Table 1 for a full list of abbreviations). Each monomer is composed of 17 intrinsic and three extrinsic sub-units, and in the latest structural model contains 35 chlorophyll a molecules, two pheophytin a molecules, 12 carotenoids, two haem molecules, one non-haem iron, two calcium ions, 1–2 chloride ions, three plastoquinones, >25 lipid molecules and a CaMn4 metal cluster that catalyses formation of dioxygen from water (Guskov et al., 2009).
Fig. 1.
Sub-unit organization of the isolated homodimeric PSII complex from Thermosynechococcus elongatus. (A) View from the cytoplasmic side of the membrane. The two monomers are separated by a black dashed line and the α-helical elements of each subunit are represented as cylinders. D1 (yellow), D2 (orange), CP43 (green), CP47 (red), cytochrome b-559 (purple) and the remaining 11 small sub-units (grey) are indicated in the monomer on the left side as well as the D1–D2–Cyt b-559 sub-complex (elliptical black dashed circle). The same colour coding system applies to the monomer on the right side where are also represented the co-factors of PSII: chlorophylls (green), carotenoids (orange), pheophytins (yellow), plastoquinones (red) and haem (blue). The co-factors are shown in stick form. (B) Two side views, differing by a rotation of 90 °, showing the lumenal subunits PsbO (dark blue), PsbV (light blue), PsbU (purple) and the large lumenal loop of CP43 interconnecting transmembrane helices e and f (green) that lies close to the CaMn4 cluster. The figure was created with the software Pymol (http://pymol.sourceforge.net, version 0·99) and the PDB files 3BZ1 and 3BZ2 (Guskov et al., 2009).
Table 1.
Abbreviations used in the text
| BN/PAGE | Blue Native/polyacrylamide gel electrophoresis |
| CAB | Chlorophyll-a/b-binding |
| CaMn4 | Calcium/manganese metal cluster of PSII that catalyses oxygen evolution |
| CM | Cytoplasmic membrane |
| CtpA | C-Terminal D1 processing protease |
| Cyt b-559 | Cytochrome b-559 |
| Cyt c-550 | Cytochrome c-550 or PsbV |
| HLIPS | High-light-induced proteins, a synonym for SCPs |
| iD1 | Intermediate form of D1 after removal of part of the C-terminal extension |
| LMM | Low molecular mass |
| NMR | Nuclear magnetic resonance |
| P680+ | Chlorophyll cation in PSII that oxidizes Yz |
| pD1 | Precursor form of D1 with complete C-terminal extension |
| PQ | Plastoquinone |
| POR | Protochlorophyllide reductase |
| QA | Bound plastoquinone electron acceptor in PSII |
| RC | PSII reaction centre-like complex containing D1, D2 and Cyt b-559 but lacking CP47 and CP43 |
| RC47 | PSII core complexes lacking CP43 |
| RCC1 | Monomeric PSII core complex |
| RCC2 | Dimeric PSII core complex |
| SCPs | Small CAB-like proteins |
| TM | Thylakoid membrane |
| Yz | A redox-active tyrosine in PSII, at position D1-Tyr161, that oxidizes the CaMn4 cluster |
At the heart of the complex are the D1 and D2 reaction centre (RC) sub-units, each containing five transmembrane α-helices, which bind the chlorophyll, pheophytin and plastoquinone co-factors involved in transmembrane light-induced charge separation (reviewed by Rappaport and Diner, 2008). On either side are CP43 and CP47, each containing six transmembrane α-helices, which bind chlorophyll a (16 molecules in CP47 and 13 in CP43) and β-carotene, and in the case of CP43 participates with D1 in the ligation of the CaMn4 cluster involved in water oxidation (Ferreira et al., 2004). Surrounding these sub-units, on the periphery of the complex, are the 13 low molecular mass (LMM) sub-units of PSII (Fig. 1A). Three extrinsic sub-units (PsbO, PsbU and PsbV) are bound on the lumenal side of the complex (Fig. 1B) and are thought to contribute to the stability of the CaMn4 cluster (reviewed by Roose et al., 2007a). In the case of chloroplast PSII, different sets of extrinsic proteins are bound to the lumenal surface of PSII depending on the species (reviewed by Enami et al., 2008).
Associated with the PSII complex in vivo are distal light-harvesting complexes, containing bound pigment, whose role it is to absorb photons of light and to pass the excitation energy to the RC. There is tremendous variability in the type of antenna system: for instance, water-soluble, extrinsic phycobilisomes in cyanobacteria and red algae, and membrane-embedded light-harvesting chlorophyll-a/b-binding (CAB) sub-units in green chloroplasts (reviewed by Green and Gantt, 2005).
PSII is also prone to various types of light-induced irreversible damage, which unless repaired would lead to an inhibition of PSII activity, sometimes referred to as chronic photoinhibition, and hence to a reduction in oxygenic photosynthesis and growth (reviewed by Adir et al., 2003). In order to maintain PSII homeostasis, a PSII repair cycle operates to replace damaged protein sub-units, mainly the D1 sub-unit, by a newly synthesized copy (reviewed by Nixon et al., 2005). In this review, we critically evaluate recent progress in understanding the mechanism of PSII assembly and repair in cyanobacteria, particularly in the thermophilic cyanobacterium T. elongatus BP-1 (hereafter called T. elongatus), which is widely used for structure/function studies, and in the mesophilic cyanobacterium Synechocystis sp. PCC 6803 (hereafter called Synechocystis 6803), which has been extensively used to study PSII biogenesis and function via mutagenesis.
ASSEMBLY OF PHOTOSYSTEM II
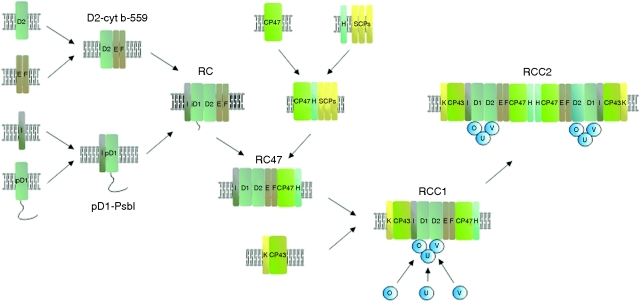
Synechocystis 6803 has proved to be an invaluable model organism to study PSII function because of the ease of generating targeted mutants and its ability to grow on glucose in the absence of PSII activity (Williams, 1988). Characterization of a range of PSII mutants lacking specific sub-units of the PSII complex plus radioactive pulse–chase experiments have provided support for the stepwise assembly of the complex involving a number of discrete PSII sub-complexes (Fig. 2). A broadly similar process is also thought to occur in chloroplasts (Aro et al., 2005). Cytochrome b-559 (Cyt b-559), which is able to accumulate in the membrane in the absence of D1 and D2 (Komenda et al., 2004), seems to act as a nucleation factor to initiate PSII assembly, to form first a D2–Cyt b-559 sub-complex (Komenda et al., 2004, 2008) and then, after addition of D1 and PsbI, possibly as a sub-complex (Dobakova et al., 2007), a PSII RC-like complex (Komenda et al., 2008) (Fig. 2). In the absence of CP47, CP43 is unable to attach stably to the PSII RC, and two types of PSII RC complexes, probably differing with respect to associated assembly factors, have been detected in membranes (Komenda et al., 2004, 2008). In the absence of CP43, CP47 is able to attach to the PSII RC sub-complex to form the so-called RC47 complex (Komenda et al., 2004). RC47 is unable to oxidize water but is still able to drive oxidation of Yz, a redox-active tyrosine in D1, at position D1-Tyr161, which is the immediate oxidant of the CaMn4 cluster (Metz et al., 1989; Rogner et al., 1991). Subsequent attachment of CP43 allows formation of the monomeric PSII core complex, RCC1, which is the starting point for light-driven assembly of the CaMn4 cluster and attachment of the lumenal extrinsic sub-units, PsbO, PsbU and PsbV, which act as a ‘cap’ to shield the cluster (Fig. 1B).
Fig. 2.
Assembly of the PSII complex in Synechocystis sp. PCC 6803. Proposed scheme for the assembly of PSII based on the analysis of PSII assembly complexes in defined mutants and radioactive pulse–chase experiments. For clarity, assembly factors and many of the low molecular mass (LMM) sub-units are not included. The LMM PsbE, PsbF, PsbH, PsbI and PsbK sub-units and the extrinsic PsbO, PsbU and PsbV sub-units are designated by the appropriate upper case letter, and the small CAB-like proteins by SCPs. Cytochome b-559 (cyt b-559) is composed of a heterodimer of the PsbE and PsbF subunits. Types of PSII complex: RC, PSII reaction centre-like complexes containing either mature D1, intermediate D1 (iD1) or precursor D1 (pD1) but lacking CP47 and CP43; RC47, PSII core complexes lacking CP43; RCC1, monomeric PSII core complex; RCC2, dimeric PSII core complex. The site of attachment of the extrinsic proteins to PSII is purely illustrative.
During the biogenesis of PSII, the D1 sub-unit, which is synthesized in most organisms as a precursor protein (pD1) with a C-terminal extension, must be cleaved to allow assembly of a functional CaMn4 cluster (Nixon et al., 1992; Anbudurai et al., 1994). During C-terminal processing, the 16 amino acid extension found in Synechocystis 6803 is cleaved by CtpA to leave an intermediate eight amino acid extension (Komenda et al., 2007a). This intermediate form of D1, designated iD1, can be differentiated from pD1 by SDS–PAGE (Inagaki et al., 2001) and is found mainly in PSII RC complexes in vivo (Komenda et al., 2004) (Fig. 2). The C-terminal extension is not required for assembly of oxygen-evolving PSII (Nixon et al., 1992; Satoh and Yamamoto, 2007). However, mutants either lacking the extension or containing a modified C-terminal extension are in general less fit than the wild type (WT) and are more prone to photoinhibition (Ivleva et al., 2000; Kuvikova et al., 2005).
Oxygen-evolving PSII is found as both a dimer (RCC2) and a monomer (RCC1) in Synechocystis 6803 and T. elongatus, with the isolated dimer more active than the monomer (Nowaczyk et al., 2006). Both the monomer and dimer complexes can be detected when gently solubilized thylakoid membranes (TMs) of Synechocystis 6803 and T. elongatus are analysed rapidly by Blue Native/PAGE (BN/PAGE) (M. Boehm, unpubl. res.). Moreover, in many mutants, for instance the psbO deletion strain of Synechocystis 6803, the dimeric form is specifically missing, regardless of the detergent concentration used for solubilization (Komenda et al., 2010). These results plus those of Watanabe et al. (2009) argue against the recent suggestion that dimeric PSII is a detergent-induced artefact and not relevant to the situation in vivo (Takahashi et al., 2009).
Role of pigments in assembly
It is clear that pigment availability is a major factor in the accumulation of PSII. Early studies suggested that β-carotene was vital for accumulation of D1 in the green alga Chlamydomonas reinhardtii (Trebst and Depka, 1997) as well as in Synechocystis 6803 (Masamoto et al., 2004). Moreover, genetic manipulation of the type of carotenoid synthesized suggested that PSII assembly required the presence of a carotenoid with at least one β-ionylidene ring, possibly to play a structural role in the early stages of assembly (Bautista et al., 2005). However, a more recent study using a Synechocystis 6803 strain completely lacking carotenoids, due to the deletion of the crtB gene encoding phytoene synthase (J. Komenda, unpubl. res.), has shown that the monomeric PSII core complex (RCC1) can actually be formed in the absence of carotenoids, although it is unstable and strongly light sensitive and can only be detected by radioactive labelling. Only the PSII assembly intermediate, RC47, is able to accumulate to a level detectable by conventional staining on a 2-D BN/PAGE gel. Interestingly, synthesis of CP47, and especially CP43, was affected by the absence of carotenoids much more than the synthesis of D2 and D1. This is in agreement with the latest PSII structural models (Guskov et al., 2009) showing that most of the β-carotene molecules are located in the vicinity of the transmembrane α-helices of CP47 and CP43. In contrast to the situation in PSII, functional PSI and Cyt b6–f complexes can assemble in the absence of carotenoid (J. Komenda, unpubl. res.).
The role of chlorophyll in the assembly of PSII has been studied in mutants of Synechocystis 6803 in which the light-independent pathway of chlorophyll biosynthesis has been blocked by mutation of chlL so that only the remaining light-dependent pathway is functional. Cells grown in the dark under so-called light-activated heterotrophic growth (LAHG) conditions contain very little chlorophyll a and fail to accumulate PSII core complexes (Wu and Vermaas, 1995). However, PSII RC complexes containing D2 and mostly iD1 can be detected in these cells (J. Komenda, unpubl. res.). Upon exposure to light, chlorophyll a is synthesized by the light-dependent pathway, and CP47 and CP43 accumulate, as do functional PSII core complexes. The co-ordination between chlorophyll availability and D1 accumulation is believed to be regulated at the level of translation (He and Vermaas, 1998). This is in contrast to the situation in barley, where chlorophyll-binding proteins are inserted into the TM in the absence of chlorophyll but are rapidly degraded (Kim et al., 1994). Perhaps surprisingly, PSII complexes in Synechocystis 6803 can bind other types of chlorophyll besides chlorophyll a, including chlorophyll b (Satoh et al., 2001; Xu et al., 2001) and di-vinyl chlorophyll a (Tomo et al., 2009).
Currently much effort is focused on understanding how chlorophyll synthesis is co-ordinated with the availability of the pigment-binding apopolypeptides, especially as free pigment in the membrane is considered to promote the generation of dangerous reactive oxygen species (Krieger-Liszkay et al., 2008). In the case of the CP47 and CP43 proteins of Synechocystis 6803, insertion of chlorophyll and carotenoid into the unassembled sub-unit can occur before incorporation into larger core complexes (M. Boehm and P. J. Nixon, unpubl. res.). It would seem logical that pigment binding occurs during or very soon after insertion of the protein into the membrane. This could be mediated by locating the chlorophyll biosynthetic machinery close to the sites of protein synthesis. Indeed Nickelsen and co-workers have recently identified a membrane protein of Synechocystis 6803, termed Pitt (CyanoBase designation Slr1644), which is able to interact with the protochlorophyllide reductase (POR) involved in light-dependent chlorophyll biosynthesis (Schottkowski et al., 2009). The phenotype of a pitt insertion mutant is consistent with a role for Pitt in the early steps of assembly of pigment–protein complexes, possibly by helping to localize POR to specific regions of the membrane.
During assembly and repair of PSII, it is likely that chlorophyll is bound transiently by specific chlorophyll carrier proteins within the membrane before being delivered to its final destination (Wu and Vermaas, 1995). The cyanobacterial small CAB-like proteins (SCPs), sometimes designated high-light-induced proteins (HLIPS) (Dolganov et al., 1995), have been speculated to play such a role (Funk and Vermaas, 1999). They are members of an extended family of proteins that includes the light-harvesting CAB sub-units, the PsbS sub-unit, the early-light induced proteins (ELIPS) and the one-helix protein (OHP) of chloroplasts (Funk and Vermaas, 1999). Four of the five annotated SCPs found in Synechocystis 6803 (ScpB–E) are predicted to contain a single transmembrane helix with strong sequence similarities to the chlorophyll-binding regions in the first and third transmembrane helices of CAB proteins. Recent analysis of single and multiple scp mutants of Synechocystis 6803 suggests that they are mainly required in stabilizing chlorophyll during PSII repair rather than during de novo assembly (Vavilin et al., 2007) and indeed SCPs are able to associate with PSII (Promnares et al., 2006; Yao et al., 2007; Kufryk et al., 2008). Analysis of isolated His-tagged ScpD complexes suggests that ScpD binds to CP47 in the vicinity of PsbH (Fig. 2) (Promnares et al., 2006). Also present in Synechocystis 6803 is a larger SCP (designated ScpA) consisting of an SCP domain fused to the C-terminus of ferrochelatase (Sobotka et al., 2008) and another homologue, termed LilA, also suggested to be involved in pigment binding/storage (Kufryk et al., 2008). SCPs are not found in chloroplasts and might be functionally replaced by the ELIPS (Promnares et al., 2006).
Once bound to a target protein, it is unclear whether chlorophyll remains permanently attached for the lifetime of the protein or is able to move from one protein to another. Early work indicated that low-level expression of a higher plant CAB sub-unit helped to increase the accumulation and insertion of chlorophyll b into the PSII complex of Synechocystis 6803 (Xu et al., 2001), suggesting that such a chaperone function might operate in vivo. Similarly, Komenda and colleagues have suggested from the analysis of PSI content in various PSII mutants that unassembled PSII sub-units within the TM, such as CP47, might have a function in chlorophyll delivery to other chlorophyll-binding proteins including PSI (Dobakova et al., 2009).
Role of the small sub-units in assembly
There are 13 sub-units with molecular masses <10 kDa in each monomer of the isolated PSII dimer from T. elongatus (Muh et al., 2008; Guskov et al., 2009). All apart from PsbZ consist of a single transmembrane α-helix. The relatively low resolution of the early structural models for PSII prevented complete assignment of all the LMM sub-units (Ferreira et al., 2004; Loll et al., 2005). However, PsbX (Katoh and Ikeuchi, 2001), PsbY (Kawakami et al., 2007) and Psb30 (also termed Ycf12) (Inoue-Kashino et al., 2008) have now been unambiguously assigned in the latest 2·9 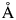 resolution structural model (Guskov et al., 2009). Inspection of the structural models suggests that one role for the LMM sub-units is to stabilize the binding of co-factors. For instance, PsbH and PsbI participate in the binding of chlorophyll and PsbK binds a Ca2+ ion (Muh et al., 2008). The PsbJ, PsbK, PsbZ and Psb30 sub-units also seem to form an interface with CP43 to aid the binding of carotenoids (Barber and Iwata, 2005). Several LMM sub-units also interact with the extrinsic proteins located in the lumen and may play a stabilizing role (Fig. 1B) (Muh et al., 2008).
resolution structural model (Guskov et al., 2009). Inspection of the structural models suggests that one role for the LMM sub-units is to stabilize the binding of co-factors. For instance, PsbH and PsbI participate in the binding of chlorophyll and PsbK binds a Ca2+ ion (Muh et al., 2008). The PsbJ, PsbK, PsbZ and Psb30 sub-units also seem to form an interface with CP43 to aid the binding of carotenoids (Barber and Iwata, 2005). Several LMM sub-units also interact with the extrinsic proteins located in the lumen and may play a stabilizing role (Fig. 1B) (Muh et al., 2008).
In general, early mutagenesis experiments failed to identify clear-cut roles for the LMM sub-units in PSII function. However, more recent detailed studies on null mutants have begun to reveal specific defects in the stabilization of the complex. For instance, PsbI is required for stable binding of CP43 within the PSII core complex (Dobakova et al., 2007), which is consistent with the close proximity of PsbI to CP43 and the N-terminal region of D1 in recent X-ray structural models (Fig. 1). The PsbM, PsbL and PsbT sub-units lie at the interface of the two monomers in the PSII dimer (Fig. 1) and indeed deletion of PsbT (Bentley et al., 2008; Henmi et al., 2008), PsbM (Kawabata et al., 2007; Bentley et al., 2008) and PsbL (Bentley et al., 2008) affects the accumulation of PSII dimers in cyanobacteria. In the case of PsbH, which binds to CP47, a detailed analysis of PSII complexes isolated from a T. elongatus psbH mutant has indicated the additional absence of the neighbouring PsbX sub-unit (Iwai et al., 2006). Likewise, the absence of PsbZ leads to loss of the neighbouring PsbK and Psb30/Ycf12 sub-units from the isolated complex (Iwai et al., 2007), with PsbK most affected (Takasaka et al., 2010). These latter two examples neatly illustrate the potential indirect effects that can sometimes arise from genetic deletion of a single sub-unit.
The sequence of events involved in the attachment of the LMM PSII sub-units is unclear and it is likely that parallel assembly pathways operate. In the case of CP47 and CP43, the recent crystal structures indicate that PsbH binds to CP47 and that PsbK and PsbZ bind to CP43 (Fig. 1). Analysis of a psbH deletion mutant and a mutant expressing His-tagged ScpD suggests that CP47 actually forms a complex with PsbH and SCPs prior to attachment to the PSII RC complex during assembly (Komenda et al., 2005; Promnares et al., 2006) (Fig. 2). CP43 also appears to form a complex with PsbK prior to attachment to RC47 as judged from the analysis of a psbK deletion mutant (J. Komenda, unpubl. res.).
Recent proteomic analysis of etiolated barley membranes has suggested that many, if not all, of the LMM sub-units of PSII are able to accumulate in the absence of chlorophyll and the major PSII sub-units CP47, CP43 and D1, and so be in a position to stabilize chlorophyll binding rapidly once synthesized (Ploscher et al., 2009). Whether this also occurs in cyanobacteria is presently unclear.
Role of the extrinsic proteins
Three extrinsic proteins are found on the lumenal surface of the PSII holoenzyme in T. elongatus: the PsbO, PsbU and PsbV (or Cyt c-550) sub-units (Fig. 1B). Analysis of null mutants has provided ample evidence that PSII assembles in their absence (although at reduced levels) but they are required for the stability of the CaMn4 cluster, both in the light and in the dark (reviewed by Roose et al., 2007a). In the dark the extrinsic proteins probably act to shield the high-valence cluster from reductant (Burnap et al., 1996; Shen et al., 1998). Additional roles for these proteins in the biogenesis and repair of PSII are also possible. Indeed mutational analysis of the two PsbO genes found in Arabidopsis thaliana suggests that the efficiency of PSII repair is dependent on the type of PsbO present (Lundin et al., 2007, 2008).
Cyanobacteria also encode homologues of the PsbP and PsbQ proteins found on the lumenal side of PSII in chloroplasts (Thornton et al., 2004). However, sequence analyses indicate that the cyanobacterial homologues, designated here as CyanoP and CyanoQ, are only distantly related to their chloroplast counterparts (De Las Rivas and Roman, 2005). Both cyanobacterial sub-units are predicted to be lipoproteins and from mutagenesis experiments neither is required for PSII activity (Thornton et al., 2004; Summerfield et al., 2005a, b). CyanoQ is present at stoichiometric amounts in Synechocystis 6803 PSII preparations (Roose et al., 2007b) but is absent in the highly active dimeric PSII preparations used to crystallize PSII from T. elongatus. These data would suggest that CyanoQ might play a regulatory role in cyanobacterial PSII in vivo (Roose et al., 2007b), as suggested from growth experiments (Summerfield et al., 2005a). In the case of CyanoP, only trace amounts (3 % of total) were found in His-tagged PSII preparations (Thornton et al., 2004). The most compelling data thus far to suggest that CyanoP does indeed have a role in PSII have come from the observation that CyanoP fails to accumulate in a mutant of Synechocystis 6803 lacking PSII (Ishikawa et al., 2005). The closest homologue to CyanoP in A. thaliana is actually not PsbP but the PsbP-like protein 1 (PLP1) which is not associated with PSII but which seems to be involved in assembly and repair (Ishihara et al., 2007). This observation would suggest a role for CyanoP in the biogenesis of PSII rather than a structural role.
Conserved PSII assembly factors
Over the years a number of proteins have been identified as being important for the assembly and repair of PSII. Some, but not all, are conserved in both cyanobacteria and chloroplasts, raising the possibility that their role is a fundamental one to PSII biogenesis and has been retained during evolution.
One of the first PSII assembly factors to be characterized was Hcf136 (also termed Ycf48), which was identified from studies on the high chlorophyll fluorescence mutant, hcf136, of A. thaliana (Meurer et al., 1998). Only trace amounts of PSII were found to accumulate in this mutant, despite WT rates of synthesis of PSII sub-units, and a role in the assembly or stability of PSII was suggested (Meurer et al., 1998). More recently the Ycf48 homologue in Synechocystis 6803 was detected in PSII RC-like assembly complexes in vivo but not in larger core complexes (Komenda et al., 2008). In addition, although PSII could still accumulate in a Synechocystis 6803 mutant lacking Ycf48, the efficiency of PSII assembly and repair was reduced dramatically (Komenda et al., 2008). A combination of mutant and yeast two-hybrid data further suggested that Ycf48 from Synechocystis 6803 binds to and stabilizes unassembled pD1 and aids formation of the PSII RC, in agreement with earlier conclusions from studies in A. thaliana (Plucken et al., 2002). Ycf48/Hcf136 is targeted to the lumen in chloroplasts and is membrane bound. Hence it is likely to bind to the lumenal side of the PSII RC. Very recently, the structure of Ycf48 from T. elongatus was determined to be a seven-bladed β-propeller by X-ray crystallography (F. Michoux, P. J. Nixon and J. Murray, unpubl. res.). Such a structural motif is ideal for making multiple contacts with different sub-units during the assembly of a larger complex. How Ycf48 interacts with the PSII RC-like complex is presently unclear, although an interaction with the C-terminal region of D1, and possibly the C-terminal region of D2, seems likely.
The Psb27, Psb28 and Psb29 sub-units were identified as sub-stoichiometric components of His-tagged CP47 preparations isolated from Synechocystis 6803 (Kashino et al., 2002). In the cases of Psb27 and Psb28, additional studies have since provided overwhelming evidence that these sub-units do indeed associate with PSII sub-complexes and were not contaminants within the original His-tagged preparation, a possibility that had not been formally excluded. In the case of Psb29, more work is needed to clarify its association with PSII.
Psb27 is an approx. 11-kDa lipoprotein and has been detected as a component of a non-oxygen-evolving PSII core complex from T. elongatus lacking PsbO, PsbU and PsbV (Nowaczyk et al., 2006). It is thought that Psb27 binds to the lumenal side of the PSII core complex and prevents binding of the three extrinsic proteins (Nowaczyk et al., 2006). A Synechocystis 6803 mutant lacking Psb27 is still able to assemble oxygen-evolving complexes, but the light-driven assembly of the CaMn4 cluster is less efficient (Roose and Pakrasi, 2008). In Arabidopsis, a mutant lacking one of the two encoded Psb27 homologues still assembles PSII but is perturbed in the repair of PSII following photodamage (Chen et al., 2006). Two groups recently determined the 3-D structure of the hydrophilic domain of cyanobacterial Psb27 by nuclear magnetic resonance spectroscopy (Cormann et al., 2009; Mabbitt et al., 2009), and found it to consist of four amphipathic α-helices in an up–down–up–down arrangement. Although a binding site on CP47 has been suggested for Psb27 based on in silico docking simulations (Cormann et al., 2009), recent biochemical data indicate a close structural relationship with CP43 (J. Komenda, unpubl. res).
BN/PAGE has revealed that Psb28 of Synechocystis 6803 is found predominantly in an unassembled state in the membrane and that some is also associated with the RC47 complex most probably by attachment to the cytoplasmic side of CP47 (Dobakova et al., 2009). Deletion of Psb28 does not affect the functional properties of PSII but does cause a decrease in the rate of synthesis of CP47 and the chlorophyll-binding PSI sub-units PsaA and PsaB (Dobakova et al., 2009). An interesting phenotype of the psb28 null mutant is the release of large quantities of the chlorophyll precursor, protoporphyrin IX, into the culture medium. Previous work had already suggested that CP47 accumulation was particularly sensitive to the availability of chlorophyll in the cell (Sobotka et al., 2005). Therefore, it is possible that Psb28 might be involved in regulating chlorophyll availability during the biogenesis of PSI and PSII. A Psb28 homologue is also found in the A. thaliana genome but has not yet been studied. In the case of Synechocystis 6803 and some other cyanobacteria, there is a second homologue of Psb28 encoded in the genome, but its role is so far unclear.
Psb29 is predicted to be a peripheral sub-unit bound to the cytoplasmic side of the TM (Keren et al., 2005b). Although there is still uncertainty about whether Psb29 associates with PSII, there is some evidence from analysis of Psb29 null mutants of Synechocystis 6803 and A. thaliana that light capture by CP47 and CP43 is perturbed. Further work is needed to clarify the defect. Previously, Wang et al. (2004) had concluded that Psb29 (designated Thf1 in their studies) was involved in the formation of the TM since the A. thaliana null mutant is variegated. However, it has been argued that the marked variegation seen in this mutant might stem from destruction of thylakoids because of enhanced production of reactive oxygen species by defective PSII (Keren et al., 2005b). More recent work on Psb29/Thf1 in Arabidopsis has revealed that it is required for normal expression of chloroplast FtsH (Zhang et al., 2009). The possible involvement of Psb29 in the assembly and repair of PSII in Synechocystis 6803 has yet to be assessed.
Ycf39 (CyanoBase designation Slr0399) was originally identified in a screen for suppressor mutants that restored the ability of a D2 mutant of Synechocystis 6803 to bind the bound plastoquinone, QA, and was suggested to be involved in delivering plastoquinone to PSII during assembly (Ermakova-Gerdes and Vermaas, 1999), although this has yet to be confirmed experimentally. Interestingly, there is an orthologue of Ycf39 in A. thaliana (AT4G35250) which is predicted to be chloroplast targeted and which therefore might play a similar role.
Cyanobacterial-specific assembly factors
PratA, which is a tetratricopeptide repeat (TPR) protein found in Synechocystis 6803, has attracted interest because of its apparent involvement in the C-terminal processing of the D1 sub-unit in vivo during the early stages of assembly of PSII (Klinkert et al., 2004). Given that PratA is found within the periplasm, the involvement of PratA in D1 processing lent support to a possible role for the cytoplasmic membrane (CM) in PSII biogenesis (Klinkert et al., 2004). More recently, a combination of yeast two-hybrid and peptide scanning experiments mapped a PratA-binding site to residues 314–328 of the D1 sub-unit (Schottkowski et al., 2009). This region, which falls within the C-terminal tail of mature D1, is folded into a short lumenally exposed α-helix in the PSII holoenzyme. Whether this binding interaction is of physiological significance remains uncertain, and, as yet, PratA has neither been identified as a component of a PSII assembly complex nor shown to interact with unassembled D1 in vivo. Nevertheless, a sub-population of PratA is membrane bound and does co-purify to some extent with membrane fragments containing precursor D1, possibly representing a distinct part of the membrane system linking the CM and TM (Schottkowski et al., 2009).
More recently, Pakrasi and colleagues have used mass spectrometry to identify low abundance proteins within a His-tagged CP47 preparation, which might play roles in the assembly of PSII. Six novel proteins, all encoded within the same region of the genome, were identified, some with putative binding sites for chlorophyll and bilins (Wegener et al., 2008). Potential effects on PSII function were analysed in a deletion mutant lacking these six genes plus three others, which together form a single transcriptional unit covering open reading frames slr0144–slr0152. The mutant cells were still able to assemble PSII and were able to evolve oxygen at 80 % of WT rates. Given the relatively minor effects on PSII activity observed in the mutant, further evidence needs to be provided to support the proposal that these gene products function as PSII assembly proteins.
A number of other Synechocystis genes have been implicated in PSII assembly from the results of suppressor screens. These include slr0286 (Kufryk and Vermaas, 2001) and slr2013 (Kufryk and Vermaas, 2003) which overcome defects in the accumulation of PSII in two different types of D2 mutant. The roles of these gene products still remain unclear.
Chloroplast-specific assembly factors
A number of factors have been shown to be important for PSII assembly or repair in chloroplasts, either in the model plant A. thaliana or in the model alga C. reinhardtii (reviewed by Mulo et al., 2008). Sequence comparisons indicate the absence of clear cyanobacterial homologues which would suggest that these factors have evolved after the divergence of chloroplasts and cyanobacteria, and therefore reflect possible differences in the PSII biogenesis between chloroplasts and prokaryotes. As PSII repair is a dominant process in higher plant chloroplasts, it remains possible that some of the assembly factors assigned a role in PSII biogenesis might actually play a more general role in the assembly of thylakoid protein complexes. For some though there are clear biochemical data to indicate a direct role in PSII assembly or repair. For instance, in the case of the intrinsic protein, LPA1, which contains two TPRs and is required for optimal assembly of PSII, yeast two-hybrid studies suggest a direct role in binding to D1 during assembly of PSII de novo (Peng et al., 2006). Rep27, which is the likely orthologue of LPA1 in the green alga C. reinhardtii has also been concluded to play a role in the synthesis of D1, but during PSII repair rather than during assembly (Dewez et al., 2009). A second factor required for normal accumulation of PSII in A. thaliana, LPA2 (Ma et al., 2007), appears to interact with CP43 and, interestingly, Alb3, which is known to facilitate insertion of proteins into the TM (Yi and Dalbey, 2005).
PHOTOSYSTEM II REPAIR
PSII damage
Damage to PSII by visible light has been studied in great detail and is sometimes referred to as chronic or irreversible photoinhibition or, more usually, PSII photoinhibition (reviewed by Adir et al., 2003; Edelman and Mattoo, 2008). On the basis of in vitro studies, two main mechanisms of PSII photoinhibition have been suggested based on the light requirement, the site of initial damage and the nature of the damaging species (reviewed by Barber and Andersson, 1992; Aro et al., 1993). In acceptor-side photoinhibition, damage is postulated to be caused by singlet oxygen generated following charge recombination reactions within PSII. This process, which involves the formation of the triplet state of the primary electron donor, P680, can potentially occur at high irradiances when the PQ pool is reduced but also at low light intensities when QB− is generated, in the so-called ‘low light syndrome’ (Keren et al., 1997). In contrast donor-side photoinhibition occurs when electron donation into the PSII RC by water oxidation is unable to match the rate of P680 oxidation so that P680+ and other oxidizing species on the donor side become relatively long lived and, because of their high oxidizing potential and reactivity, can damage nearby residues. The mechanism of PSII photodamage in vivo is currently under intense debate (discussed by Vass and Cser, 2009) following recent evidence suggesting that the CaMn4 cluster itself is the initial site of damage (Hakala et al., 2005; Ohnishi et al., 2005).
The PSII repair cycle
Despite uncertainty about the precise mechanism of photoinhibition in vivo, it is generally agreed that the D1 sub-unit is the main PSII sub-unit damaged during photoinhibition in vivo (reviewed by Edelman and Mattoo, 2008) and that the rapid synthesis and degradation (or turnover) of D1 observed in vivo in radioactive pulse–chase experiments reflects the operation of a PSII repair cycle in which damaged D1 sub-units are replaced by newly synthesized copies (Ohad et al., 1984). The current data suggest that PSII repair in cyanobacteria, like that in chloroplasts, involves partial disassembly of PSII, degradation of the damaged sub-unit, incorporation of a newly synthesized sub-unit into the sub-complex and reassembly of the holoenzyme (discussed by Nixon et al., 2005). Because irreversible damage to PSII occurs at all light intensities, PSII repair is an important photoprotective mechanism in all oxygenic photosynthetic organisms (discussed by Edelman and Mattoo, 2008). Importantly, the repair cycle itself is also sensitive to various types of environmental stress, further exacerbating the sensitivity of PSII to photoinhibition (Takahashi and Murata, 2008).
D1 degradation in vitro
Much of the early work directed at studying the mechanism of D1 degradation during PSII repair has focused on the characterization of light-induced D1 fragments generated either in isolated PSII complexes (e.g. Shipton and Barber, 1992) or in plant material in which PSII repair was either unable to match the high rates of photodamage (e.g. Canovas and Barber, 1993) or was potentially affected by the presence of cytoplasmic protein synthesis inhibitors (e.g. Greenberg et al., 1987). Given that similar sized D1 fragments could also be generated in the dark by reactive oxygen species (Miyao, 1994; Lupinkova and Komenda, 2004), the suspicion remains that many of the D1 breakdown fragments described in the literature might actually reflect the products of chemical cleavage reactions rather than intermediates in a physiologically relevant enzymatic process.
D1 degradation in vivo
Members of the FtsH and Deg protease families have long been considered to be prime candidates for the degradation of damaged D1 during PSII repair in higher plant chloroplasts (reviewed by Adam and Clarke, 2002). FtsH proteases consist of an N-terminal transmembrane domain followed by a large hydrophilic region containing an AAA+-module (ATPase associated with various cellular activities) and a Zn2+ metalloprotease domain (reviewed by Ito and Akiyama, 2005). Deg proteases are membrane-associated serine proteases, with some capable of forming extremely large homo-oligomeric complexes (reviewed by Ortega et al., 2009).
Early work based on in vitro studies suggested that degradation of photodamaged D1 in chloroplasts was a two-step process involving first the cleavage of a stromally exposed loop connecting transmembrane helices d and e of D1 (Fig. 1A) by the Deg2 protease, followed by FtsH-mediated removal of the N-terminal fragment (Spetea et al., 1999; Haussuhl et al., 2001). Whether this actually occurs in vivo has been examined more recently in defined mutants in both cyanobacteria and chloroplasts (reviewed by Nixon et al., 2005).
Interpretation of mutant data is potentially complicated by the fact that there are multiple copies of the FtsH and Deg proteases with possibility for overlap of function. For instance, in the case of Synechocystis 6803 there are four FtsH proteases and three Deg proteases (Nixon et al., 2005), and in A. thaliana chloroplasts there are possibly nine FtsH proteases and at least four Deg proteases (reviewed by Adam et al., 2005; Sakamoto, 2006). In addition, other proteases might be able to compensate to some degree for the loss of a particular protease. Thus, while unimpaired PSII repair in a protease null mutant would indicate that the protease is not vital for PSII repair, it does not necessarily mean that the protease does not participate in PSII repair in vivo. In the case of a mutation that results in a defect, further evidence is needed to clarify whether the effect is direct or indirect.
One direct approach is to test the ability of the purified protease to cleave damaged D1 in vitro. In this case care has to be taken to ensure that the enzyme is used at physiologically relevant levels, that the cleavage site in the D1 sub-unit is appropriate for the known location of the protease (lumenal side or stromal side in chloroplasts) and that the PSII complex has not been potentially damaged by other factors such as high salt and heat stress. Other biochemical lines of evidence are physical interaction between the protease and PSII and co-purification of the protease with PSII.
Role of FtsH in PSII repair
It is now clear that FtsH proteases play a more crucial role in D1 degradation than originally hypothesized. Mutants lacking FtsH2 (CyanoBase designation Slr0228) in Synechocystis 6803 (Silva et al., 2003; Komenda et al., 2006) and FtsH2 and FtsH5 in A. thaliana (Bailey et al., 2002; Kato et al., 2009) show impaired rates of D1 degradation. Importantly, damaged D1 is stabilized in these mutants, and there is no accumulation of smaller fragments of D1. These observations are consistent with a role for FtsH at an early stage of D1 degradation and not just in the removal of D1 breakdown products as proposed in the earlier two-step model (Haussuhl et al., 2001). D1 degradation is not totally blocked in the Arabidopsis mutants (Kato et al., 2009) and in mutants of Synechocystis 6803 that display extremely high rates of D1 degradation (Komenda et al., 2010). These observations would suggest the involvement of additional proteases in D1 degradation, possibly other FtsH proteases, although with a much reduced activity.
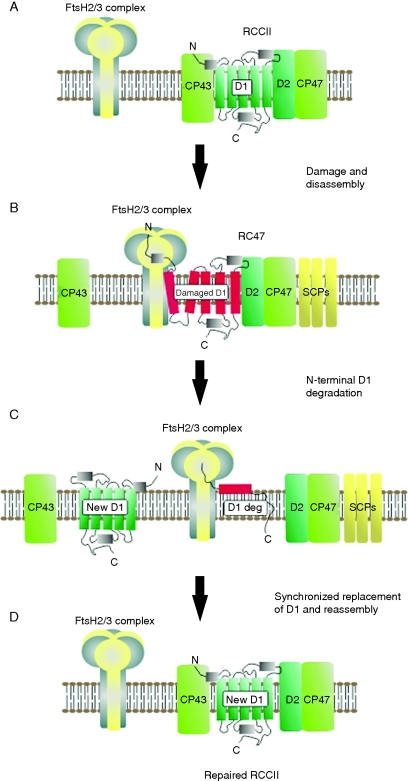
In the case of Synechocystis 6803, analysis of a triple mutant lacking all three Deg proteases found in this organism revealed that rapid and selective turnover of D1 was still operational (Barker et al., 2006). This evidence, together with the earlier data on the ftsH2 mutant and biochemical detection of FtsH in PSII preparations, led to the proposal that rapid and selective D1 turnover might be mediated by FtsH alone and need not require prior cleavage by a Deg protease (Silva et al., 2003; Nixon et al., 2005). Data from N-terminal truncation mutants of Synechocystis 6803 support a model in which FtsH degrades damaged D1 in a highly processive reaction starting from the N-terminus (Fig. 3) (Komenda et al., 2007b). Because FtsH is known to be a weak unfoldase, it seems likely that damaged D1 needs to be in a destabilized state to allow its degradation. This could be accomplished by oxidative damage to amino acid residues, damage and loss of co-factors such as pigments and metal ions, and partial disassembly of the complex to form the RC47 complex (Nixon et al., 2005).
Fig. 3.
Selective synchronized replacement of damaged D1 during PSII repair according to the FtsH-only model (Nixon et al., 2005). (A and B) Light-induced irreversible damage to D1 triggers partial disassembly of the dimeric PSII complex (RCCII) to form the monomeric RC47 complex containing damaged D1. For clarity the extrinsic proteins have been omitted. The hexameric FtsH2/FtsH3 hetero-oligomeric complex engages with the N-terminal tail of the damaged D1 sub-unit. SCPs might store chlorophyll during repair. (C) Damaged D1 is removed from the membrane by the FtsH complex in a process driven by ATP hydrolysis and is degraded in a highly processive reaction at the Zn2+ protease domain. Degradation of damaged D1 is synchronized at the transmembrane helix level with insertion of the ‘new’ D1 subunit into the RC47 complex to minimize destabilization of the RC47 complex. (D) CP43 reattaches to form a non-oxygen-evolving complex which then reassembles the CaMn4 cluster, binds the extrinsic subunits and dimerizes to form the fully functional RCCII complex. For clarity, assembly factors are not included.
FtsH2 of Synechocystis 6803 is also important for the rapid degradation of D1 following heat stress (Kamata et al., 2005) or following donor-side photoinhibition induced by UV-B damage (Cheregi et al., 2007) or by illumination of cells in the presence of ammonia (Drath et al., 2008), or by genetic deletion of the extrinsic PsbO and PsbV sub-units (Komenda et al., 2010). These observations are consistent with a model in which degradation of damaged D1 is triggered not by a precise damaging event but by its overall degree of structural destabilization. FtsH2 is also important for the degradation of unassembled PSII sub-units so appears to play a general role in removing damaged, misassembled and unassembled PSII sub-units from the TM (Komenda et al., 2006). What role FtsH has in remodelling the protein content of the TM in response to changes in physiological conditions, e.g. nutrient depletion rather than damage, has not been investigated.
So far the vast majority of the data implicating FtsH at an early stage of D1 degradation have come from analysis of mutants. Whether FtsH can degrade full-length D1 in a reconstituted system has not yet been demonstrated, although there are data to indicate that Escherichia coli-expressed chloroplast FtsH1 is able to degrade a 23 kDa N-terminal D1 fragment, albeit at a low rate (Lindahl et al., 2000). Effective degradation of D1 will probably require the development of a membrane-based system as used to investigate FtsH-mediated degradation of E. coli membrane proteins (Akiyama and Ito, 2003).
Given the multiplicity of FtsH sub-units in chloroplasts and cyanobacteria, current research is directed at understanding their possible physical interaction. We have recently constructed and isolated from Synechocystis 6803 a glutathione S-transferase (GST)-tagged derivative of the FtsH2 protease and have found that it co-purifies with FtsH3 (Barker et al., 2008). Furthermore, preliminary negative stain electron microscopy suggests a hexameric complex. This result provides important direct evidence that FtsH complexes in Synechocystis 6803 are hetero-oligomeric, which opens up new questions regarding substrate specificity and regulation. Large hetero-oligomeric complexes, consisting of two types of FtsH sub-units, have also been proposed for chloroplasts based on co-immunoprecipitation experiments (Sakamoto et al., 2003). In yeast mitochondria and E. coli, even larger FtsH supercomplexes, of ill-defined function, have been isolated containing FtsH and specific members of the Band 7 superfamily of proteins (Ito and Akiyama, 2005). However, there is currently no evidence to support the formation of an equivalent type of FtsH/Band 7 protein supercomplex in cyanobacteria, although potential instability of the complex upon disruption of the membrane means that their presence cannot yet be totally ruled out (Boehm et al., 2009).
Role of the Deg proteases
A number of groups have argued in favour of an important physiological role for Deg proteases in D1 degradation, especially in chloroplasts (discussed by Edelman and Mattoo, 2008; Huesgen et al., 2009). In the case of A. thaliana, one stromally exposed (Deg2) and three lumenally exposed (Deg1, 5 and 8) Deg proteases of the TM have so far been characterized. Of these, Deg2 is not required for D1 degradation in planta (Huesgen et al., 2006) although it is able to cleave D1 in vitro (Haussuhl et al., 2001). Radioactive pulse–chase experiments have revealed that the rapid turnover of D1 is unimpaired in deg5 and deg8 single mutants as well as the deg5/deg8 double mutant under permissive growth conditions (Sun et al., 2007). This fact argues strongly against the obligatory role for Deg5 and Deg8 in D1 turnover. At higher light intensities, D1 degradation is impaired in the deg5/deg8 mutants, consistent with a possible auxiliary role in D1 degradation under conditions where the FtsH pathway is insufficient to cope with high levels of damage to PSII. However, the deg5/deg8 mutants grow poorly at high light, so an indirect effect of these mutations on D1 degradation, such as through a limitation in ATP availability for FtsH-mediated cleavage, cannot yet be excluded. Although the lumenal Deg1 protease has also been suggested to be involved in D1 degradation (Kapri-Pardes et al., 2007), D1 turnover has not yet been assessed in Deg1 knock-down mutants. Overall, the current mutant data support the hypothesis that the chloroplast Deg proteases are not obligatory for D1 degradation in vivo but might play a role at higher light intensities possibly to facilitate and accelerate the FtsH-mediated processive degradation of D1, presumably by cleaving interhelical loops, thereby preventing the formation of dangerous reactive oxygen species.
DYNAMICS OF ASSEMBLY AND REPAIR
Cellular location
Early studies, based on biochemical fractionation and immunoblotting, indicated that D1, D2, Cyt b-559 and PsbO were found in the CM as well as the TM of Synechocystis 6803 (Smith and Howe, 1993; Zak et al., 2001), whereas CP47 and CP43 were located exclusively in the TM fraction. Subsequently, Pakrasi and colleagues provided evidence that D1 and D2 in the CM were part of a PSII RC-like complex that was able to perform charge separation but not water oxidation (Keren et al., 2005a). The CtpA protease, which cleaves the C-terminal extension of pD1 and which is necessary for assembly of the CaMn4 cluster, was also found in the CM fraction. Based on these data it was proposed that the CM was the initial site of assembly of a D1–D2–Cyt b-559 complex which then migrated to the TM to complete its biogenesis into the holoenzyme (Zak et al., 2001).
Concerns have been raised about the purity of the CM and TM fractions generated following fractionation (Komenda et al., 2008). This fact notwithstanding, the question arises as to how PSII migrates from the CM to the TM. There is no evidence for a direct connection to allow the rapid diffusion of proteins from one membrane to the other (Liberton et al., 2006; Schneider et al., 2007). One possibility is that membrane vesicles might be involved in transporting PSII and other proteins from the CM to the TM (discussed by Schneider et al., 2007). Interestingly, ultrastructural studies have also revealed that TMs seem to converge at defined regions, called ‘thylakoid centres’, close to the CM (Kunkel, 1982; van de Meene et al., 2006). The composition and function of these ‘thylakoid centres’ are unknown, but they might play a role in gating the transport of proteins and lipid from the CM to the TM or even, conceivably, be the site of assembly and repair of PSII. More work is needed to test these ideas.
Where damaged PSII is repaired is also still a matter of debate. The immunochemical localization of the D1 C-terminal processing protease, CtpA, argues in favour of repair in the CM (Zak et al., 2001). However, both FtsH2, the protease implicated in the removal of damaged D1, and RC47, the PSII sub-complex that is thought to be the substrate for removal of damaged D1, are found in the TM (Komenda et al., 2006). One explanation to reconcile these differing viewpoints is the possibility that CtpA is actually present in the TM but escaped detection in earlier studies because of its relatively low abundance.
What happens to the pigment during repair?
Recent work from the Vermaas group has indicated that chlorophyll does not turn over at the same rate as the D1 sub-unit (Vavilin et al., 2005). This means that chlorophyll must be temporarily stored during PSII repair. Vermaas and colleagues have suggested, based on isotopic labelling of chlorophyll, that the phytol tail of chlorophyll a is removed during PSII repair and that the tetrapyrrole headgroup is reused (Vavilin and Vermaas, 2007). Taking into consideration the weak unfoldase activity of the FtsH protease, it is possible that the phytyl chains of the bound chlorophyll molecules may represent an important obstacle during the proposed processive degradation of D1 and must be removed to allow the pulling out of the D1 polypeptide chain from the membrane. Interestingly, the identity of the phytol-removing enzyme remains unknown as a gene encoding the homologue of the ‘classical’ plant chlorophyllase is missing from the genome of Synechocystis 6803 (Vavilin and Vermaas, 2007).
Under high light conditions PSII complexes become associated with SCPs (Promnares et al., 2006), which has led to the latter being regarded as excellent candidates to play a role in binding chlorophyll during PSII repair and, indeed, chlorophyll turnover is increased in their absence (Vavilin et al., 2007). However, the efficiency of repair and the rate of D1 turnover under physiological conditions do not seem to be compromised in a mutant of Synechocystis 6803 lacking the SCPs. Only when the mutant cells are exposed to extremely high irradiances is oxidative damage to PSII observed (J. Komenda, unpubl. res.). Accordingly, SCPs appear to act as ‘emergency’ scavengers of chlorophyll molecules that escape the normal control mechanisms of PSII repair. We assume that the key components involved in transient chlorophyll binding during selective D1 degradation still await discovery. It is even possible that FtsH proteases may execute this function.
Recent studies on a variety of Synechocystis 6803 mutants have also revealed that the overall level of cellular chlorophyll is inversely correlated to the rate of D1 synthesis during PSII repair. This finding raises the possibility that intensive repair may limit the availability of chlorophyll for the assembly of other chlorophyll-binding proteins such as PSI (J. Komenda, unpubl. res.).
Disassembly of PSII during repair and synchronizing the replacement of damaged D1
For both cyanobacteria and chloroplasts, disassembly of PSII is thought to involve detachment of the lumenal extrinsic sub-units and CP43 (Fig. 3). The resulting RC47 complex is a pivotal complex in PSII biogenesis as it seems to be involved in PSII assembly and the replacement of damaged D1 during repair (Figs 2 and 3) (Nixon et al., 2005). Key questions that remain to be answered include the mechanism that triggers monomerization of damaged PSII and partial disassembly to form the RC47 complex, and the nature of the degradation signal that is used to target damaged PSII sub-units such as D1 for degradation.
The recent crystal structures have revealed that the PsbO sub-unit from one PSII monomer interacts with the CP47 sub-unit of the second monomer in the dimeric complex (Guskov et al., 2009). Thus detachment or structural reorganization of PsbO on the lumenal side following damage to PSII could be a trigger for disassembly of the dimer to form the monomer. Consistent with this, a mutant of Synechocystis 6803 lacking PsbO fails to accumulate dimeric PSII (Komenda et al., 2010).
Given that CP43 plays a role in ligating the CaMn4 cluster, it is conceivable that damage to D1 and the cluster, coupled to detachment of PsbO, leads to detachment of CP43 to form the RC47 complex. According to the FtsH-only model of PSII repair it has been speculated that the N-terminus of D1 (and possibly of other thylakoid proteins destined for replacement) plays a key role in engaging with the FtsH protease, possibly by a conformational change induced by damage or partial disassembly (Fig. 3). Initial interactions between FtsH and PSII might occur within the membrane as well as the lumenal side of the membrane.
Recent structural studies have also identified a belt of lipid molecules around the D1 sub-unit in the PSII holoenzyme which might facilitate the extraction of damaged D1 from the RC47 complex (Loll et al., 2007). The structural work has emphasized the fact that PSII is a true lipoprotein complex and that bound lipid might play an important functional and structural role as suggested from analysis of mutants defective in lipid biosynthesis. For example, phosphatidylglycerol (PG) is important for optimizing electron transfer on the acceptor (Gombos et al., 2002) and donor sides (Sakurai et al., 2007) of the complex, for the stable attachment of CP43 and for accumulation of the PSII dimer (Laczko-Dobos et al., 2008). Both PG and digalactosyldiacylglycerol (DGDG) are needed for efficient repair of PSII, possibly at the level of forming the active dimeric complex (Sakurai et al., 2003; Mizusawa et al., 2009). Lipids also play a role in stabilizing the isolated dimeric PSII complex in chloroplasts (Kruse et al., 2000). Whether lipases play a role in the disassembly processes involved in PSII repair is presently unclear.
How the newly synthesized D1 sub-unit is integrated into the RC47 complex is unknown. The Synechocystis 6803 ftsH2 mutants impaired in PSII repair accumulate unassembled pD1, iD1 and mature D1 in the membrane (Komenda et al., 2006), which would suggest that a post-translation mechanism might be involved. In contrast, studies in chloroplasts have led to the conclusion that replacement occurs co-translationally (Zhang et al., 1999). In the case of a post-translational mechanism, Ycf48 appears to play a role in stabilizing unassembled pD1 in the membrane (Komenda et al., 2008).
Two lines of evidence support the idea that degradation of damaged D1 is synchronized to the availability of a replacement D1 sub-unit. First the rate of turnover of radiolabelled D1 in pulse–chase assays is reduced in the presence of an inhibitor of protein synthesis (Komenda and Barber, 1995). Secondly, mutants of Synechocystis 6803 that have reduced levels of psbA mRNA, and reduced rates of D1 synthesis, also show a reduced rate of degradation even in the absence of an inhibitor of protein synthesis (Komenda et al., 2000).
Because the various PSII assembly and repair complexes are incapable of water oxidation, they are potentially highly vulnerable to donor-side photoinhibition. Consequently one would anticipate that many of the mechanisms invoked in the past to protect non-oxygen-evolving core complexes against photoinhibition are actually highly relevant for assembly complexes. These mechanisms include changes to the redox potential of QA so that singlet oxygen production is reduced (Vass and Cser, 2009), and the operation of side-path electron donors ChlZ and Cyt b-559 in PSII (Faller et al., 2005). In addition, recent work has highlighted the possibility that RC47 is ‘switched off’ in vivo by spillover of excitation energy to PSI (Shimada et al., 2008).
CONCLUSIONS AND FUTURE OUTLOOK
The combination of molecular biology, biochemistry and structural biology is proving a successful approach to understanding the molecular details of the assembly and repair of PSII. In the short term it is highly likely that there will be additional information on the structures and binding sites for the various assembly factors. Conceivably it might even be possible, using a suitable engineered strain of T. elongatus, to isolate defined PSII assembly complexes with associated assembly factors for crystallization studies.
In a broader context, very little is known about how large membrane-bound complexes are assembled and repaired (Daley, 2008). The structural information available now for PSII and the relatively high rate of repair of PSII make it a suitable model complex to study fundamental aspects of the biogenesis of membrane complexes, including the mechanism of insertion of pigments and other co-factors and the dynamic exchange of a single sub-unit in a multisub-unit complex.
Another main area of study will be to investigate the dynamics of the assembly and repair processes in the cyanobacterial cell (Sarcina et al., 2006) and how sub-complexes are trafficked around the membrane system. Evidence is emerging that the TM and CM are more heterogeneous than previously thought, with proteins possibly segregated into specific regions (Srivastava et al., 2006; Nevo et al., 2007; Vermaas et al., 2008). Various types of advanced fluorescent microscopy techniques, coupled with inducible gene expression systems, might allow the movement of PSII assembly complexes to be visualized (Mullineaux and Sarcina, 2002; Vermaas et al., 2008).
The recent advances in identifying the various proteins involved in PSII assembly and repair also mean that experiments can now be designed to increase their expression in the chloroplast or cyanobacterium to test whether repair can be made more efficient and photosynthesis made more resistant to visible light stress. However, it should be borne in mind that the capacity for PSII repair is dependent on a number of factors including the availability of chlorophyll and ATP, and might require an integrated systems biology approach.
ACKNOWLEDGEMENTS
P.J.N. and J.K. are grateful to all their collaborators, past and present, for their scientific and personal support. P.J.N. is also extremely grateful to the New Zealand Society of Plant Biologists for the kind invitation to address its annual meeting in Dunedin in December 2008 and for the generous hospitality of its members. This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic [project no. MSM6007665808], by the Czech Academy of Sciences [projects no. AV0Z50200510 and IAA400200801] and by the Biotechnology and Biological Sciences Research Council.
LITERATURE CITED
- Adam Z, Clarke AK. Cutting edge of chloroplast proteolysis. Trends in Plant Science. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- Adam Z, Zaltsman A, Sinvany-Villalobo G, Sakamoto W. FtsH proteases in chloroplasts and cyanobacteria. Physiologia Plantarum. 2005;123:386–390. [Google Scholar]
- Adir N, Zer H, Shochat S, Ohad I. Photoinhibition – a historical perspective. Photosynthesis Research. 2003;76:343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Ito K. Reconstitution of membrane proteolysis by FtsH. Journal of Biological Chemistry. 2003;278:18146–18153. doi: 10.1074/jbc.M302152200. [DOI] [PubMed] [Google Scholar]
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB. The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem-II reaction-center complex. Proceedings of the National Academy of Sciences, USA. 1994;91:8082–8086. doi: 10.1073/pnas.91.17.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. Photoinhibition of photosystem 2 – inactivation, protein damage and turnover. Biochimica et Biophysica Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, et al. Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. Journal of Experimental Botany. 2005;56:347–356. doi: 10.1093/jxb/eri041. [DOI] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, et al. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. Journal of Biological Chemistry. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B. Too much of a good thing – light can be bad for photosynthesis. Trends in Biochemical Sciences. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- Barber J, Iwata S. Refined X-ray structure of photosystem II and its implications. In: Wydrzynski T, Satoh K, editors. Photosystem II: the light-driven water:plastoquinone oxidoreductase. The Netherlands: Springer; 2005. pp. 469–489. [Google Scholar]
- Barker M, de Vries R, Nield J, Komenda J, Nixon PJ. The Deg proteases protect Synechocystis sp PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. Journal of Biological Chemistry. 2006;281:30347–30355. doi: 10.1074/jbc.M601064200. [DOI] [PubMed] [Google Scholar]
- Barker M, Boehm M, Nixon PJ, Nield J. Structural analysis of an FtsH2/FtsH3 complex isolated from Synechocystis sp. PCC 6803. In: Allen JF, Gantt E, Golbeck JH, Osmond B, editors. Photosynthesis. Energy from the sun. 14th International Congress on Photosynthesis. The Netherlands: Springer; 2008. pp. 738–740. [Google Scholar]
- Bautista JA, Tracewell CA, Schlodder E, Cunningham FX, Brudvig GW, Diner BA. Construction and characterization of genetically modified Synechocystis sp. PCC 6803 photosystem II core complexes containing carotenoids with shorter pi-conjugation than beta-carotene. Journal of Biological Chemistry. 2005;280:38839–38850. doi: 10.1074/jbc.M504953200. [DOI] [PubMed] [Google Scholar]
- Bentley FK, Luo H, Dilbeck P, Burnap RL, Eaton-Rye JJ. Effects of inactivating psbM and psbT on photodamage and assembly of photosystem II in Synechocystis sp PCC 6803. Biochemistry. 2008;47:11637–11646. doi: 10.1021/bi800804h. [DOI] [PubMed] [Google Scholar]
- Boehm M, Nield J, Zhang PP, Aro EM, Komenda J, Nixon PJ. Structural and mutational analysis of Band 7 proteins in the cyanobacterium Synechocystis sp strain PCC 6803. Journal of Bacteriology. 2009;191:6425–6435. doi: 10.1128/JB.00644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Qian M, Pierce C. The manganese-stabilizing protein of photosystem II modifies the in vivo deactivation and photoactivation kinetics of the H2O oxidation complex in Synechocystis sp. PCC6803. Biochemistry. 1996;35:874–882. doi: 10.1021/bi951964j. [DOI] [PubMed] [Google Scholar]
- Canovas PM, Barber J. Detection of a 10 kDa breakdown product containing the c-terminus of the D1-protein in photoinhibited wheat leaves suggests an acceptor side mechanism. FEBS Letters. 1993;324:341–344. doi: 10.1016/0014-5793(93)80147-m. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang DY, Guo JK, et al. A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Molecular Biology. 2006;61:567–575. doi: 10.1007/s11103-006-0031-x. [DOI] [PubMed] [Google Scholar]
- Cheregi O, Sicora C, Kos PB, Barker M, Nixon PJ, Vass I. The role of the FtsH and Deg proteases in the repair of UV-B radiation-damaged photosystem II in the cyanobacterium Synechocystis PCC 6803. Biochimica et Biophysica Acta. 2007;1767:820–828. doi: 10.1016/j.bbabio.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Cormann KU, Bangert JA, Ikeuchi M, Rogner M, Stoll R, Nowaczyk MM. Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair. Biochemistry. 2009;48:8768–8770. doi: 10.1021/bi9012726. [DOI] [PubMed] [Google Scholar]
- Daley DO. The assembly of membrane proteins into complexes. Current Opinion in Structural Biology. 2008;18:420–424. doi: 10.1016/j.sbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- De Las Rivas J, Roman A. Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photochemical and Photobiological Sciences. 2005;4:1003–1010. doi: 10.1039/b506874f. [DOI] [PubMed] [Google Scholar]
- Dewez D, Park S, Garcia-Cerdan JG, Lindberg P, Melis A. Mechanism of REP27 protein action in the D1 protein turnover and photosystem II repair from photodamage. Plant Physiology. 2009;151:88–99. doi: 10.1104/pp.109.140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobakova M, Tichy M, Komenda J. Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp PCC 6803. Plant Physiology. 2007;145:1681–1691. doi: 10.1104/pp.107.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobakova M, Sobotka R, Tichy M, Komenda J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp PCC 6803. Plant Physiology. 2009;149:1076–1086. doi: 10.1104/pp.108.130039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganov NAM, Bhaya D, Grossman AR. Cyanobacterial protein with similarity to the chlorophyll a/b binding-proteins of higher-plants – evolution and regulation. Proceedings of the National Academy of Sciences, USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drath M, Kloft N, Batschauer A, Marin K, Novak J, Forchhammer K. Ammonia triggers photodamage of photosystem II in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiology. 2008;147:206–215. doi: 10.1104/pp.108.117218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman M, Mattoo AK. D1-protein dynamics in photosystem II: the lingering enigma. Photosynthesis Research. 2008;98:609–620. doi: 10.1007/s11120-008-9342-x. [DOI] [PubMed] [Google Scholar]
- Enami I, Okumura A, Nagao R, Suzuki T, Iwai M, Shen JR. Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynthesis Research. 2008;98:349–363. doi: 10.1007/s11120-008-9343-9. [DOI] [PubMed] [Google Scholar]
- Ermakova-Gerdes S, Vermaas W. Inactivation of the open reading frame slr0399 in Synechocystis sp PCC 6803 functionally complements mutations near the Q(A) niche of photosystem II – a possible role of slr0399 as a chaperone for quinone binding. Journal of Biological Chemistry. 1999;274:30540–30549. doi: 10.1074/jbc.274.43.30540. [DOI] [PubMed] [Google Scholar]
- Faller P, Fufezan C, Rutherford AW. Side-path electron donors: cytochrome b559, chlorophyll Z and β-carotene. In: Wydrzynski T, Satoh K, editors. Photosystem II: the light-driven water:plastoquinone oxidoreductase. The Netherlands: Springer; 2005. pp. 347–365. [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- Funk C, Vermaas W. A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry. 1999;38:9397–9404. doi: 10.1021/bi990545+. [DOI] [PubMed] [Google Scholar]
- Gombos Z, Varkonyi Z, Hagio M, et al. Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone QB in the photosystem II reaction center. Biochemistry. 2002;41:3796–3802. doi: 10.1021/bi011884h. [DOI] [PubMed] [Google Scholar]
- Green BR, Gantt E. Distal and extrinsic photosystem II antennas. In: Wydrzynski T, Satoh K, editors. Photosystem II: the light-driven water:plastoquinone oxidoreductase. The Netherlands: Springer; 2005. pp. 23–44. [Google Scholar]
- Greenberg BM, Gaba V, Mattoo AK, Edelman M. Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO Journal. 1987;6:2865–2869. doi: 10.1002/j.1460-2075.1987.tb02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2·9-angstrom resolution and the role of quinones, lipids, channels and chloride. Nature Structural and Molecular Biology. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Hakala M, Tuominen I, Keränen M, Tyystjärvi T, Tyystjärvi E. Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochimica et Biophysica Acta. 2005;1706:68–80. doi: 10.1016/j.bbabio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Haussuhl K, Andersson B, Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO Journal. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QF, Vermaas W. Chlorophyll a availability affects psbA translation and D1 precursor processing in vivo in Synechocystis sp. PCC 6803. Proceedings of the National Academy of Sciences, USA. 1998;95:5830–5835. doi: 10.1073/pnas.95.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henmi T, Iwai M, Ikeuchi M, Kawakami K, Shen JR, Kamiya N. X-ray crystallographic and biochemical characterizations of a mutant photosystem II complex from Thermosynechococcus vulcanus with the psbTc gene inactivated by an insertion mutation. Journal of Synchrotron Radiation. 2008;15:304–307. doi: 10.1107/S0909049508002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I. Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Letters. 2006;580:6929–6932. doi: 10.1016/j.febslet.2006.11.058. [DOI] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I. Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Research in Microbiology. 2009;160:726–732. doi: 10.1016/j.resmic.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Inagaki N, Yamamoto Y, Satoh K. A sequential two-step proteolytic process in the carboxyl-terminal truncation of precursor D1 protein in Synechocystis sp PCC6803. FEBS Letters. 2001;509:197–201. doi: 10.1016/s0014-5793(01)03180-5. [DOI] [PubMed] [Google Scholar]
- Inoue-Kashino N, Takahashi T, Ban A, et al. Evidence for a stable association of Psb30 (Ycf12) with photosystem II core complex in the cyanobacterium Synechocystis sp PCC 6803. Photosynthesis Research. 2008;98:323–335. doi: 10.1007/s11120-008-9340-z. [DOI] [PubMed] [Google Scholar]
- Ishihara S, Takabayashi A, Ido K, Endo T, Ifuku K, Sato F. Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiology. 2007;145:668–679. doi: 10.1104/pp.107.105866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Schroder WP, Funk C. Functional analysis of the PsbP-like protein (sll1418) in Synechocystis sp PCC 6803. Photosynthesis Research. 2005;84:257–262. doi: 10.1007/s11120-005-0477-8. [DOI] [PubMed] [Google Scholar]
- Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annual Review of Microbiology. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Shestakov SV, Pakrasi HB. The carboxyl-terminal extension of the precursor D1 protein of photosystem II is required for optimal photosynthetic performance of the cyanobacterium Synechocystis sp PCC 6803. Plant Physiology. 2000;124:1403–1411. doi: 10.1104/pp.124.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Katayama M, Ikeuchi M. Absence of the psbH gene product destabilizes the photosystem II complex and prevents association of the photosystem II-X protein in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. Photosynthesis Research. 2006;87:313–322. doi: 10.1007/s11120-005-9013-0. [DOI] [PubMed] [Google Scholar]
- Iwai M, Suzuki T, Dohmae N, Inoue Y, Ikeuchi M. Absence of the PsbZ subunit prevents association of PsbK and Ycf12 with the PSII complex in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. Plant and Cell Physiology. 2007;48:1758–1763. doi: 10.1093/pcp/pcm148. [DOI] [PubMed] [Google Scholar]
- Kamata T, Hiramoto H, Morita N, Shen JR, Mann NH, Yamamoto Y. Quality control of photosystem II: an FtsH protease plays an essential role in the turnover of the reaction center D1 protein in Synechocystis PCC 6803 under heat stress as well as light stress conditions. Photochemical and Photobiological Sciences. 2005;4:983–990. doi: 10.1039/b506068k. [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. The Plant Cell. 2007;19:1039–1047. doi: 10.1105/tpc.106.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, et al. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–8012. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiology. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Ikeuchi M. Targeted disruption of psbX and biochemical characterization of photosystem II complex in the thermophilic cyanobacterium Synechococcus elongatus. Plant and Cell Physiology. 2001;42:179–188. doi: 10.1093/pcp/pce024. [DOI] [PubMed] [Google Scholar]
- Kawabata Y, Henmi T, Iwai M, et al. Crystal structure analysis of a mutant photosystem II complex lacking PsbM from Thermosynechococcus vulcanus. Plant and Cell Physiology. 2007;48:S97. [Google Scholar]
- Kawakami K, Iwai M, Ikeuchi M, Kamiya N, Shen JR. Location of PsbY in oxygen-evolving photosystem II revealed by mutagenesis and X-ray crystallography. FEBS Letters. 2007;581:4983–4987. doi: 10.1016/j.febslet.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Keren N, Berg A, Vankan PJM, Levanon H, Ohad I. Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: the role of back electron flow. Proceedings of the National Academy of Sciences, USA. 1997;94:1579–1584. doi: 10.1073/pnas.94.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Liberton M, Pakrasi HB. Photochemical competence of assembled photosystem II core complex in cyanobacterial plasma membrane. Journal of Biological Chemistry. 2005a;280:6548–6553. doi: 10.1074/jbc.M410218200. [DOI] [PubMed] [Google Scholar]
- Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB. Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. The Plant Cell. 2005b;17:2768–2781. doi: 10.1105/tpc.105.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Eichacker LA, Rudiger W, Mullet JE. Chlorophyll regulates accumulation of the plastid-encoded chlorophyll proteins P700 and D1 by increasing apoprotein stability. Plant Physiology. 1994;104:907–916. doi: 10.1104/pp.104.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B, Ossenbuhl F, Sikorski M, Berry S, Eichacker L, Nickelsen J. PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp PCC 6803. Journal of Biological Chemistry. 2004;279:44639–44644. doi: 10.1074/jbc.M405393200. [DOI] [PubMed] [Google Scholar]
- Komenda J, Barber J. Comparison of PsbO and PsbH deletion mutants of Synechocystis PCC-6803 indicates that degradation of D1 protein is regulated by the Q(B) site and dependent on protein-synthesis. Biochemistry. 1995;34:9625–9631. doi: 10.1021/bi00029a040. [DOI] [PubMed] [Google Scholar]
- Komenda J, Hassan HAG, Diner BA, Debus RJ, Barber J, Nixon PJ. Degradation of the photosystem II D1 and D2 proteins in different strains of the cyanobacterium Synechocystis PCC 6803 varying with respect to the type and level of psbA transcript. Plant Molecular Biology. 2000;42:635–645. doi: 10.1023/a:1006305308196. [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Muller BC, Dobakova M, Granvogl B, Eichacker LA. Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. Journal of Biological Chemistry. 2004;279:48620–48629. doi: 10.1074/jbc.M405725200. [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichy M, Eichacker LA. The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant and Cell Physiology. 2005;46:1477–1483. doi: 10.1093/pcp/pci159. [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuvikova S, et al. The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp PCC 6803. Journal of Biological Chemistry. 2006;281:1145–1151. doi: 10.1074/jbc.M503852200. [DOI] [PubMed] [Google Scholar]
- Komenda J, Kuvikova S, Granvogl B, Eichacker LA, Diner BA, Nixon PJ. Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of photosystem II in Synechocystis PCC 6803. Biochimica et Biophysica Acta. 2007a;1767:829–837. doi: 10.1016/j.bbabio.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichy M, Prasil O, et al. The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. The Plant Cell. 2007b;19:2839–2854. doi: 10.1105/tpc.107.053868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichy M, Prasil O, Eichacker LA, Nixon PJ. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp PCC 6803. Journal of Biological Chemistry. 2008;283:22390–22399. doi: 10.1074/jbc.M801917200. [DOI] [PubMed] [Google Scholar]
- Komenda J, Knoppová J, Krynická V, Nixon PJ, Tichý M. Role of FtsH2 in the repair of Photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochimica et Biophysica Acta, in press. 2010 doi: 10.1016/j.bbabio.2010.02.006. doi:10.1016/j.bbabio.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynthesis Research. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- Kruse O, Hankamer B, Konczak C, et al. Phosphatidylglycerol is involved in the dimerization of photosystem II. Journal of Biological Chemistry. 2000;275:6509–6514. doi: 10.1074/jbc.275.9.6509. [DOI] [PubMed] [Google Scholar]
- Kufryk GI, Vermaas WFJ. A novel protein involved in the functional assembly of the oxygen-evolving complex of photosystem II in Synechocystis sp PCC 6803. Biochemistry. 2001;40:9247–9255. doi: 10.1021/bi0026526. [DOI] [PubMed] [Google Scholar]
- Kufryk GI, Vermaas WFJ. Slr2013 is a novel protein regulating functional assembly of photosystem II in Synechocystis sp strain PCC 6803. Journal of Bacteriology. 2003;185:6615–6623. doi: 10.1128/JB.185.22.6615-6623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufryk G, Hernandez-Prieto MA, Kieselbach T, Miranda H, Vermaas W, Funk C. Association of small CAB-like proteins (SCPs) of Synechocystis sp PCC 6803 with photosystem II. Photosynthesis Research. 2008;95:135–145. doi: 10.1007/s11120-007-9244-3. [DOI] [PubMed] [Google Scholar]
- Kunkel DD. Thylakoid centers: structures associated with the cyanobacterial photosynthetic membrane system. Archives of Microbiology. 1982;133:97–99. [Google Scholar]
- Kuvikova S, Tichy M, Komenda J. A role of the C-terminal extension of the photosystem II D1 protein in sensitivity of the cyanobacterium Synechocystis PCC 6803 to photoinhibition. Photochemical and Photobiological Sciences. 2005;4:1044–1048. doi: 10.1039/b506059a. [DOI] [PubMed] [Google Scholar]
- Laczko-Dobos H, Ughy B, Toth SZ, et al. Role of phosphatidylglycerol in the function and assembly of photosystem II reaction center, studied in a cdsA-inactivated PAL mutant strain of Synechocystis sp. PCC6803 that lacks phycobilisomes. Biochimica et Biophysica Acta. 2008;1777:1184–1194. doi: 10.1016/j.bbabio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Liberton M, Berg RH, Heuser J, Roth R, Pakrasi HB. Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp strain PCC 6803. Protoplasma. 2006;227:129–138. doi: 10.1007/s00709-006-0145-7. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. The Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3·0 angstrom resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Lipids in photosystem II: interactions with protein and cofactors. Biochimica et Biophysica Acta. 2007;1767:509–519. doi: 10.1016/j.bbabio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Lundin B, Hansson M, Schoefs B, Vener AV, Spetea C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. The Plant Journal. 2007;49:528–539. doi: 10.1111/j.1365-313X.2006.02976.x. [DOI] [PubMed] [Google Scholar]
- Lundin B, Nurmi M, Rojas-Stuetz M, Aro EM, Adamska I, Spetea C. Towards understanding the functional difference between the two PsbO isoforms in Arabidopsis thaliana – insights from phenotypic analyses of psbO knockout mutants. Photosynthesis Research. 2008;98:405–414. doi: 10.1007/s11120-008-9325-y. [DOI] [PubMed] [Google Scholar]
- Lupinkova L, Komenda J. Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochemistry and Photobiology. 2004;79:152–162. doi: 10.1562/0031-8655(2004)079<0152:omotpi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ma JF, Peng LW, Guo JK, Lu QT, Lu CM, Zhang LX. LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. The Plant Cell. 2007;19:1980–1993. doi: 10.1105/tpc.107.050526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mabbitt PD, Rautureau GJP, Day CL, Wilbanks SM, Eaton-Rye JJ, Hinds MG. Solution structure of Psb27 from cyanobacterial photosystem II. Biochemistry. 2009;48:8771–8773. doi: 10.1021/bi901309c. [DOI] [PubMed] [Google Scholar]
- Masamoto K, Hisatomi S, Sakurai I, Gombos Z, Wada H. Requirement of carotene isomerization for the assembly of photosystem II in Synechocystis sp PCC 6803. Plant and Cell Physiology. 2004;45:1325–1329. doi: 10.1093/pcp/pch144. [DOI] [PubMed] [Google Scholar]
- van de Meene AML, Hohmann-Marriott MF, Vermaas WFJ, Roberson RW. The three-dimensional structure of the cyanobacterium Synechocystis sp PCC 6803. Archives of Microbiology. 2006;184:259–270. doi: 10.1007/s00203-005-0027-y. [DOI] [PubMed] [Google Scholar]
- Metz JG, Nixon PJ, Rogner M, Brudvig GW, Diner BA. Directed alteration of the D1 polypeptide of photosystem II – evidence that tyrosine-161 is the redox component, Z, connecting the oxygen-evolving complex to the primary electron-donor, P680. Biochemistry. 1989;28:6960–6969. doi: 10.1021/bi00443a028. [DOI] [PubMed] [Google Scholar]
- Meurer J, Plucken H, Kowallik KV, Westhoff P. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO Journal. 1998;17:5286–5297. doi: 10.1093/emboj/17.18.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao M. Involvement of active oxygen species in degradation of the D1 protein under strong illumination in isolated subcomplexes of photosystem II. Biochemistry. 1994;33:9722–9730. doi: 10.1021/bi00198a043. [DOI] [PubMed] [Google Scholar]
- Mizusawa N, Sakurai I, Sato N, Wada H. Lack of digalactosyldiacylglycerol increases the sensitivity of Synechocystis sp PCC 6803 to high light stress. FEBS Letters. 2009;583:718–722. doi: 10.1016/j.febslet.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Muh F, Renger T, Zouni A. Crystal structure of cyanobacterial photosystem II at 3·0 angstrom resolution: a closer look at the antenna system and the small membrane-intrinsic subunits. Plant Physiology and Biochemistry. 2008;46:238–264. doi: 10.1016/j.plaphy.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Sarcina M. Probing the dynamics of photosynthetic membranes with fluorescence recovery after photobleaching. Trends in Plant Science. 2002;7:237–240. doi: 10.1016/s1360-1385(02)02283-5. [DOI] [PubMed] [Google Scholar]
- Mulo P, Sirpio S, Suorsa M, Aro EM. Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynthesis Research. 2008;98:489–501. doi: 10.1007/s11120-008-9320-3. [DOI] [PubMed] [Google Scholar]
- Nevo R, Charuvi D, Shimoni E, et al. Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria. EMBO Journal. 2007;26:1467–1473. doi: 10.1038/sj.emboj.7601594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Trost JT, Diner BA. Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp PCC 6803 – assembly requires a free carboxyl group at C-terminal position 344. Biochemistry. 1992;31:10859–10871. doi: 10.1021/bi00159a029. [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Barker M, Boehm M, de Vries R, Komenda J. FtsH-mediated repair of the photosystem II complex in response to light stress. Journal of Experimental Botany. 2005;56:357–363. doi: 10.1093/jxb/eri021. [DOI] [PubMed] [Google Scholar]
- Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, Rogner M. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. The Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I, Kyle DJ, Arntzen CJ. Membrane-protein damage and repair – removal and replacement of inactivated 32-kilodalton polypeptide in chloroplast membranes. Journal of Cell Biology. 1984;99:481–485. doi: 10.1083/jcb.99.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Allakhverdiev SI, Takahashi S, et al. Two-step mechanism of photodamage to photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry. 2005;44:8494–8499. doi: 10.1021/bi047518q. [DOI] [PubMed] [Google Scholar]
- Ortega J, Iwanczyk J, Jomaa A. Escherichia coli DegP: a structure-driven functional model. Journal of Bacteriology. 2009;191:4705–4713. doi: 10.1128/JB.00472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LW, Ma JF, Chi W, et al. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. The Plant Cell. 2006;18:955–969. doi: 10.1105/tpc.105.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploscher M, Granvogl B, Zoryan M, Reisinger V, Eichacker LA. Mass spectrometric characterization of membrane integral low molecular weight proteins from photosystem II in barley etioplasts. Proteomics. 2009;9:625–635. doi: 10.1002/pmic.200800337. [DOI] [PubMed] [Google Scholar]
- Plucken H, Muller B, Grohmann D, Westhoff P, Eichacker LA. The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Letters. 2002;532:85–90. doi: 10.1016/s0014-5793(02)03634-7. [DOI] [PubMed] [Google Scholar]
- Promnares K, Komenda J, Bumba L, Nebesarova J, Vacha F, Tichy M. Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. Journal of Biological Chemistry. 2006;281:32705–32713. doi: 10.1074/jbc.M606360200. [DOI] [PubMed] [Google Scholar]
- Rappaport F, Diner BA. Primary photochemistry and energetics leading to the oxidation of the (Mn)4Ca cluster and to the evolution of molecular oxygen in photosystem II. Coordination Chemistry Reviews. 2008;252:259–272. [Google Scholar]
- Rogner M, Chisholm DA, Diner BA. Site-directed mutagenesis of the psbC gene of photosystem II – isolation and functional characterization of CP43-less photosystem II core complexes. Biochemistry. 1991;30:5387–5395. doi: 10.1021/bi00236a009. [DOI] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB. The Psb27 protein facilitates manganese cluster assembly in photosystem II. Journal of Biological Chemistry. 2008;283:4044–4050. doi: 10.1074/jbc.M708960200. [DOI] [PubMed] [Google Scholar]
- Roose JL, Wegener KM, Pakrasi HB. The extrinsic proteins of photosystem II. Photosynthesis Research. 2007a;92:369–387. doi: 10.1007/s11120-006-9117-1. [DOI] [PubMed] [Google Scholar]
- Roose JL, Kashino Y, Pakrasi HB. The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proceedings of the National Academy of Sciences, USA. 2007b;104:2548–2553. doi: 10.1073/pnas.0609337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. Protein degradation machineries in plastids. Annual Review of Plant Biology. 2006;57:599–621. doi: 10.1146/annurev.arplant.57.032905.105401. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. The Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Hagio M, Gombos Z, Tyystjarvi T, Paakkarinen V, Aro EM, Wada H. Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiology. 2003;133:1376–1384. doi: 10.1104/pp.103.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Mizusawa N, Ohashi S, Kobayashi M, Wada H. Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiology. 2007;144:1336–1346. doi: 10.1104/pp.107.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarcina M, Bouzovitis N, Mullineaux CW. Mobilization of photosystem II induced by intense red light in the cyanobacterium Synechococcus sp PCC7942. The Plant Cell. 2006;18:457–464. doi: 10.1105/tpc.105.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Yamamoto Y. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynthesis Research. 2007;94:203–215. doi: 10.1007/s11120-007-9191-z. [DOI] [PubMed] [Google Scholar]
- Satoh S, Ikeuchi M, Mimuro M, Tanaka A. Chlorophyll b expressed in cyanobacteria functions as a light harvesting antenna in photosystem I through flexibility of the proteins. Journal of Biological Chemistry. 2001;276:4293–4297. doi: 10.1074/jbc.M008238200. [DOI] [PubMed] [Google Scholar]
- Schneider D, Fuhrmann E, Scholz I, Hess WR, Graumann PL. Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biology. 2007;8:39. doi: 10.1186/1471-2121-8-39. doi:10.1186/1471-2121-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottkowski M, Gkalympoudis S, Tzekova N, et al. Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp PCC 6803. Journal of Biological Chemistry. 2009;284:1813–1819. doi: 10.1074/jbc.M806116200. [DOI] [PubMed] [Google Scholar]
- Shen J-R, Qian M, Inoue Y, Burnap RL. Functional characterization of Synechocystis sp. PCC 6803 ΔpsbU and ΔpsbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry. 1998;37:1551–1558. doi: 10.1021/bi971676i. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Tsuchiya T, Akimoto S, et al. Spectral properties of the CP43-deletion mutant of Synechocystis sp PCC 6803. Photosynthesis Research. 2008;98:303–314. doi: 10.1007/s11120-008-9350-x. [DOI] [PubMed] [Google Scholar]
- Shipton CA, Barber J. Characterization of photoinduced breakdown of the D1-polypeptide in isolated reaction centers of photosystem-II. Biochimica et Biophysica Acta. 1992;1099:85–90. [PubMed] [Google Scholar]
- Silva P, Thompson E, Bailey S, et al. FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. The Plant Cell. 2003;15:2152–2164. doi: 10.1105/tpc.012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Howe CJ. The distribution of photosystem-I and photosystem-II polypeptides between the cytoplasmic and thylakoid membranes of cyanobacteria. FEMS Microbiology Letters. 1993;110:341–347. [Google Scholar]
- Sobotka R, Komenda J, Bumba L, Tichy M. Photosystem II assembly in CP47 mutant of Synechocystis sp PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. Journal of Biological Chemistry. 2005;280:31595–31602. doi: 10.1074/jbc.M505976200. [DOI] [PubMed] [Google Scholar]
- Sobotka R, McLean S, Zuberova M, Hunter CN, Tichy M. The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. Journal of Bacteriology. 2008;190:2086–2095. doi: 10.1128/JB.01678-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetea C, Hundal T, Lohmann F, Andersson B. GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proceedings of the National Academy of Sciences, USA. 1999;96:6547–6552. doi: 10.1073/pnas.96.11.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Battchikova N, Norling B, Aro EM. Plasma membrane of Synechocystis PCC 6803: a heterogeneous distribution of membrane proteins. Archives of Microbiology. 2006;185:238–243. doi: 10.1007/s00203-006-0086-8. [DOI] [PubMed] [Google Scholar]
- Summerfield TC, Shand JA, Bentley FK, Eaton-Rye JJ. PsbQ (Sll1638) in Synechocystis sp PCC 6803 is required for photosystem II activity in specific mutants and in nutrient-limiting conditions. Biochemistry. 2005a;44:805–815. doi: 10.1021/bi048394k. [DOI] [PubMed] [Google Scholar]
- Summerfield TC, Winter RT, Eaton-Rye JJ. Investigation of a requirement for the PsbP-like protein in Synechocystis sp PCC 6803. Photosynthesis Research. 2005b;84:263–268. doi: 10.1007/s11120-004-6431-3. [DOI] [PubMed] [Google Scholar]
- Sun XW, Wang LY, Zhang LX. Involvement of DEG5 and DEG8 proteases in the turnover of the photosystem II reaction center D1 protein under heat stress in Arabidopsis thaliana. Chinese Science Bulletin. 2007;52:1742–1745. [Google Scholar]
- Takahashi S, Murata N. How do environmental stresses accelerate photoinhibition? Trends in Plant Science. 2008;13:178–182. doi: 10.1016/j.tplants.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Inoue-Kashino N, Ozawa S, Takahashi Y, Kashino Y, Satoh K. Photosystem II complex in vivo is a monomer. Journal of Biological Chemistry. 2009;284:15598–15606. doi: 10.1074/jbc.M109.000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaka K, Iwai M, Umena Y, et al. Structural and functional studies on Ycf12 (Psb30) and PsbZ-deletion mutants from a thermophilic cyanobacterium. Biochimica et Biophysica Acta. 2010;1797:278–284. doi: 10.1016/j.bbabio.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Thornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, Pakrasi HB. Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. The Plant Cell. 2004;16:2164–2175. doi: 10.1105/tpc.104.023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomo T, Akimoto S, Ito H, et al. Replacement of chlorophyll with di-vinyl chlorophyll in the antenna and reaction center complexes of the cyanobacterium Synechocystis sp PCC 6803: characterization of spectral and photochemical properties. Biochimica et Biophysica Acta. 2009;1787:191–200. doi: 10.1016/j.bbabio.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Trebst A, Depka B. Role of carotene in the rapid turnover and assembly of photosystem II in Chlamydomonas reinhardtii. FEBS Letters. 1997;400:359–362. doi: 10.1016/s0014-5793(96)01419-6. [DOI] [PubMed] [Google Scholar]
- Vass I, Cser K. Janus-faced charge recombinations in photosystem II photoinhibition. Trends in Plant Science. 2009;14:200–205. doi: 10.1016/j.tplants.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Vavilin D, Vermaas W. Continuous chlorophyll degradation accompanied by chlorophyllide and phytol reutilization for chlorophyll synthesis in Synechocystis sp PCC 6803. Biochimica et Biophysica Acta. 2007;1767:920–929. doi: 10.1016/j.bbabio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Vavilin D, Brune DC, Vermaas W. N-15-labeling to determine chlorophyll synthesis and degradation in Synechocystis sp PCC 6803 strains lacking one or both photosystems. Biochimica et Biophysica Acta. 2005;1708:91–101. doi: 10.1016/j.bbabio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Vavilin D, Yao D, Vermaas W. Small Cab-like proteins retard degradation of photosystem II-associated chlorophyll in Synechocystis sp PCC 6803 – kinetic analysis of pigment labeling with N-15 andC-13. Journal of Biological Chemistry. 2007;282:37660–37668. doi: 10.1074/jbc.M707133200. [DOI] [PubMed] [Google Scholar]
- Vermaas WF, Timlim JA, Jones HD, et al. In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proceedings of the National Academy of Sciences, USA. 2008;105:4050–4055. doi: 10.1073/pnas.0708090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sullivan RW, Kight A, et al. Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiology. 2004;136:3594–3604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Iwai M, Narikawa R, Ikeuchi M. Is the photosystem II complex a monomer or a dimer? Plant and Cell Physiology. 2009;50:1674–1680. doi: 10.1093/pcp/pcp112. [DOI] [PubMed] [Google Scholar]
- Wegener KM, Welsh EA, Thornton LE, et al. High sensitivity proteomics assisted discovery of a novel operon involved in the assembly of photosystem II, a membrane protein complex. Journal of Biological Chemistry. 2008;283:27829–27837. doi: 10.1074/jbc.M803918200. [DOI] [PubMed] [Google Scholar]
- Williams JGK. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic-engineering methods in Synechocystis 6803. Methods in Enzymology. 1988;167:766–778. [Google Scholar]
- Wu QG, Vermaas WFJ. Light-dependent chlorophyll a biosynthesis upon chlL deletion in wild-type and photosystem I-less strains of the cyanobacterium Synechocystis sp PCC 6803. Plant Molecular Biology. 1995;29:933–945. doi: 10.1007/BF00014967. [DOI] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Vermaas W. Chlorophyll b can serve as the major pigment in functional photosystem II complexes of cyanobacteria. Proceedings of the National Academy of Sciences, USA. 2001;98:14168–14173. doi: 10.1073/pnas.251530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Kieselbach T, Komenda J, et al. Localization of the small CAB-like proteins in photosystem II. Journal of Biological Chemistry. 2007;282:267–276. doi: 10.1074/jbc.M605463200. [DOI] [PubMed] [Google Scholar]
- Yi L, Dalbey RE. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (Review) Molecular Membrane Biology. 2005;22:101–111. doi: 10.1080/09687860500041718. [DOI] [PubMed] [Google Scholar]
- Zak E, Norling B, Maitra R, Huang F, Andersson B, Pakrasi HB. The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proceedings of the National Academy of Sciences, USA. 2001;98:13443–13448. doi: 10.1073/pnas.241503898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LG, Wei Q, Wu WJ, et al. Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the FtsH-mediated inhibition of chloroplast development in Arabidopsis. The Plant Journal. 2009;58:1041–1053. doi: 10.1111/j.1365-313X.2009.03843.x. [DOI] [PubMed] [Google Scholar]
- Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM. Co-translational assembly of the D1 protein into photosystem II. Journal of Biological Chemistry. 1999;274:16062–16067. doi: 10.1074/jbc.274.23.16062. [DOI] [PubMed] [Google Scholar]