Abstract
Podosphaera is a genus of the powdery mildew fungi belonging to the tribe Cystotheceae of the Erysiphaceae. Among the host plants of Podosphaera, 86 % of hosts of the section Podosphaera and 57 % hosts of the subsection Sphaerotheca belong to the Rosaceae. In order to reconstruct the phylogeny of Podosphaera and to determine evolutionary relationships between Podosphaera and its host plants, we used 152 ITS sequences and 69 28S rDNA sequences of Podosphaera for phylogenetic analyses. As a result, Podosphaera was divided into two large clades: clade 1, consisting of the section Podosphaera on Prunus (P. tridactyla s.l.) and subsection Magnicellulatae; and clade 2, composed of the remaining member of section Podosphaera and subsection Sphaerotheca. Because section Podosphaera takes a basal position in both clades, section Podosphaera may be ancestral in the genus Podosphaera, and the subsections Sphaerotheca and Magnicellulatae may have evolved from section Podosphaera independently. Podosphaera isolates from the respective subfamilies of Rosaceae each formed different groups in the trees, suggesting a close evolutionary relationship between Podosphaera spp. and their rosaceous hosts. However, tree topology comparison and molecular clock calibration did not support the possibility of co-speciation between Podosphaera and Rosaceae. Molecular phylogeny did not support species delimitation of P. aphanis, P. clandestina, P. ferruginea, P. spiraeae and P. tridactyla in their current circumscriptions, which suggests the need for revision of these species.
Keywords: 28S rDNA, evolution, ITS, molecular clock, phylogeny, powdery mildew fungi, Rosaceae
INTRODUCTION
The Erysiphaceae is a group of obligate biotrophic fungi that cause powdery mildew disease on about 1 × 104 angiosperm species (Amano 1986), and it consists of 16 genera and approximately 650 species (Braun & Takamatsu 2000, Braun et al. 2002, Takamatsu et al. 2005a, b, Liberato et al. 2006). The host range of this fungal group is strictly confined to angiosperms and the fungi have never been reported to infect ferns or gymnosperms (Amano 1986). Molecular phylogenetic analyses demonstrated that the Erysiphaceae form a distinct monophyletic group (Mori et al. 2000b, Lutzoni et al. 2004, Takamatsu 2004, Wang et al. 2006). Thus, the family Erysiphaceae is derived from a single ancestral taxon that may have acquired parasitism just once. Molecular clock calibration suggested that the fungi originated in the late Cretaceous (Mori et al. 2000b, Takamatsu & Matsuda 2004), which is consistent with the hypothesis of Heluta (1992) and the fact that their host range is restricted to angiosperms.
The Erysiphaceae are divided into five tribes and two basal genera. Both tree-parasitic and herb-parasitic fungi are included in three of the five tribes: Cystotheceae, Erysipheae and Phyllactinieae. Tree-parasitic fungi usually take basal positions in these tribes and herb-parasitic fungi have derived positions. These observations, as well as the fact that the most basal genera of the Erysiphaceae, i.e. Parauncinula and Caespitotheca, infect trees, suggest that the early host plants of the Erysiphaceae were trees (Mori et al. 2000a). Multiple host shifts from trees to herbs may have then occurred during the Tertiary (Takamatsu 2004).
The powdery mildews belonging to the tribe Cystotheceae have both herbaceous and woody plants as hosts and consist of three genera, Cystotheca, Podosphaera and Sawadaea, of which the host ranges of the genera Cystotheca and Sawadaea are restricted to a narrow range of host families, i.e. all hosts of Cystotheca belong to the Fagaceae, and Sawadaea mostly occurs on Aceraceae. All hosts of these two genera are trees. The genus Podosphaera consists of two sections, Podosphaera and Sphaerotheca. The section Podosphaera (formerly the genus Podosphaera) parasitizes woody plants, and about 90 % of its hosts belong to the Rosaceae. The section Sphaerotheca (formerly the genus Sphaerotheca) is further divided into the subsections Sphaerotheca and Magnicellulatae, each of which forms a separate monophyletic clade derived from different ancestors (Takamatsu et al. 2000). More than 50 % of the hosts of the subsection Sphaerotheca are woody or herbaceous plants belonging to the Rosaceae. On the other hand, all hosts of the subsection Magnicellulatae are herbaceous plants scattered among 40 plant families that do not include the Rosaceae. Thus, although the genus Podosphaera has a close affinity to Rosaceae, subsection Magnicellulatae is unique in having no rosaceous plant as host. This may be supported by the fact that Magnicellulatae has its own conidial germination type, i.e. the Magnicellulatae type, which differs from the Fibroidium type of other taxa of Podosphaera (Cook & Braun 2009).
Phylogenetic relationships within the tribe Cystotheceae were previously reported by Takamatsu et al. (2000). They reported that host shifts from trees to herbs occurred at least twice in this tribe and that the subtribes Sphaerotheca and Magnicellulatae evolved independently from separate ancestral taxa belonging to tree-parasitic Podosphaera species. However, they used only 26 ITS sequences in the analysis, which was inadequate to determine the phylogenetic relationships within the genus Podosphaera. In this study, we conducted comprehensive phylogenetic analyses by combining newly determined sequences with sequences reported in Takamatsu et al. (2000) and retrieved from DNA databases.
The aims of this study were
to reconstruct the phylogeny of the genus Podosphaera;
to discuss the evolution of Podosphaera with special reference to host relationships; and
to consider species delimitation of Podosphaera from the perspective of molecular phylogeny.
We included sequences of the subtribe Magnicellulatae in the present analyses, but we did not address them in the discussion because this fungal group is too large and distinct to be analyzed in this paper. Therefore, the phylogeny of Magnicellulatae has been discussed elsewhere (Ito & Takamatsu 2010).
MATERIALS AND METHODS
DNA extraction and amplification
The collection and location of host plants and the accession numbers for the nucleotide sequence databases (DDBJ, EMBL and GenBank) are provided in Table 1. Whole-cell DNA was isolated from chasmothecia or mycelia via the chelex method (Walsh et al. 1991, Hirata & Takamatsu 1996). The ITS region including the 5.8S rDNA, and the 5′ end of the 28S rDNA including the variable domains D1 and D2 were amplified separately by two sequential PCR reactions using partially nested primer sets. The PCR reactions were conducted using TaKaRa Taq DNA polymerase (TaKaRa, Tokyo, Japan) in a TP-400 thermal cycler (TaKaRa) under the following thermal cycling conditions: an initial denaturation step of 2 min at 95 °C, 30 cycles of 30 s at 95 °C, followed by 30 s at 52 °C for annealing, and 30 s at 72 °C for extension, and a final extension step of 7 min at 72 °C. A negative control that lacked template DNA was included in each set of reactions. The PCR products were subjected to electrophoresis in a 1.5 % agarose gel in TAE buffer, excised from the ethidium bromide-stained gel, and purified using the JETSORB Kit (Genomed, Oeynhausen, Germany) according to the manufacturer’s protocol. The nucleotide sequences of the PCR products were obtained for both strands using direct sequencing in a CEQ2000XL DNA sequencer (Beckman Coulter, Fullerton, CA, USA). The sequence reactions were conducted using the CEQ Dye Terminator Cycle Sequencing Kit (Beckman Coulter) according to the manufacturer’s instructions.
Table 1.
Sources of Podosphaera material sequenced in this study and DNA database accession numbers.
| Host | Fungal species | Location and year | Designation1 | Database ID no.2 |
|---|---|---|---|---|
| Asteraceae | ||||
| Calendula officinalis | fusca | Bariloche, Argentina; 2001 | MUMH 1933 | AB525914 |
| Calendula officinalis | fusca | Bariloche, Argentina; 2004 | MUMH 2432 | AB525915 |
| Taraxacum officinale | fusca | Bariloche, Argentina; 2004 | MUMH 2440 | AB525916 |
| Cannabaceae | ||||
| Humulus lupulus | macularis | Nagano, Japan; 2003 | MUMH 2926 | AB525917 |
| Caricaceae | ||||
| Carica papaya | caricae-papayae | Chiang Rai, Thailand; 2002 | MUMH 1853 | AB525918 |
| Escalloniaceae | ||||
| Escallonia rubra | negeri | Lago Curruhue, Argentina; 2001 | MUMH 1478 | AB525919 |
| Escallonia rubra | negeri | Bariloche, Argentina; 2001 | MUMH 1479 | AB525920 |
| Escallonia virgata | negeri | Cerro Tronador, Argentina; 2004 | MUMH 2515 | AB525921 |
| Geraniaceae | ||||
| Geranium thunbergii | fugax | Nara, Japan; 1997 | MUMH 343 | AB525922 |
| Gunneraceae | ||||
| Gunnera magellanica | gunnerae | Tierra del Fuego, Argentina; 1999 | BCRU 03874 (MUMH 1480) | AB525923 |
| Gunnera magellanica | gunnerae | Tierra del Fuego, Argentina; 1999 | BCRU 03890 (MUMH 1481) | AB525924 |
| Linaceae | ||||
| Linum usitatissimum | lini | Switzerland; 1998 | MUMH 1392 | AB525925 |
| Onagraceae | ||||
| Epilobium ciliatum | epilobii | Chall Huaco, Argentina; 2001 | MUMH 1873 | AB525926 |
| Rosaceae | ||||
| Amelanchier laevis | clandestina | Halle (Saale), Germany; 2009 | MUMH 4968 | AB525927 |
| Aria alnifolia | curvispora | Toyama, Japan; 2001 | MUMH 3266 | AB525928 |
| Cerasus incana | salatai | Tbilisi, Georgia; 2001 | MUMH 2595 | AB525929 |
| Crataegus monogyna | clandestina | Bariloche, Argentina; 2001 | MUMH 2429 | AB525930 |
| Crataegus oxyacantha | clandestina | Lago Lacar, Argentina; 2001 | MUMH 1869 | AB525931 |
| Crataegus sp. | clandestina | Bariloche, Argentina; 2001 | MUMH 1868 | AB525932 |
| Fragaria chiloensis | aphanis | Lago Gutierrez, Argentina; 2001 | MUMH 1871 | AB525933 |
| Photinia serratifolia | sp. | Aichi, Japan; 1997 | MUMH 407 | AB525934 |
| Pyracantha aff. crenatoserrata | Oidium sp.3 | Bariloche, Argentina; 2004 | MUMH 2450 | AB525935 |
| Pyracantha crenulata | Oidium sp.3 | Bariloche, Argentina; 2001 | MUMH 1870 | AB525936 |
| Rosa rubiginosa | pannosa | Lago Gutierrez, Argentina; 2001 | MUMH 1476 | AB525937 |
| Rosa rubiginosa | pannosa | Lago Lacar, Argentina; 2001 | MUMH 1872 | AB525938 |
| Rosa maltiflora | pannosa | Yamanashi, Japan; 2000 | MUMH 819 | AB525939 |
| Spiraea cantoniensis | spiraeae | Villa La Angostura, Argentina; 2004 | MUMH 2490 | AB525940 |
| Spiraea japonica | clandestina | Buenos Aires, Argentina; 2004 | MUMH 2535 | AB525941 |
| Stachyurus praecox | sp. | Mie, Japan; 1999 | MUMH 830 | AB525942 |
| Stephanandra incisa | stephanandrae | Mie, Japan; 1999 | MUMH 831 | AB525943 |
| Verbenaceae | ||||
| Diostea juncea | Oidium sp.3 | Cerro Tronador, Argentina; 2004 | MUMH 2498 | AB525944 |
| Diostea juncea | Oidium sp.3 | Bariloche, Argentina; 2009 | MUMH 4937 | AB525945 |
| Diostea juncea | Oidium sp.3 | Bariloche, Argentina; 2009 | MUMH 4938 | AB525946 |
| Violaceae | ||||
| Viola maculata | O. maculatae3 | Bariloche, Argentina; 2002 | BCRU 04343 (MUMH 3050) | AB525947 |
| Viola maculata | O. maculatae3 | Bariloche, Argentina; 2002 | BCRU 04345 (MUMH 3051) | AB525948 |
1 BCRU: Institutional Herbarium of Centro Regional Universitario Bariloche, San Carlos de Bariloche, Argentina; MUMH: Mie University, Mycological Herbarium, Japan.
2 DDBJ, EMBL, and GenBank database accession number of nucleotide sequence data.
3 Oidium.
For amplification of the ITS region, primers ITS5 (White et al. 1990) and P3 (Kusaba & Tsuge 1995) were used for the first amplification. One microlitre of the first reaction mixture was used for the second amplification, along with the partially nested primer sets ITS5 and ITS4 (White et al. 1990). The ITS5/ITS4 fragment was subjected to cycle sequencing using the primers ITS1, ITS4, T3 and T4 (Hirata & Takamatsu 1996). For amplification of the 28S rDNA, primers PM3 (Takamatsu & Kano 2001) and TW14 (Mori et al. 2000a), and NL1 (Mori et al. 2000a) and TW14 were used for the first and second amplifications, respectively. Primers NL1, NL2, NL3 and NLP2 (Mori et al. 2000a) were used for cycle-sequencing.
Phylogenetic analysis
The sequences were initially aligned using the Clustal X package (Thompson et al. 1997). The alignment was then visually refined with a word processing program using colour-coded nucleotides. The alignments were deposited in TreeBASE (http://www.treebase.org/) under the accession number S2604. Phylogenetic trees were obtained from the data using the maximum parsimony (MP) method in PAUP v4.0 (Swofford 2001) and a Bayesian analysis in MrBayes v3.1.1 (Ronquist & Huelsenbeck 2003). MP analyses were performed with the heuristic search option using the ‘tree-bisection-reconstruction’ (TBR) algorithm with the stepwise addition option set as simple and maximum tree number as 1 × 104. All sites were treated as unordered and unweighted, with gaps treated as missing data. The strength of the internal branches of the resulting trees was tested with bootstrap (BS) analyses (Felsenstein 1985) using 1 000 replications with the stepwise addition option set as simple and maximum tree number as 10 to save analysis time. BS values of 70 % or higher were shown. We also constructed MP trees with the parsimony ratchet method (Nixon 1999) in PAUP and PAUPRat v1 (Sikes & Lewis 2001) to confirm that the MP tree generated by the MP analysis is not the result of a local optimum.
For Bayesian phylogenetic analyses, the best-fit evolutionary model was determined for each dataset by comparing different evolutionary models via the Akaike information criterion (AIC) using PAUP and MrModeltest v2.2 (Nylander 2004). MrBayes was launched with random starting trees and Markov chains were sampled every 100 generations. To ensure that the Markov chain did not become trapped in local optima, we used the MCMCMC algorithm and performed the estimation with four incrementally heated Markov chains. The average standard deviation of split frequencies (ASDSF) was observed to verify that the values dropped below 0.01. Support for individual nodes was tested by Bayesian posterior probabilities (Bpp) obtained from a 50 % majority rule consensus. Bpp values of 0.95 or higher are shown.
Host plants
The host plant data were extracted from the database ‘Host plants of the powdery mildew fungi v1.0’ (available from the corresponding author upon request), which was based on the table ‘Host plants of powdery mildew fungi and their distribution by country’ (Amano 1986).
RESULTS
Phylogeny inferred from ITS sequences
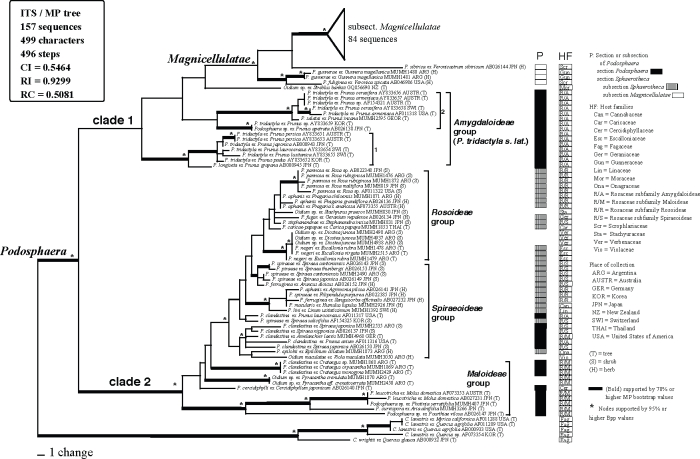
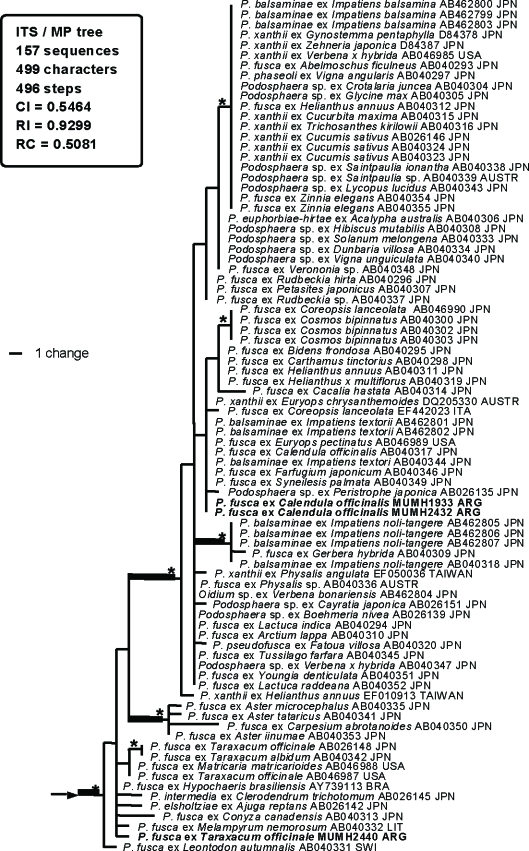
A total of 157 ITS sequences, including 33 sequences newly determined in this study, were used for the current analysis. Cystotheca spp. were used as outgroup taxa based on Mori et al. (2000a). The dataset consisted of 499 characters, of which 179 characters were variable and 140 characters were informative for parsimony analysis. A total of 1 × 104 MP trees with 496 steps (CI = 0.5464; RI = 0.9299; RC = 0.5081) were constructed by the MP analysis. One of the 1 × 104 MP trees, excluding subtribe Magnicellulatae (shown in Appendix 2), is shown in Fig. 1. Parsimony ratchet analysis generated trees with the same tree length and similar tree topologies. Therefore, we concluded that the tree shown in Fig. 1 is not the result of a local optimum. MrModeltest selected the GTR+I+Γ model as the best for this dataset. Using this evolution model, MrBayes was run for 1 × 107 generations, resulting in approximately 1 × 105 sampling trees. The first 59 390 trees were discarded (burn-in) because ASDSF dropped below 0.01. The remaining 40 611 trees were summarised in a majority-rule consensus tree, yielding the probability of each clade being monophyletic. The tree topology generated by the Bayesian analysis was almost identical to the MP tree, and thus the tree is not shown.
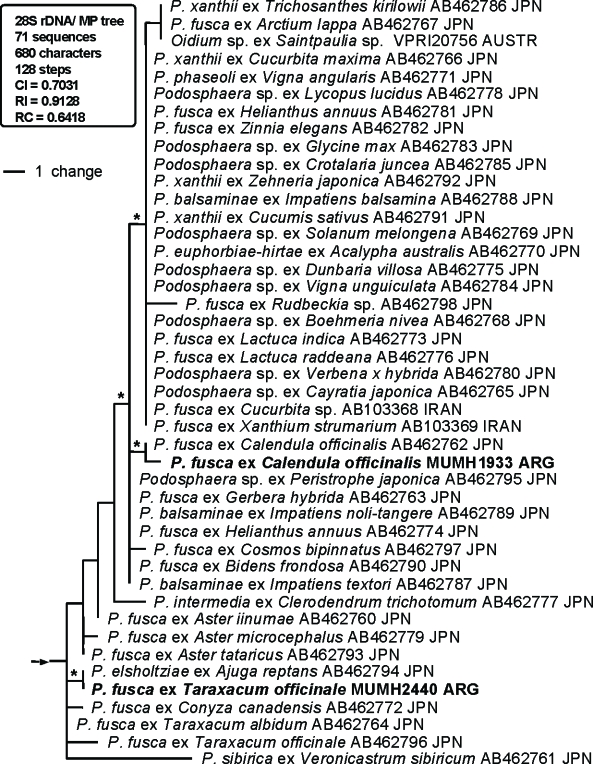
Fig. 1.
Phylogenetic analysis of the nucleotide sequences of the ITS region including 5.8S rDNA for 157 sequences from Podosphaera and Cystotheca used as outgroup taxa. The tree is a phylogram of one of the 1 x 104 MP trees with 496 steps, which was obtained by a heuristic search employing the random stepwise addition option of PAUP. Gaps were treated as missing data. Horizontal branch lengths are proportional to the number of nucleotide substitutions that were inferred to have occurred along a particular branch of the tree.
The 152 ITS sequences of Podosphaera species were divided into two large clades, clade 1 and clade 2. Clade 1 appeared in the MP strict consensus tree, although statistical support of this clade was low in both BS and Bpp analyses. Clade 1 consisted of 15 sequences of Podosphaera tridactyla s.l. (section Podosphaera) on Prunus spp. (Rosaceae), 89 sequences of the isolates belonging to the subsection Magnicellulatae of section Sphaerotheca, and one sequence of Oidium sp. on Streblus banksii. The basal nodes of clade 1 were occupied by the sequences of Podosphaera tridactyla s.l. and the sequences of subsection Magnicellulatae formed a distinct clade (BS = 61 %; Bpp = 1.0) at a derived position. Clade 2 (BS = 95 %; Bpp = 1.0) consisted of the sequences of the section Podosphaera parasitizing the subfamilies Maloideae (apple subfamily) and Spiraeoideae of the Rosaceae and all sequences of the subsection Sphaerotheca of section Sphaerotheca. The sequences of the section Podosphaera occupied a basal position of clade 2. The sequences of the subsection Sphaerotheca of section Sphae- rotheca were placed at a derived position but did not form a distinct monophyletic group.
Phylogeny inferred from the 28S rDNA region
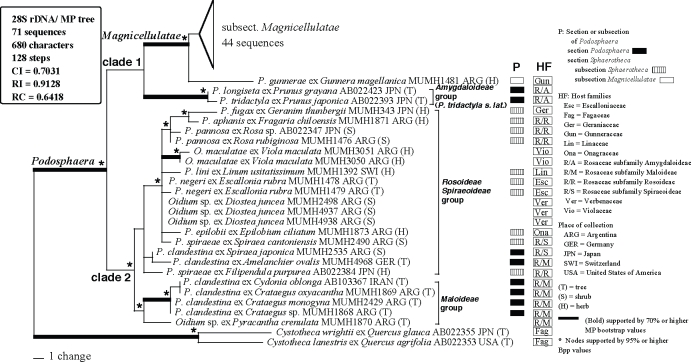
A total of 71 28S rDNA sequences, including 23 sequences newly determined in this study, were used for the current analysis. Cystotheca spp. were used as outgroup taxa based on Mori et al. (2000a). The dataset consisted of 680 characters, of which 78 characters were variable and 47 characters were informative for parsimony analysis. A total of 1 × 104 MP trees with 128 steps (CI = 0.7031; RI = 0.9128; RC = 0.6418) were constructed by the MP analysis. One of the 1 × 104 MP trees, excluding subtribe Magnicellulatae (shown in Appendix 1), is shown in Fig. 2. Parsimony ratchet analysis generated trees with the same tree length and similar tree topologies. Therefore, we concluded that the tree shown in Fig. 2 is not the result of a local optimum. MrModeltest selected the GTR+I+Γ model as the best for this dataset. Using this evolution model, MrBayes was run for 5 × 106 generations, resulting in 50 001 sampling trees. The first 14 650 trees were discarded (burn-in) because ASDSF dropped below 0.01. The remaining 35 351 trees were summarised in a majority-rule consensus tree, yielding the probability of each clade being monophyletic. The tree topology produced by the Bayesian analysis was almost identical to the MP tree and thus the tree is not shown. The tree topology of the 28S rDNA sequences was almost identical to that of the ITS tree, but the statistical support for the major clades was lower than that of the ITS tree.
Fig. 2.
Phylogenetic analysis of the divergent domains D1 and D2 sequences of the 28S rDNA for 71 sequences from Podosphaera and Cystotheca used as outgroup taxa. The tree is a phylogram of one of the 1 x 104 MP trees with 128 steps, which was obtained by a heuristic search employing the random stepwise addition option of PAUP. Gaps were treated as missing data. Horizontal branch lengths are proportional to the number of nucleotide substitutions that were inferred to have occurred along a particular branch of the tree.
Timing of evolutionary events
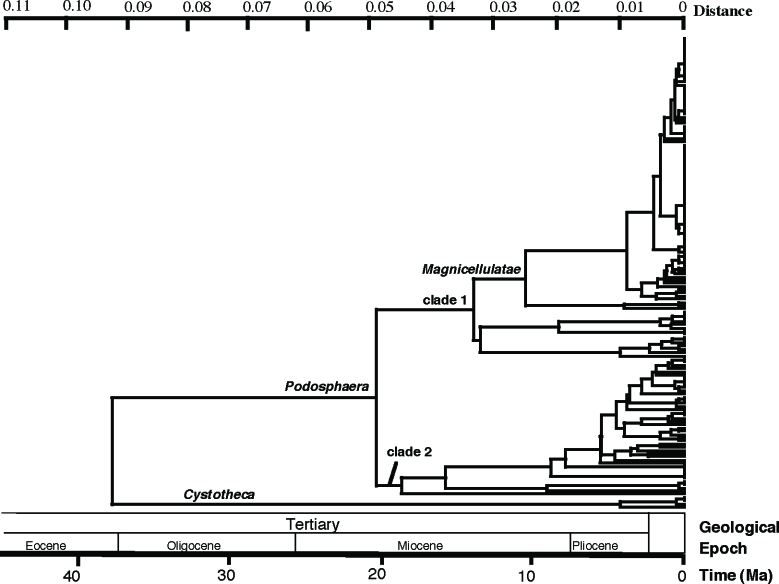
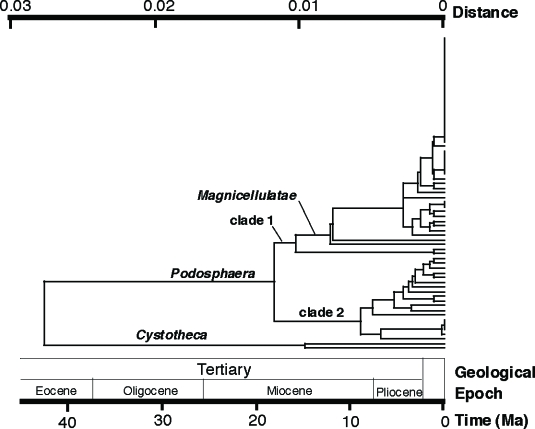
Because a likelihood ratio test (LRT) significantly rejected the molecular clock of the ITS dataset of 157 sequences, five sequences with extremely long or short terminal branches were removed from the dataset. The molecular clock hypothesis of the reduced dataset consisting of 152 ITS sequences was not rejected by the LRT and thus was used to construct a UPGMA tree. The LRT did not reject the molecular clock hypothesis of the 28S rDNA dataset consisting of 71 sequences. Thus, the dataset was used to construct a UPGMA tree. The Kimura two-parameter model (Kimura 1980) was used to calculate genetic distances. The molecular clocks of the ITS (2.52 × 10−9 substitutions per site per year, ssy) and the D1/D2 domain of the 28S rDNA region (6.5 × 10−10 ssy) of the Erysiphales (Takamatsu & Matsuda 2004) were used to calculate evolutionary timing. The molecular clocks suggested that Podosphaera split from Cystotheca about 40 million years ago (Ma) at the end of the Eocene, and the split of clade 1 and clade 2 occurred about 20 Ma in the Miocene (Fig. 3, 4).
Fig. 3.
Estimated dates of divergence of major clades of Podosphaera based on the nucleotide sequences of the rDNA ITS region and nucleotide substitution rate of Erysiphaceae (2.52 × 10−9 substitutions per site per year) reported by Takamatsu & Matsuda (2004). Ma, million years ago.
Fig. 4.
Estimated dates of divergence of major clades of Podosphaera based on the divergent domains D1 and D2 sequences of the 28S rDNA and the nucleotide substitution rate of Erysiphaceae (6.5 × 10−10 substitutions per site per year) reported by Takamatsu & Matsuda (2004). Ma, million years ago.
DISCUSSION
Host relationships
Host plants of Podosphaera
The number of host plant species of Podosphaera, arranged by plant families, is shown in Table 2. Podosphaera is divided into two sections, Podosphaera (formerly genus Podosphaera) and Sphaerotheca (formerly genus Sphaerotheca). The latter section is further divided into two subsections, Sphaerotheca and Magnicellulatae, respectively. The subsection Magnicellulatae forms a distinct monophyletic group that diverged from an ancestral fungus by a host shift from Prunus (Rosaceae) to herbaceous plants (Takamatsu et al. 2000). Magnicellulatae is unique in its host range and morphological characteristics compared with other Podosphaera species, suggesting that this subsection evolved independently from other Podosphaera taxa. Thus, we discuss the evolution of this fungal group elsewhere (Hirata et al. 2000, Ito & Takamatsu 2010). There are 250 host species of the section Podosphaera spanning 10 orders and 13 families, of which 216 host species (86.4 %) belong to the Rosaceae. The subsection Sphaerotheca of section Sphaerotheca has 806 host species covering 15 orders and 28 families, of which 456 (56.6 %) belong to the Rosaceae. Of the non-Rosaceae hosts, 70 belong to the Euphorbiaceae, 67 to the Geraniaceae, 65 to the Onagraceae and 54 to the Hydrangeaceae. Thus, the Rosaceae has the highest number of host species for both section Podosphaera and subsection Sphaerotheca. On the other hand, Magnicellulatae has 1 110 host species spanning 40 families. About half (45 %) of the hosts belong to the Asteraceae and none belong to the Rosaceae.
Table 2.
Number of host plant species of the genus Podosphaera.
| Number of host species |
||||
|---|---|---|---|---|
| Plant order & family | Sect. Podosphaera | Sect. Sphaerotheca subsect. Sphaerotheca | Total | |
| EUROSIDS I | ||||
| Rosales | Ulmaceae | 1 | 2 | 3 |
| Moraceae | 0 | 4 | 4 | |
| Urticaceae | 0 | 2 | 2 | |
| Rosaceae | 216 | 456 | 672 | |
| Elaeagnaceae | 0 | 3 | 3 | |
| Fagales | Betulaceae | 1 | 0 | 1 |
| Celastrales | Celastraceae | 0 | 3 | 3 |
| Malpighiales | Salicaceae | 3 | 0 | 3 |
| Linaceae | 0 | 1 | 1 | |
| Euphorbiaceae | 0 | 70 | 70 | |
| EUROSIDS | ||||
| Sapindales | Anacardiaceae | 0 | 8 | 8 |
| Myrtales | Myrtaceae | 0 | 18 | 18 |
| Onagraceae | 0 | 65 | 65 | |
| Punicaceae | 0 | 1 | 1 | |
| Geraniales | Geraniaceae | 0 | 67 | 67 |
| Saxifragales | Saxifragaceae | 0 | 54 | 54 |
| Hamamelidaceae | 4 | 0 | 4 | |
| Cercidiphyllaceae | 2 | 0 | 2 | |
| EUASTERIDS I & II | ||||
| Solanales | Convolvulaceae | 0 | 1 | 1 |
| Lamiales | Oleaceae | 2 | 2 | 4 |
| Verbenaceae | 0 | 2 | 2 | |
| Gentianales | Hydrophyllaceae | 0 | 6 | 6 |
| Asclepiadaceae | 1 | 0 | 1 | |
| Apiales | Apiaceae | 1 | 1 | 2 |
| Dipsacales | Caprifoliaceae | 4 | 1 | 5 |
| EUDICOTS | ||||
| Ericales | Ebenaceae | 2 | 0 | 2 |
| Ericaceae | 12 | 11 | 23 | |
| Polemoniaceae | 0 | 21 | 21 | |
| Cornales | Cornaceae | 1 | 0 | 1 |
| Caryophyllales | Polygonaceae | 0 | 1 | 1 |
| Phytolaccaceae | 0 | 1 | 1 | |
| Tamaricaceae | 0 | 1 | 1 | |
| Plumbaginaceae | 0 | 1 | 1 | |
| Ranunculales | Ranunculaceae | 0 | 2 | 2 |
| Papaveraceae | 0 | 1 | 1 | |
| Total | 250 | 806 | 1056 | |
Powdery mildews of Rosaceae
The number of host species of powdery mildew fungi reported on Rosaceae is shown in Table 3 and is organised by plant subfamilies and fungal genera. Five powdery mildew genera have been reported to occur on the Rosaceae. Of these, the genus Podosphaera has the highest number of host species in the Rosaceae (672). Podosphaera occurs on all four subfamilies of Rosaceae, and the highest number of host species is in the subfamily Rosoideae (418 species). Phyllactinia is the genus having the next highest number of host species in Rosaceae (110), and finally Erysiphe has 68 host species. Although all three sections of Erysiphe occur on Rosaceae, the number of host species differs depending on the plant subfamilies and fungal sections. Although Golovinomyces and Leveillula also occur on Rosaceae, the number of host species is less than 10 in both genera. Therefore, Rosaceae is the most important plant family as host for Podosphaera and vice versa.
Table 3.
Number of host species of the powdery mildew fungi in Rosaceae.
| Subfamily of Rosaceae | Podosphaera | Phyllactinia |
Erysiphe |
Golovinomyces | Leveillula | ||||
|---|---|---|---|---|---|---|---|---|---|
| Erysiphe | Microsphaera | Uncinula | Total | ||||||
| Amygdaloideae | 96 | 21 | 0 | 1 | 13 | 14 | 2 | 1 | |
| Maloideae | 108 | 76 | 3 | 6 | 3 | 12 | 0 | 0 | |
| Rosoideae | 418 | 11 | 32 | 1 | 6 | 39 | 6 | 1 | |
| Spiraeoideae | 46 | 2 | 1 | 2 | 0 | 3 | 1 | 1 | |
| Unknown | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 672 | 110 | 36 | 10 | 22 | 68 | 9 | 3 | |
Relationships between host plants and phylogeny of Podosphaera
Phylogenetic trees of Podosphaera excluding Magnicellulatae are shown in Fig. 1 and 2. Podosphaera taxa are divided into four groups according to the subfamilies of Rosaceae: the Amygdaloideae group, the Maloideae group, the Spiraeoideae group and the Rosoideae group. The Amygdaloideae group (= P. tridactyla s.l.) is an assemblage that splits from the other three groups first and forms a large group (clade 1) with the fungi belonging to Magnicellulatae. The first node of clade 1 was shared by the isolates from Amygdaloideae, which suggests that the Amygdaloideae group is ancestral in clade 1 and the Magnicellulatae diverged from an ancestor that was parasitic to the Amygdaloideae. This group is further divided into two small groups with large genetic divergence. Subgroup 1 (BS = 95 %; Bpp = 0.95) split from other clade 1 groups first and consists of isolates from the subgenera Amygdalus, Laurocerasus and Padus of Prunus. Subgroup 2 (BS = 58 %; Bpp = 0.84), sister to Magnicellulatae, consists of isolates from the subgenera Cerasus and Prunus. Thus, the phylogeny of the Amygdaloideae group is closely related to the subgeneric-level taxonomy of Prunus, which suggests the possibility of co-speciation between Prunus and the Amygdaloideae group. However, in the phylogeny of the plant genus, subgenus Amygdalus groups with Prunus, and the remaining three subgenera Cerasus, Laurocerasus and Padus form another group (Bortiri et al. 2001, Lee & Wen 2001), which is not consistent with the grouping of powdery mildew fungi on Prunus. Because we used only 15 sequences in the present analysis, more sequence data will be required to clarify the phylogenetic relationships within P. tridactyla s.l.
Powdery mildews on the subfamilies Maloideae, Spiraeoideae and Rosoideae formed a large clade (clade 2: BS = 95 %; Bpp = 1.0) distinct from the Amygdaloideae group. Because the first split of clade 2 occurred within the Maloideae group, taxa belonging to the Maloideae group may be the most ancestral in clade 2. The Maloideae group was further divided into two groups that are parasitic to the tribes Maleae and Cratageae, respectively. The former group split at the base of clade 2 and was sister to the other groups. The fungi parasitic to the tribe Cratageae formed a clade with the Spiraeoideae and Rosoideae groups and occupied a basal position in the clade. Podosphaera cercidiphylli on Cercidiphyllum japonicum (Cercidiphyllaceae) was included in the Maloideae group in the phylogeny.
The Spiraeoideae group formed a clade together with the Rosoideae group (BS < 50 %; Bpp = 0.98). Although the two groups did not form independent clades, they were generally situated at different places in the tree (Fig. 1). Podosphaera epilobii on Epilobium (Onagraceae) and Oidium maculatae on Viola (Violaceae) were included in the Spiraeoideae group. Podosphaera clandestina on Prunus (Amygdaloideae, Rosaceae) was also included in this group. Podosphaera lini on Linum (Linaceae), P. macularis on Humulus (Cannabaceae), P. caricae-papayae on Papaya (Caricaceae), P. negeri on Escallonia (Escalloniaceae), and P. fugax on Geranium (Geraniaceae) were included in the Rosoideae group. A fungus on Diostea juncea (Verbenaceae) was identified as ‘Sphaerotheca verbenae’ (= Podosphaera xanthii) by Havrylenko (1997). However, ‘Sphaerotheca verbenae’ belongs to subsection Magnicellulatae of section Sphaerotheca. In this study, this fungus has a DNA sequence identical to that of P. negeri in both ITS and 28S rDNA regions and belongs to the Rosoideae group. We thus treated this fungus as Oidium sp. in this study.
The present study revealed that more than 50 % of the hosts of Podosphaera (excluding Magnicellulatae) belong to the Rosaceae, and phylogenetic relationships of Podosphaera have close affinity with the taxonomy of the Rosaceae. These results strongly suggest a close evolutionary relationship between Podosphaera and Rosaceae. The Rosaceae may have been the first host family for Podosphaera, and host shifts from the Rosaceae to other plant families may have occurred spontaneously during the evolution of Podosphaera. The Rosaceae belongs to the Rosales of EUROSIDS I (APG II 2003). Of the host families analysed in this study, all plant families, excepting Rosaceae, Cannabaceae and Moraceae, do not belong to the Rosales. These results suggest that Podosphaera tend to expand their hosts to closely related plant species when expanding within a single plant family. Conversely, inter-family level host shifts seem to occur independently of host phylogeny. Similar phenomena have been also found in host expansions of Golovinomyces and Phyllactinia of the Erysiphaceae (Matsuda & Takamatsu 2003, Takamatsu et al. 2008).
Is there any co-speciation between Podosphaera and Rosaceae?
The above results suggest a close evolutionary relationship between Podosphaera and Rosaceae, but did co-speciation occur between Podosphaera and Rosaceae? At least two items should be evaluated to determine whether co-speciation between organisms has occurred. One is similar or identical phylogeny between the organisms concerned, and another is timing of divergence of the organisms. Of the four subfamilies of the Rosaceae, the subfamilies Amygdaloideae, Maloideae and Rosoideae were supported as monophyletic groups by the rbcL sequence phylogeny, but Spiraeoideae was shown to be polyphyletic (Morgan et al. 1994, Potter et al. 2002). Podosphaera isolates were mostly divided into different groups according to the subfamilies of Rosaceae, suggesting that the phylogeny of Podosphaera is mostly consistent with the subfamily-level taxonomy of Rosaceae. Phylogenetic analyses using rbcL and matK sequences showed that the first split is shared by the Rosoideae, suggesting that Rosoideae is the most ancestral of the Rosaceae (Morgan et al. 1994, Potter et al. 2002). In Podosphaera phylogeny, the first split occurs between isolates from the Amygdaloideae and Maloideae, and the isolates from the Rosoideae are placed at a derived part of the trees (Fig. 1, 2). Thus, the phylogeny of Podosphaera does not conform to that of the Rosaceae. Evolutionary timing of Podosphaera divergence, as calculated by the molecular clocks of ITS and 28S rDNA regions, suggested that the split of Cystotheca and Podosphaera occurred in the late Eocene (c. 40 Ma) and that the split of clade 1 and clade 2 occurred in the mid-Miocene (c. 20 Ma). Fossil records suggest that the traditional subfamilies Amygdaloideae and Maloideae are found in the Eocene (DeVore & Pigg 2007). Therefore, divergence of Podosphaera may have occurred later than the divergence of the Rosaceae. In conclusion, there is no evidence that co-speciation occurred between Podosphaera and Rosaceae.
Taxonomic implications
Sections and subsections
The genus Podosphaera is divided into two sections, the section Podosphaera, with appendages dichotomously branched at the apex, and the section Sphaerotheca, with hypha-like simple appendages. Section Sphaerotheca is further divided into two subsections, Sphaerotheca and Magnicellulatae, by the size of the peridium cells of the chasmothecia. These sections and subsections were introduced by Braun & Takamatsu (2000) as morphological, i.e., non-monophyletic groups. The present analysis revealed that the two subsections do not share a common ancestor. Therefore, the section Sphaerotheca is polyphyletic, consisting of two different groups derived from different ancestral taxa. Section Podosphaera is a paraphyletic group situated at the basal part of clade 1 and clade 2. The present analysis indicates that the section Podosphaera is ancestral in the genus Podosphaera, and the subsections Sphaerotheca and Magnicellulatae were derived from the Maloideae group and Amygdaloideae group, respectively. Therefore, the genus Podosphaera is a natural unit supported by molecular phylogeny, but the sections Podosphaera and Sphaerotheca are artificial, morphological units that are not supported by phylogeny, as already stated by Braun & Takamatsu (2000).
Species supported by phylogeny
The genus Gunnera consists of herbaceous plants distributed in the Southern Hemisphere and tropical regions. Podosphaera gunnerae was first described as a powdery mildew of Gunnera by Havrylenko & Braun (1998). This species has been reported only in Argentinian Patagonia. Both ITS and 28S rDNA sequences clearly indicated that P. gunnerae forms an independent clade. This clade was situated at the basal part of Magnicellulatae and was sister to P. fuliginea. This suggests that P. gunnerae split from the other fungi in the early stage of the evolution of Magnicellulatae. This species is the only powdery mildew species described for the genus Gunnera.
Podosphaera negeri was first described as Sphaerotheca spiralis in 1907 (Braun 1987) and revised as P. negeri by Braun et al. (2006). This species has a unique characteristic with coiled appendages, infects Escallonia (Escalloniaceae) and has been reported only in Argentinian Patagonia. Both ITS and 28S rDNA sequences clearly indicated that P. negeri forms an independent clade. Oidium sp. found on Diostea juncea has a sequence identical to that of P. negeri in both ITS and 28S rDNA regions. Podosphaera fugax found on Geranium was sister to P. negeri, but this was supported neither by BS nor by Bpp values.
Oidium maculatae was first described on Viola maculata (Havrylenko & Takamatsu 2005). Both ITS and 28S rDNA sequences showed that this species forms a unique clade.
Species not supported by phylogeny
Podosphaera species parasitic to Prunus have been mostly classified as P. tridactyla. Cunnington et al. (2005) reported that this species has large genetic variation and is divided into several groups, which was confirmed by the present analysis. These results indicate that P. tridactyla is a species complex composed of several biological species. Prunus s.l. has been divided into 5 to 6 subgenera, which were sometimes treated as separate genera (Bortiri et al. 2001, Lee & Wen 2001). Groups of P. tridactyla are mostly consistent with the delimitation of the subgenera. Podosphaera tridactyla may have specialised to the respective host groups along with the genetic divergence of Prunus. Some segregated species like P. longiseta (Sawada 1951) and P. salatai (Heluta et al. 2005) have been proposed to lie within P. tridactyla s.l., which is supported by molecular analysis. Comprehensive revision of P. tridactyla s.l. is required.
Podosphaera clandestina is a well known species described in 1851 and parasitizes 14 genera of the Rosaceae containing Crataegus, Prunus and Spiraea as hosts (Braun 1987). Nine ITS sequences of P. clandestina from Amelanchier, Crataegus, Prunus and Spiraea used in this study revealed that these sequences do not form a single clade. In particular, the sequences of the isolates from Crataegus formed a distinct clade distantly related to other P. clandestina on Amelanchier, Spiraea and Prunus. 28S rDNA sequence of P. clandestina on Cydonia was identical to that of isolates from Crataegus. Isolates from Amelanchier, Prunus and Spiraea were closely related to each other, but there were some nucleotide substitutions among them. A taxonomic re-evaluation of this species is necessary. The fungus on Pyracantha was reported as P. clandestina by Amano (1986) and Delhey et al. (2003). However, as far as we know, a teleomorph of this fungus has not yet been found. The two ITS sequences and one 28S rDNA sequence of this fungus determined in this study showed that the fungus on Pyracantha forms an independent lineage different from other P. clandestina groups.
The five ITS sequences of P. spiraeae on Spiraea formed a clade but contained some nucleotide substitutions among the sequences. A sequence from P. spiraeae collected in Korea, AF011317, did not belong to this clade and was sister to a sequence of P. clandestina on Prunus avium. Zhao (1981) proposed a new species, Sphaerotheca filipendulae (= Podosphaera filipendulae), for the fungus on Filipendula. Braun (1987) regarded S. filipendulae as a synonym of S. spiraeae (= P. spiraeae). The present analysis shows that the fungus on Filipendula purpurea does not belong to the clade of P. spiraeae on Spiraea, but to the clade consisting of P. aphanis on Agrimonia, P. ferruginea on Sanguisorba, and P. macularis on Humulus. The ITS sequence of the fungus on F. purpurea was identical to that of P. macularis on Humulus. Spiraea belongs to the subfamily Spiraeoideae and Filipendula to Rosoideae. Considering the close relationship between the subfamilies of Rosaceae and the phylogeny of Podosphaera, the result of the present phylogenetic analysis seems to be acceptable.
Podosphaera ferruginea was introduced for the fungus on Sanguisorba (Junell 1965). Braun (1987) assigned the fungus on Aruncus to this species. The present analysis revealed that the fungus on Aruncus belongs to a clade different from the fungus on Sanguisorba. Because Saguisorba belongs to the subfamily Rosoideae and Aruncus to Spiraeoideae, the result of phylogenetic analysis seems to be acceptable.
Thirteen genera of the Rosaceae and Eucalyptus (Myrtaceae) have been listed as hosts of P. aphanis (Braun 1987). Of these, the fungi on Fragaria and Agrimonia were used in this analysis. The three sequences from Fragaria grouped together. The sequence of the fungus on Agrimonia did not belong to this clade but to the clade composed of the fungi on Filipendula, Humulus and Sanguisorba. Taxonomic revision of this species may be required.
Appendix 1.
Phylogeny of subsection Magnicellulatae of the genus Podosphaera inferred from the nucleotide sequences of the D1/D2 domains of the 28S rDNA. This is a part of the phylogenetic tree of Podosphaera, but was not shown in Fig. 2. A bold line means that the node was supported by an MP bootstrap value of 70 % or higher. An asterisk means that the node was supported by a Bayesian posterior probability value of 0.95 or higher. Taxon name shown by bold type is the sequence determined in this study.
Appendix 2.
Phylogeny of subsection Magnicellulatae of the genus Podosphaera inferred from nucleotide sequences of rDNA ITS region. This is a part of the phylogenetic tree of Podosphaera, but was not shown in Fig. 1. A bold line means that the node was supported by an MP bootstrap value of 70 % or higher. An asterisk means that the node was supported by a Bayesian posterior probability value of 0.95 or higher. Taxon name shown by bold type is the sequence determined in this study.
Acknowledgments
We thank Dr Peter Johnston for kindly providing ITS sequence of Oidium sp. on Streblus banksii.
REFERENCES
- Amano K. 1986. Host range and geographical distribution of the powdery mildew fungi Japan Scientific Societies, Tokyo, Japan: . [Google Scholar]
- APG II (Angiosperm Phylogeny Group) . 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Botanical Journal of the Linnean Society 141: 399 – 436 . [Google Scholar]
- Bortiri E, Oh SH, Jiang JG, Baggett S, Granger A, Weeks C, Buckingham M, Potter D, Parfitt DE. 2001. Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL-trnF spacer DNA. Systematic Botany 26: 797 – 807 . [Google Scholar]
- Braun U. 1987. A monograph of the Erysiphales (powdery mildews). Beihefte zur Nova Hedwigia 89: 1 – 700 . [Google Scholar]
- Braun U, Cook RTA, Inman AJ, Shin HD. 2002. The taxonomy of the powdery mildew fungi. In: Bélanger R, Bushnell WR, Dik AJ, Carver TLW. (eds), The powdery mildews: a comprehensive treatise: 13–55 APS Press, St. Paul, USA: . [Google Scholar]
- Braun U, Delhey R, Dianese JC, Hosagoudar VB. 2006. Miscellaneous notes on biotrophic micromycetes. Schlechtendalia 14: 85 – 97 . [Google Scholar]
- Braun U, Takamatsu S. 2000. Phylogeny of Erysiphe, Microsphaera, Uncinula (Erysipheae) and Cystotheca, Podosphaera, Sphaerotheca (Cystotheceae) inferred from rDNA ITS sequences – some taxonomic consequences. Schlechtendalia 4: 1 – 33 . [Google Scholar]
- Cook RTA, Braun U. 2009. Conidial germination patterns in powdery mildews. Mycological Research 113: 616 – 636 . [DOI] [PubMed] [Google Scholar]
- Cunnington JH, Lawrie AC, Pascoe IG. 2005. Genetic variation within Podosphaera tridactyla reveals a paraphyletic species complex with biological specialization towards specific Prunus subgenera. Mycological Research 109: 357 – 362 . [DOI] [PubMed] [Google Scholar]
- Delhey R, Braun U, Kiehr M. 2003. Some new records of powdery mildew fungi from Argentina (2). Schlechtendalia 10: 79 – 90 . [Google Scholar]
- DeVore ML, Pigg KB. 2007. A brief review of the fossil history of the family Rosaceae with a focus on the Eocene Okanogan Highlands of eastern Washington State, USA, and British Columbia, Canada. Plant Systematics and Evolution 266: 45 – 57 . [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783 – 791 . [DOI] [PubMed] [Google Scholar]
- Havrylenko M. 1997. Erysiphales de la region Andino-Patagonica. PhD thesis, Centro Regional Universitario Bariloche, Universidad Nacional del Comahue, Argentina: . [Google Scholar]
- Havrylenko M, Braun U. 1998. A new species of Sphaerotheca (Erysiphales) from Argentina. Nova Hedwigia 66: 173 – 175 . [Google Scholar]
- Havrylenko M, Takamatsu S. 2005. Notes on Erysiphales (Ascomycetes) from Patagonia (Argentina). Mycoscience 46: 32 – 38 . [Google Scholar]
- Heluta VP. 1992. Hypothesis on the origin and migrations of fungi of Erysiphales order. Ukranian Botanical Journal 49: 5 – 13 . [Google Scholar]
- Heluta VP, Braun U, Gvritishvili MN. 2005. Podosphaera salatai sp. nov. (Erysiphales) from Georgia. Fungal Diversity 18: 89 – 94 . [Google Scholar]
- Hirata T, Cunnington JH, Paksiri U, Limkaisang S, Shishkoff N, Grigaliunaite B, Sato Y, Takamatsu S. 2000. Evolutionary analysis of subsection Magnicellulatae of Podosphaera section Sphaerotheca (Erysiphales) based on the rDNA ITS sequences with special reference to host plants. Canadian Journal of Botany 78: 1521 – 1530 . [Google Scholar]
- Hirata T, Takamatsu S. 1996. Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37: 283 – 288 . [Google Scholar]
- Ito M, Takamatsu S. 2010. Molecular phylogeny and evolution of subsection Magnicellulatae (Erysiphaceae: Podosphaera) with special reference to host plants. Mycoscience 51: 34 – 43 . [Google Scholar]
- Junell L. 1965. Nomenclatural remarks on some species of Erysiphaceae. Transactions of the British Mycological Society 48: 539 – 548 . [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111 – 120 . [DOI] [PubMed] [Google Scholar]
- Kusaba M, Tsuge T. 1995. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Current Genetics 28: 491 – 498 . [DOI] [PubMed] [Google Scholar]
- Lee S, Wen J. 2001. A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of nuclear ribosomal DNA. American Journal of Botany 88: 150 – 160 . [PubMed] [Google Scholar]
- Liberato JR, Barreto RW, Niinomi S, Takamatsu S. 2006. Queirozia turbinata (Phyllactinieae, Erysiphaceae): a powdery mildew with a dematiaceous anamorph. Mycological Research 110: 567 – 574 . [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G , et al. 2004. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany 91: 1446 – 1480 . [DOI] [PubMed] [Google Scholar]
- Matsuda S, Takamatsu S. 2003. Evolution of host–parasite relationships of Golovinomyces (Ascomycete: Erysiphaceae) inferred from nuclear rDNA sequences. Molecular Phylogenetics and Evolution 27: 314 – 327 . [DOI] [PubMed] [Google Scholar]
- Morgan DR, Soltis DE, Robertson KR. 1994. Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. American Journal of Botany 81: 890 – 903 . [Google Scholar]
- Mori Y, Sato Y, Takamatsu S . 2000a. Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia 92: 74 – 93 . [Google Scholar]
- Mori Y, Sato Y, Takamatsu S. 2000b. Molecular phylogeny and radiation time of Erysiphales inferred from the nuclear ribosomal DNA sequences. Mycoscience 41: 437 – 447 . [Google Scholar]
- Nixon KC. 1999. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 15: 407 – 414 . [DOI] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Evolutionary Biology Centre, Uppsala University; Program distributed by the author . [Google Scholar]
- Potter D, Gao F, Bortiri PE, Oh SH, Baggett S. 2002. Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data. Plant Systematics and Evolution 231: 77 – 89 . [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572 – 1574 . [DOI] [PubMed] [Google Scholar]
- Sawada K. 1951. Researches on fungi in the Tohoku District of Japan I. Erysiphaceae. Bulletin of the Government Forest Experiment Station 50: 97 – 140 . [Google Scholar]
- Sikes DS, Lewis PO. 2001. Beta software, version 1. PAUPRat: PAUP* implementation of the parsimony ratchet Distributed by the authors Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, USA: . [Google Scholar]
- Swofford DL. 2001. PAUP*: Phylogenetic analysis using parsimony (* and other methods) 4.0b8 Sinauer, Sunderland, MA: . [Google Scholar]
- Takamatsu S. 2004. Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience 45: 147 – 157 . [Google Scholar]
- Takamatsu S, Braun U, Limkaisang S . 2005a. Phylogenetic relationships and generic affinity of Uncinula septata inferred from nuclear rDNA sequences. Mycoscience 46: 9 – 16 . [Google Scholar]
- Takamatsu S, Hirata T, Sato Y. 2000. A parasitic transition from trees to herbs occurred at least twice in tribe Cystotheceae (Erysiphaceae): Evidence from nuclear ribosomal DNA. Mycological Research 104: 1304 – 1311 . [Google Scholar]
- Takamatsu S, Inagaki M, Niinomi S, Khodaparast SA, Shin HD, Grigaliunaite B, Havrylenko M. 2008. Comprehensive molecular phylogenetic analysis and evolution of the genus Phyllactinia (Ascomycota: Erysiphales) and its allied genera. Mycological Research 112: 299 – 315 . [DOI] [PubMed] [Google Scholar]
- Takamatsu S, Kano Y. 2001. PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fungi. Mycoscience 42: 135 – 139 . [Google Scholar]
- Takamatsu S, Matsuda S. 2004. Estimation of molecular clocks for ITS and 28S rDNA in Erysiphales. Mycoscience 45: 340 – 344 . [Google Scholar]
- Takamatsu S, Niinomi S, Cabrera de Álvarez MG, Álvarez RE, Havrylenko M, Braun U . 2005b. Caespitotheca gen. nov., an ancestral genus in the Erysiphales. Mycological Research 109: 903 – 911 . [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876 – 4882 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10: 506 – 513 . [PubMed] [Google Scholar]
- Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS. 2006. Phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98: 1066 – 1076 . [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds). PCR protocols: a guide to methods and applications: 315–322 Academic, San Diego, California: . [Google Scholar]
- Zhao ZY. 1981. Taxonomic studies on the genus Sphaerotheca of China IV. New species and new combination on Rosaceae and Labiatae. Acta Microbiologica Sinica 21: 438 – 442 . [Google Scholar]